Abstract
The mitochondrial intermembrane space contains a protein complex essential for cell viability, the Tim9-Tim10 complex. This complex is required for the import of hydrophobic membrane proteins, such as the ADP/ATP carrier (AAC), into the inner membrane. Different views exist about the role played by the Tim9-Tim10 complex in translocation of the AAC precursor across the outer membrane. For this report we have generated a new tim10 yeast mutant that leads to a strong defect in AAC import into mitochondria. Thereby, for the first time, authentic AAC is stably arrested in the translocase complex of the outer membrane (TOM), as shown by antibody shift blue native electrophoresis. Surprisingly, AAC is still associated with the receptors Tom70 and Tom20 when the function of Tim10 is impaired. The nonessential Tim8-Tim13 complex of the intermembrane space is not involved in the transfer of AAC across the outer membrane. These results define a two-step mechanism for translocation of AAC across the outer membrane. The initial insertion of AAC into the import channel is independent of the function of Tim9-Tim10; however, completion of translocation across the outer membrane, including release from the TOM complex, requires a functional Tim9-Tim10 complex.
Most proteins of mitochondria must be imported from the cytosol. Two major classes of precursor preproteins have been characterized: (i) hydrophilic proteins with cleavable amino-terminal signal sequences (presequences) and (ii) noncleavable membrane proteins with multiple internal targeting signals, of which the inner-membrane metabolite carriers form the major representatives (4, 23, 29, 46, 58). While cleavable preproteins are translocated as linear polypeptide chains and are directed by the presequence through the import channels of the translocase of the outer membrane (TOM) and the presequence translocase of the inner membrane (TIM23 complex), noncleavable carrier proteins follow a distinct import pathway. Five stages of carrier protein import into mitochondria have been defined by using the most abundant carrier, the ADP/ATP carrier (AAC) (47, 48, 50). The precursor of AAC is escorted through the cytosol by molecular chaperones (stage I). AAC is then bound by multiple molecules of the receptor Tom70, involving recognition of several internal targeting signals of AAC (stage II) (61). With the help of additional receptors, including Tom20, AAC is translocated through the general import pore (GIP) complex of the TOM machinery and interacts with soluble intermembrane space Tim protein subunits which form the Tim9-Tim10 complex (stage III) (14, 26, 28, 50, 56, 61). The membrane potential (Δψ)-dependent insertion of AAC into the inner membrane occurs at the carrier translocase, TIM22 complex (stage IV), and is followed by assembly of AAC into a functional dimer in the inner membrane (stage V) (24, 26, 50, 55, 56). The translocation of carrier proteins through the GIP complex, binding by the Tim9-Tim10 complex, and insertion into the inner membrane, i.e., stages III and IV, do not occur in the form of linear polypeptide chains. Rather, these stages apparently involve loop formation and an interplay of the multiple internal targeting signals of the precursor, similar to the events shown for stage II binding to Tom70 (8, 14, 61). Several components of the carrier import pathway are strictly essential for cell viability of the yeast Saccharomyces cerevisiae: the outer-membrane channel-forming protein Tom40, the hetero-oligomeric TIM complex consisting of Tim9 and Tim10 in the intermembrane space, and the subunits Tim22 and Tim12 of the inner-membrane TIM22 complex (1, 3, 21, 26, 28, 32, 33, 55, 56).
Different views have been reported describing the role that the essential intermembrane space Tim proteins play in translocation of carrier proteins across the outer membrane. Protease accessibility assays initially indicated that in mutant mitochondria with a defective Tim9-Tim10 complex, the precursor of AAC accumulated at the outer membrane, although a direct association of the accumulated precursor with the GIP complex was not observed (1, 26, 56). When a fusion protein consisting of AAC and a tightly folded passenger protein (dihydrofolate reductase [DHFR] to which methotrexate was bound) was used, the precursor was stably arrested in the GIP complex. Blue native-polyacrylamide gel electrophoresis (BN-PAGE) of detergent-lysed mitochondria demonstrated that the arrested fusion protein was directly associated with the GIP complex, while chemical cross-linking revealed a close proximity of the fusion protein to Tim10 (50, 61), supporting the view that transfer of the carrier precursor across the outer membrane involved an interaction with the Tim9-Tim10 complex. However, two recent studies challenged a relevant role of the essential Tim proteins in carrier translocation across the outer membrane. First, purified outer-membrane vesicles devoid of Tim proteins were able to stably accumulate the intermediate of the AAC-DHFR fusion protein in the GIP complex (40). The AAC-DHFR-GIP complex showed the same mobility on BN-PAGE, independently of whether mitochondria (with Tim9 and Tim10) or outer-membrane vesicles (without Tim9 and Tim10) were used. Second, a detailed study using mutant forms of the intermembrane space Tim proteins did not yield evidence for a carrier translocation intermediate associated with the TOM complex but rather showed defects in carrier insertion into the inner membrane (43). It was thus concluded that the critical role of the soluble intermembrane space Tim proteins for import of carrier proteins is not precursor translocation across the outer membrane but insertion into the inner membrane. Moreover, Curran et al. (8) reported that the Tim9-Tim10 complex preferentially bound to hydrophobic segments of the AAC precursor, supporting the view that the Tim9-Tim10 complex possessed chaperone-like properties to guide hydrophobic proteins through the aqueous intermembrane space.
Analysis of the mechanism of AAC transfer across the mitochondrial outer membrane has been limited because it has not been possible so far to demonstrate a stable interaction of authentic AAC, i.e., without artificial domains attached, with the GIP complex. In fact, when authentic AAC was accumulated in mitochondria in the absence of a membrane potential, i.e., in association with the Tim9-Tim10 complex (stage III), BN-PAGE showed a mobility in the low-molecular-weight range (50), suggesting that this AAC was not associated with the 400,000-molecular-weight (400K) GIP complex of the TOM machinery. This raised the question of whether the import of authentic AAC involves a stage of stable association with the GIP complex.
For this report we have generated a new mutant of TIM10 with a strong defect in carrier translocation across the outer mitochondrial membrane. We found that authentic AAC stably interacted with the GIP complex. Surprisingly, upon inactivation of Tim10, AAC was still associated with the two primary surface receptors Tom70 and Tom20. These results suggest a functional cooperation between the essential Tim proteins of the intermembrane space and the Tom proteins on the mitochondrial surface. We propose a model that explains the different results reported so far. According to our model, insertion of AAC into the GIP complex occurs independently of the Tim9-Tim10 complex, while completion of translocation across the outer membrane and release from the TOM complex depend on the interaction of AAC with a functional Tim9-Tim10 complex.
MATERIALS AND METHODS
Yeast strains and plasmids.
The S. cerevisiae strains used in this study are shown in Table 1. Standard procedures were used for yeast genetics and manipulation (7, 16).
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| YPH499 (wild type) | MATaade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 | 54 |
| GB100 | MATaade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 tim10::ADE2 pGB5184(URA3)-TIM10 | This study |
| GB101 | MATaade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 tim10::ADE2 pGB5183(TRP1)-tim10-2 | This study |
| GB102 | MATaade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 tim10::tim10-2 | This study |
| NWY50 | MATaade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 tim13::kanMX4 | This study |
| PRY34 | MATaade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 tim13::kanMX4 tim8::TRP1 | This study |
Plasmids used were generated as follows. A PCR product (∼300 bp) containing the TIM10 open reading frame (ORF) was amplified from yeast genomic DNA using primers T10-III (5′-CGCGGATCCATGTCTTTCTTAGGTTTCGGTGGTGGT-3′) and T10-IV (5′-CGGGTCGACCTAAAACTTACCGGCTGCGTTAAATGA-3′), digested with BamHI and SalI, and then inserted between the MET25 promoter and CYC1 terminator in the 2μm URA3-containing plasmid YEp352 (20) to yield pGB5184.
Plasmid pGB5192, used to disrupt genomic TIM10 with the ADE2 gene by homologous recombination, was generated as follows. The 5′ and 3′ untranslated regions (UTRs) of TIM10 were amplified by PCR from genomic DNA by using oligonucleotides T10-I (5′-GCCGAATTCGTTGGTAAGGCGCCACACTAG-3′) and T10-V (5′-TCCCGGGGTCAGATCTCGTTTTTCTAAGTATGATAGTTCCTTC-3′) and oligonucleotides T10-II (5′-GAAACTGCAGCGGTGAAATAACACGAAGATGCG-3′) and T10-VI (5′-GAGATCTGACCCCGGGAGTGCATTAAAGCAGTAATGATAAGGAC-3′), respectively. The promoter- and terminator-carrying PCR fragments, identical at their 3′ and 5′ ends (17-bp), were fused and amplified by PCR using primers T10-I and T10-II. Restriction sites SmaI and BglII were engineered into the junction site of the two UTRs to facilitate subsequent gene insertion. The 600-bp fragment obtained was digested with EcoRI and PstI and was inserted into similarly digested pFL39 (which carries the TRP1 gene [6]) and pUC19 (60) to generate pGB5183 and pGB5182, respectively. A DNA fragment carrying the ADE2 gene was inserted into the BglII site of pGB5182, generating pGB5192.
For TIM10 disruption by homologous recombination, an EcoRI-PstI DNA fragment containing tim10::ADE2 was released from pGB5192 and transformed into YPH499, carrying plasmid pGB5184, thus generating strain GB100. Disruption of the TIM10 ORF with the ADE2 gene was verified by PCR analysis.
Random mutations in TIM10 were generated using a low-fidelity PCR technique. PCR was performed with primers T10-I and T10-II by using genomic DNA as a template. The PCR products (∼900 bp) containing putative TIM10 mutant ORFs and their 5′ and 3′ UTRs were cotransformed with the linearized plasmid pGB5183 into strain GB100. Trp+ transformants were selected at 24°C and screened for the ability to grow at 24°C, but not at 37°C, on minimal glucose media containing the appropriate growth supplements and 5-fluoroorotic acid (5-FOA), indicating loss of the URA3- and TIM10-containing plasmid pGB5184. A plasmid containing the mutant gene tim10-2 [pGB5183(TRP1)-tim10-2] was rescued and retransformed into strain GB100, thereby reestablishing the mutant phenotype. This strain containing plasmid-borne tim10-2 was designated GB101(YPHBG10-7-1).
The isolated tim10-2 mutant was integrated at the TIM10 locus by homologous recombination. The EcoRI-PstI DNA fragment containing the Tim10-2 ORF and its 5′ and 3′ flanking regions was used to transform strain GB100. Transformants were screened for growth at 24°C on minimal glucose media containing 5-FOA and for loss of the ADE2 marker. A temperature-sensitive phenotype and PCR analysis of the genomic DNA of the mutant strain confirmed that the copy of tim10::ADE2 had been replaced by tim10-2, yielding strain GB102(YPH499 10-71-1), used for biochemical analysis.
Deletion of the TIM13 ORF in the wild-type strain YPH499 was generated by transformation of a PCR product (38) containing the KanMX4 gene flanked by regions complementary to TIM13 UTRs and subsequent selection of Kan+ transformants. The deletion cassette was generated by PCR utilizing genomic DNA isolated from a TIM13 disruption strain (Euroscarf accession number Y14811) (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 tim13::KanMX4) and primers 5′-CATTAAACCATACCCTTGACG-3′ and 5′-AACCCAACCTCATAAAGGCAC-3′. The resulting strain was termed NWY50. To generate a tim8Δ tim13Δ double-deletion strain (PRY34), a cassette containing the TRP1 gene flanked by regions complementary to the TIM8 ORF was prepared by PCR using primers 5′-GTATTGAAGAAGAGGTAAAAAGGAAAACAAATTTACAAACAACAAAGAATGCGTACGCTGCAGGTCGAC-3′ and 5′-TATTAAGGTAAAGAGAATGACTCGGAGAGATAAATCGGTTTCATATTAATCGATGAATTCGAGCTCG-3′ and was used to transform strain NWY50, yielding PRY34. Deletion of both the TIM8 and TIM13 ORFs was confirmed by PCR using genomic DNA and Western blotting of isolated mitochondria.
Growth of yeast and isolation conditions.
Mitochondria isolated from yeast strains grown at either 24 or 30°C (for the tim8Δ tim13Δ yeast) in YPG medium (1% [wt/vol] yeast extract, 2% [wt/vol] Bacto Peptone, and 3% [vol/vol] glycerol) (9, 18) were adjusted to a protein concentration of 10 mg/ml with SEM (250 mM sucrose, 1 mM EDTA, 10 mM morpholinepropanesulfonic acid [MOPS]-KOH [pH 7.2]) and stored at −80°C. Growth of the tim10-2 yeast strain at 24°C on YPG was slightly impaired relative to that of the wild-type strain YPH499.
Import of radiolabeled precursor proteins into isolated mitochondria.
Radiolabeled mitochondrial precursor proteins were generated by in vitro transcription from cloned genes using SP6 polymerase and subsequent in vitro translation using rabbit reticulocyte lysate supplemented with [35S]methionine-cysteine (2, 51, 57). S. cerevisiae AAC2 was used in this study. Prior to in vitro import, isolated yeast mitochondria were resuspended in import buffer (3% [wt/vol] bovine serum albumin, 250 mM sucrose, 5 mM MgCl2, 80 mM KCl, 5 mM methionine, 10 mM MOPS-KOH [pH 7.2]) containing 2 mM (each) ATP and NADH. For dissipation of the mitochondrial membrane potential, 8 μM antimycin A, 20 μM oligomycin, and 1 μM valinomycin were added prior to import. Import was initiated by addition of rabbit reticulocyte lysate containing 35S-labeled preproteins (4 to 10% [vol/vol] of the import reaction mixture) to mitochondria (25 to 50 μg of protein) and incubation at 25°C for the times indicated. To stop import, valinomycin (1 μM) was added. Where indicated, mitochondria were treated with proteinase K (50 μg/ml) for 15 min on ice. Protease digestion was terminated with 1 mM phenylmethylsulfonyl fluoride. Mitochondria were reisolated by centrifugation and washed once with SEM. Then detergent-solubilized mitochondrial proteins were separated by sodium dodecyl sulfate (SDS)-PAGE or BN-PAGE.
BN-PAGE and antibody shift BN-PAGE.
BN-PAGE (11, 53) was performed as follows. Essentially, mitochondrial pellets (50 to 75 μg of protein) were resuspended in 45 μl of ice-cold digitonin buffer (1% [wt/vol] digitonin, 20 mM Tris-HCl [pH 7.4], 0.1 mM EDTA, 50 mM NaCl, 10% [vol/vol] glycerol, 1 mM phenylmethylsulfonyl fluoride) and incubated on ice for 15 min, then immediately centrifuged at 12,000 × g and 4°C for 15 min. Sample buffer (5 μl; 5% [wt/vol] Coomassie brilliant blue G-250, 100 mM Bis-Tris [pH 7.0], 500 mM ɛ-amino-n-caproic acid) was added to the solubilized mitochondrial proteins. Soluble protein fractions were separated on 6-to-13% or 6-to-16.5% polyacrylamide gradient gels at 4°C. Gels were destained, dried, and subjected to digital autoradiography (Molecular Dynamics) for detection of radiolabeled imported proteins. Second-dimension analysis using 16.5% separating gels (Tris-Tricine-buffered SDS-polyacrylamide gels [52]) was performed by setting gel strips representing single lanes from the first-dimension BN-PAGE in a stacking gel on top of individual SDS separating gels. Following electrophoresis, proteins were transferred directly to polyvinylidene difluoride (PVDF) membranes by use of a semidry transfer system with transfer buffer (20 mM Tris base, 150 mM glycine, 20% [vol/vol] methanol, 0.08% [wt/vol] SDS) and were then analyzed by immunodecoration with specific polyclonal antibodies.
For antibody shift BN-PAGE of in vitro-imported radiolabeled proteins, mitochondrial pellets were resuspended in 100 μl of SEM; then purified immunoglobin G (IgG) in import buffer was added and samples were incubated on ice for 30 min with occasional mixing. Mitochondria with bound IgG were reisolated and washed once with SEM prior to preparation for BN-PAGE as above. Antibody depletion of the endogenous GIP complex was performed as follows. Mitochondria were lysed with digitonin buffer; then purified IgG in import buffer was added and samples were incubated on ice for 30 min with occasional mixing. Protein A-Sepharose (bed volume, 20 μl) preequilibrated with digitonin buffer was then added, and samples were incubated on ice for 30 min to remove the bulk of IgG, which would otherwise interfere with the subsequent immunodecoration. Following a low-speed centrifugation to remove the Sepharose beads, proteins were separated by BN-PAGE and then subjected to Western blot analysis.
Chemical cross-linking and immunoprecipitation assays.
Mitochondria, in import buffer minus bovine serum albumin, were pretreated with 1 μM valinomycin and 20 μM oligomycin to dissipate the membrane potential. After addition of 2 mM ATP, 100 μg of creatine kinase/ml, and 5 mM creatine phosphate, the import reaction was initiated by addition of 35S-labeled AAC and the reaction mixture was incubated for 20 min at 25°C. For cross-linking, the import mix was cooled on ice and incubated with 0.8 mM ethylene glycol-bis(succinimidylsuccinate) (EGS) for 30 min. The cross-linking was quenched with 50 mM Tris-HCl, pH 7.4, for 5 min. The reisolated mitochondria were resuspended in lysis buffer (150 mM NaCl, 10 mM Tris-HCl [pH 7.4], 0.5% [vol/vol] Triton X-100) containing 1% (wt/vol) SDS, incubated for 5 min at 95°C, diluted with 15 volumes of lysis buffer, and subjected to immunoprecipitation using specific antibodies covalently attached to protein A-Sepharose. The antigens were eluted with 100 mM glycine, pH 2.5.
Miscellaneous.
Polyclonal antibodies to Tim54 were affinity purified from an antiserum by using the purified antigen Tim54sd (32) immobilized to CNBr-activated-Sepharose (17). IgG used in antibody shift BN-PAGE was isolated from an antiserum by using protein A-Sepharose, lyophilized, and then resuspended in import buffer (57). SDS-PAGE and urea-SDS-PAGE were performed according to the work of Laemmli (35) and Ito et al. (22), respectively. For some figures, nonrelevant gel lanes were excised by digital processing. Immune complexes were detected by enhanced chemiluminescence (Amersham). An assessment of the mitochondrial membrane potential was obtained by using 3,3′-dipropylthiadicarbocyanine iodide [DiSC3(5)] essentially as described previously (15).
RESULTS
A new tim10 mutant with a strong defect in AAC import.
Yeast mutants of TIM10 were generated by in vitro mutagenesis of the TIM10 ORF by use of low-fidelity PCR. The chromosomal copy of TIM10 was deleted in a haploid yeast strain, while a plasmid containing wild-type TIM10 and the marker gene URA3 was present. The resulting strain, GB100, was transformed with a plasmid containing mutant tim10, and the loss of the URA3 plasmid (with wild-type TIM10) was selected by growth on 5-FOA. A mutant strain defective in growth at 37°C was selected. The mutant tim10-2 ORF was integrated into the original TIM10 chromosomal locus, yielding strain GB102 (′tim10-2′) used in this study.
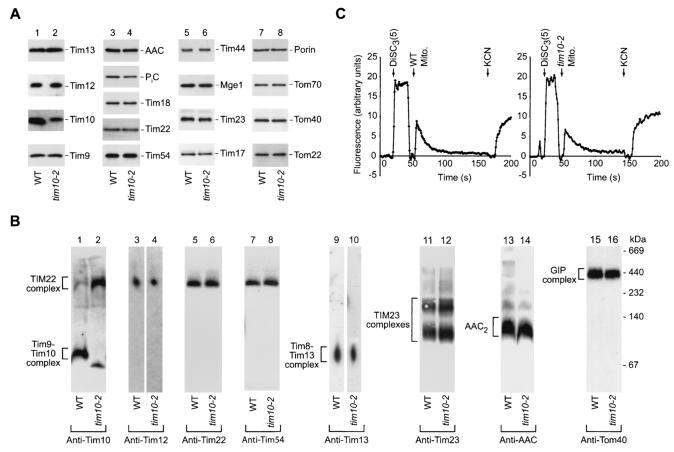
Studies were performed using mitochondria isolated from tim10-2 or the corresponding wild-type cells grown at 24°C on a nonfermentable carbon source. The steady-state levels of a selection of proteins representing all four mitochondrial subcompartments were determined by Western blot analysis with specific antibodies (Fig. 1A). The small Tim proteins, Tim13, Tim12, and Tim9, were present in similar amounts in wild-type and tim10-2 mitochondria (Fig. 1A, lanes 1 and 2). The levels of the mutant protein Tim10-2 detected by Western blot analysis with different polyclonal antisera were reduced to different degrees compared to the level of wild-type Tim10, at least in part reflecting loss of antigen recognition due to the substitution of several amino acid residues in Tim10-2. Additionally the stability of the Tim10-2 mutant protein might also be altered. The steady state levels of other mitochondrial proteins tested were not significantly affected, including the integral components of the TIM22 complex, Tim18, Tim22, and Tim54 (Fig. 1A, lanes 3 and 4); components of the presequence translocase, Tim23, Tim17, Tim44, and the matrix cochaperone Mge1 (Fig. 1A, lanes 5 and 6); and several outer membrane proteins, porin, Tom70, Tom40, and Tom22 (Fig. 1A, lanes 7 and 8). Steady-state levels of the phosphate carrier (PiC) and possibly also AAC, substrates of the Tim9-Tim10 carrier import pathway, were slightly reduced in tim10-2 mitochondria (Fig. 1A, lanes 3 and 4). While the tim10 mutant yeast strains used previously had strongly reduced steady-state levels of other essential components of the carrier import pathway, including Tim22 (26, 43), the tim10-2 mutant strain has the advantage that the function of Tim10 is selectively impaired while all other components of the carrier import pathway are present in normal amounts.
FIG. 1.
The Tim9-Tim10 complex is selectively modified in tim10-2 mitochondria. (A) Immunodecoration of purified mitochondria demonstrating the protein composition in tim10-2 compared to wild-type (WT) mitochondria. Proteins (25 μg/lane) were separated by SDS-PAGE, transferred to a PVDF membrane by Western blotting, and then decorated with antibodies as indicated. (B) Analysis of mitochondrial protein complexes by immunodecoration following separation of detergent-solubilized mitochondria by BN-PAGE and, for lanes 3, 4, 9, and 10, additional separation by a second-dimension SDS-PAGE. Isolated wild-type and tim10-2 mitochondria (50 to 75 μg of protein/lane) were solubilized in 1% digitonin-containing buffer. Following either first-dimension BN-PAGE or second-dimension SDS-PAGE, proteins were blotted to a PVDF membrane and then immunodecorated with specific antibodies as indicated. AAC2, assembled AAC dimer. (C) Comparison of the membrane potential of mitochondria isolated from tim10-2 yeast cells to that of control wild-type mitochondria. Estimation of the membrane potential was performed at 25°C using the fluorescent dye DiSC3(5). The difference in fluorescence prior to and following addition of potassium cyanide (KCN) yields a relative assessment of the membrane voltage.
Using BN-PAGE and Western blotting, proper assembly of the translocase complexes was analyzed. The Tim9-Tim10 complex, which is normally present as a soluble 70-kDa complex in the intermembrane space (Fig. 1B, lane 1), was mainly associated with the large TIM22 inner-membrane complex in tim10-2 mitochondria (Fig. 1B, lane 2). A minor amount of the Tim9-Tim10 complex still migrated in the low-molecular-mass range (the small shift in the apparent mobility compared to that of the wild-type complex is probably due to the amino acid substitutions in Tim10-2) (Fig. 1B, lane 2). Apart from the change in Tim10 localization, no obvious alterations were found for the TIM22 complex (Fig. 1B, lanes 3 to 8), the other soluble intermembrane space complex comprising Tim8 and Tim13 (Fig. 1B, lanes 9 and 10), the TIM23 complexes (Fig. 1B, lanes 11 and 12), or the GIP complex (Fig. 1B, lanes 15 and 16). The mature AAC dimer was present in tim10-2 cells but with perhaps a slight reduction in steady-state levels (Fig. 1B, lanes 13 and 14).
Since import of all cleavable and noncleavable precursor proteins into the inner membrane is dependent on the mitochondrial membrane potential, we investigated the integrity of this aspect of mitochondrial function in tim10-2 mitochondria. An assessment of membrane potential using the quenching of the fluorescent dye DiSC3(5) found no major difference in generation of a membrane potential between wild-type and tim10-2 mitochondria (Fig. 1C).
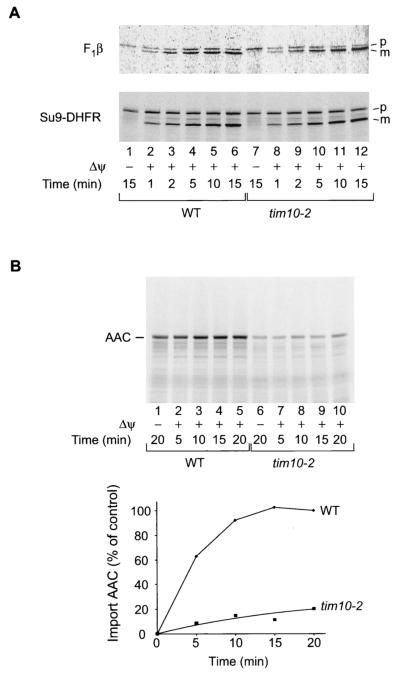
To test the import competence of tim10-2 mitochondria, 35S-labeled mitochondrial precursor proteins synthesized in rabbit reticulocyte lysates were incubated with isolated mitochondria. The import rates of the presequence-containing proteins, F1-ATPase subunit β (F1β) and the model protein Su9-DHFR (presequence of Fo-ATPase subunit 9 fused to DHFR), as revealed by formation of the mature proteins, were very similar in wild-type and tim10-2 mitochondria (Fig. 2A). However, import of AAC, analyzed by transport to a protease-protected location, was dramatically reduced, with a 10-fold decrease in the import rate and a 5-fold decrease in the import yield after 20 min (Fig. 2B). Since the three mitochondrial translocase complexes TOM, TIM23, and TIM22 are properly assembled and the import of presequence-containing preproteins is largely unaffected, we conclude that the tim10-2 mutation selectively leads to a strong defect in the carrier import pathway.
FIG. 2.
Selective impairment of the import of carrier proteins, but not of presequence-containing preproteins, in tim10-2 mitochondria. (A) Radiolabeled matrix targeted preproteins F1β and Su9-DHFR were imported into isolated wild-type (WT) or tim10-2 mitochondria in the presence or absence of a membrane potential (Δψ) for the indicated times. Precursor (p) and mature (m) proteins were detected by digital autoradiography following separation of reisolated mitochondrial proteins by urea-SDS-PAGE. (B) Radiolabeled AAC was imported into isolated wild-type or tim10-2 mitochondria in the presence or absence of a Δψ for the indicated times, followed by proteinase K treatment; then proteins were separated by urea-SDS-PAGE. Quantification of imported (non-protease-accessible) AAC is shown in the graph below the gel.
AAC accumulates at the outer membrane in tim10-2 mitochondria.
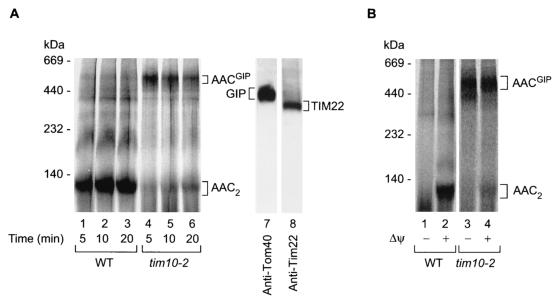
To determine the stage at which the import of AAC was inhibited, we made use of BN-PAGE following import of the radiolabeled precursor. Upon import into isolated energized mitochondria, the precursor of AAC assembled to the mature dimer when wild-type yeast mitochondria were used (Fig. 3A, lanes 1 to 3). However, when tim10-2 mitochondria were used, formation of the mature AAC dimer was strongly inhibited (Fig. 3A, lanes 4 to 6), in agreement with the result of the protease protection experiment shown in Fig. 2B. Instead, the AAC precursor was found in the high-molecular-mass range around 500 to 600 kDa (Fig. 3A, lanes 4 to 6). Two large translocase complexes exist on the carrier import pathway, the TOM or GIP complex of the outer membrane (∼400 to 500 kDa) and the TIM22 complex of the inner membrane (∼300 kDa), at which the AAC precursor could accumulate (Fig. 3A, lanes 7 and 8) (1, 12, 25, 28, 30, 32, 34, 40, 55, 56, 59). The complex containing the accumulated, radiolabeled AAC precursor should be larger than the translocase complex detected by Western blotting, since the molecular mass of AAC will add to the translocase complex. Since the radiolabeled precursor protein is present in only minute amounts, it does not add to the mass of the bulk of translocase complexes analyzed by Western blotting. Since the size of the TOM complex is closer to the 500- to 600-kDa range, we suspected that the AAC precursor may have been arrested in the TOM/GIP complex, and thus we tentatively referred to this putative GIP intermediate as AACGIP. Translocation of AAC across the outer membrane is independent of the membrane potential across the inner membrane, while insertion into the inner membrane via Tim22 requires a membrane potential (1, 14, 24, 26, 32, 48, 50, 55, 56). We thus tested if the accumulation of AACGIP required the presence of a membrane potential. Figure 3B demonstrates that tim10-2 mitochondria accumulated the AAC precursor (AACGIP) independently of whether a membrane potential was present (lane 4) or not (lane 3).
FIG. 3.
In tim10-2 mitochondria, imported AAC accumulates as a high-molecular-weight intermediate. (A) (Left panel) Radiolabeled AAC was imported into wild-type (WT) or tim10-2 mitochondria for the times indicated. The mitochondria were solubilized in 1% digitonin-containing buffer and separated by BN-PAGE. AACGIP, accumulated radiolabeled AAC intermediate. (Right panel) Immunodecoration indicating the relative positions of the endogenous GIP complex and TIM22 complex in tim10-2 mitochondria. (B) Accumulation of the outer-membrane AAC translocation intermediate (AACGIP) is independent of the membrane potential (Δψ). Radiolabeled AAC was incubated (for 10 min at 25°C) with isolated wild-type or tim10-2 mitochondria that were either pretreated with valinomycin to dissipate the membrane potential (lanes 1 and 3) or left untreated (lanes 2 and 4). Mitochondria were solubilized with 1% digitonin-containing buffer and subjected to BN-PAGE and digital autoradiography.
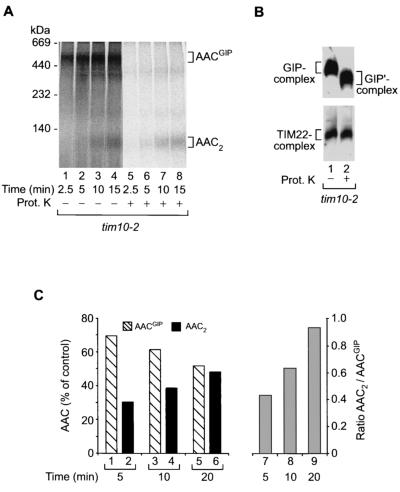
To obtain further experimental evidence in order to discern at which of the translocase complexes the AAC precursor was accumulated, we analyzed the protease accessibility of the precursor protein in the 500- to 600K complex. Upon accumulation of AACGIP in tim10-2 mitochondria, the samples were split equally and half were treated with externally added proteinase K (Fig. 4A, lanes 5 to 8). The accumulated precursor protein was completely digested, supporting the view that it was arrested in the TOM complex. Western blot analysis of the TOM and TIM22 complexes of tim10-2 mitochondria following proteinase K treatment confirmed that only proteins accessible to the outside of mitochondria were digested by the protease, as indicated by the shift in the molecular mass of the TOM complex (Fig. 4B, upper panel) while the inner-membrane TIM22 complex remained intact (Fig. 4B; lower panel). The TIM22 complex was digested only after rupturing of the outer membrane (data not shown). Thus, digestion of AACGIP by proteinase K in tim10-2 mitochondria is a consequence of its location at the outer side of the outer membrane, most likely in association with the GIP complex.
FIG. 4.
AACGIP is located at the mitochondrial outer membrane and represents a translocation intermediate on an import pathway. (A) AACGIP is protease sensitive in whole mitochondria. Radiolabeled AAC was imported into tim10-2 mitochondria for the indicated times. Following import, samples were split in half and either left untreated (lanes 1 to 4) or treated with proteinase K (Prot. K) (lanes 5 to 8). Detergent-solubilized (1% digitonin) mitochondrial proteins were then separated by BN-PAGE and subjected to digital autoradiography. (B) Outer membrane integrity is maintained in tim10-2 mitochondria following protease treatment. As a control tim10-2 mitochondria were left untreated (lane 1) or treated with proteinase K (lane 2) under experimental conditions identical to those for panel A. Following solubilization in 1% digitonin and separation by BN-PAGE, mitochondrial proteins were transferred to a PVDF membrane and immunodecorated with anti-Tom40 and anti-Tim22 antibodies. GIP′-complex, core of GIP in which surface receptors have been removed by protease treatment. (C) (Left graph) Quantitation of AAC complexes in tim10-2 mitochondria during import by digital autoradiography (the experiment was performed as described in the legend to Fig. 3A), indicating the transition of AACGIP to the fully imported species AAC2 with time. The 100% control is the sum of the AACGIP and AAC2 radioactive signals. (Right graph) Ratio of AAC2 to AACGIP during the course of the import reaction.
Is the AACGIP complex of tim10-2 mitochondria a dead-end product or a real intermediate in the import pathway of AAC? Quantitation of the import and assembly kinetics of AAC in tim10-2 mitochondria shows that at longer import times the amount of AACGIP complex slowly declines (Fig. 4C, bars 1, 3, and 5) concomitantly with the slow increase of mature, dimeric AAC (Fig. 4C, bars 2, 4, and 6), a finding also indicated by the significant increase in the ratio of dimeric AAC to AACGIP (Fig. 4C, bars 7 to 9). This suggests that AACGIP does not represent an irreversibly blocked precursor protein but is an intermediate in the import reaction whose transfer to the next stage is strongly delayed in tim10-2 mitochondria compared to that in wild-type mitochondria. Indeed, reinspection of a large number of experiments on the import of AAC into wild-type mitochondria revealed the presence of small amounts of AACGIP (see, e.g., Fig. 3A, lanes 1 to 3, and below, Fig. 5, lane 8, and Fig. 7, lanes 1 and 4). Apparently due to the rapid kinetic transfer of AAC to subsequent import stages in wild-type mitochondria (5, 13, 19, 26, 41, 59), the intensity of the AACGIP band is low and dependent on the overall background of the blue native gel.
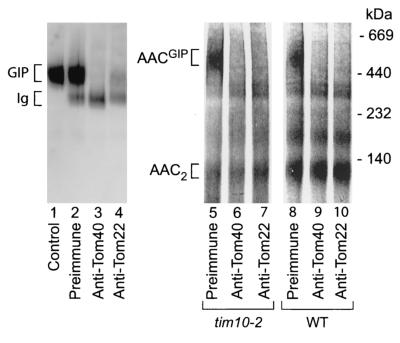
FIG. 5.
AAC arrests in the GIP complex. (Left panel) Mitochondria were resuspended in SEM buffer and either left untreated (lane 1) or treated with preimmune IgG (lane 2), anti-Tom40 IgG (lane 3), or anti-Tom22 IgG (lane 4) prior to solubilization in digitonin buffer and separation of protein complexes by BN-PAGE. The GIP complex was detected by immunodecoration using an anti-Tom40 antiserum following transfer of proteins to a PVDF membrane by Western blotting. (Right panel) Radiolabeled AAC was imported into tim10-2 and wild-type (WT) mitochondria for 15 min to allow accumulation of the AAC intermediate; then mitochondria were washed, resuspended in SEM, and treated with preimmune IgG (lanes 5 and 8), anti-Tom40 IgG (lanes 6 and 9), or anti-Tom22 IgG (lanes 7 and 10) prior to solubilization in digitonin buffer and separation of protein complexes by BN-PAGE. Radiolabeled AAC was visualized by digital autoradiography.
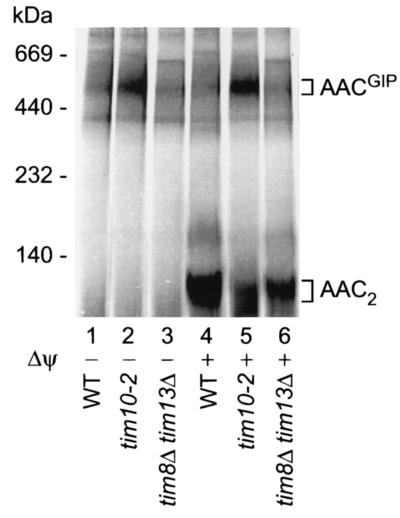
FIG. 7.
The Tim8-Tim13 complex in the intermembrane space is not required for transition of the AAC precursor from GIP-associated species to subsequent import steps. Radiolabeled AAC was incubated with isolated wild-type (WT), tim10-2, or tim8Δ tim13Δ mitochondria with (lanes 1 to 3) or without (lanes 4 to 6) dissipation of the membrane potential prior to import at 25°C for 15 min. Radiolabeled AAC was visualized by digital autoradiography following separation of digitonin (1%)-solubilized proteins by BN-PAGE.
Taken together, these results suggest that the functional impairment of the intermembrane space-located Tim9-Tim10 complex by the tim10-2 mutant leads to the accumulation of an import intermediate of AAC in the translocase of the outer membrane.
Accumulated AAC is associated with the GIP complex.
To directly determine where the AAC precursor was accumulated in tim10-2 mitochondria, its association with the GIP complex was investigated. Under defined digitonin solubilization conditions the GIP complex of S. cerevisiae consists of Tom40, Tom22, and the small Tom proteins (12, 40, 59). We used a new method of antibody shift BN-PAGE to analyze AACGIP. The large sizes of antibodies, together with their inherent high-affinity interactions with their antigens, result in significant size shifts in the BN-PAGE profiles of protein complexes when one or more antibody molecules are bound to a specific complex. Antibodies were applied to tim10-2 mitochondria in which radiolabeled AAC had previously accumulated; the mitochondria were then solubilized with digitonin, and protein complexes were separated by BN-PAGE. When an anti-Tom40 or anti-Tom22 antibody was added prior to BN-PAGE analysis, the radioactive signal of AACGIP was no longer visible (Fig. 5, lanes 6 and 7), presumably due to the shift in the size of the complex to a very-high-molecular-mass species not resolved by the gel system employed. Antibodies from preimmune serum did not result in a shift of radiolabeled AAC (Fig. 5, lane 5). When the antibodies were added to wild-type mitochondria upon import of radiolabeled AAC, the mobility of the mature AAC dimer on BN-PAGE gels was not affected (Fig. 5, lanes 8 to 10), indicating the specificity of the assay. The small amount of AAC observed in the GIP complex range in wild-type mitochondria (Fig. 5, lane 8) was shifted by the anti-Tom40 and anti-Tom22 antibodies (Fig. 5, lanes 9 and 10), providing direct evidence that wild-type mitochondria were also able to form AACGIP. As an additional control for the method, we investigated whether antibodies directed against components of the GIP complex could indeed specifically deplete solubilized mitochondria of the endogenous 400K GIP complex. Preincubation of mitochondria with an anti-Tom40 or anti-Tom22 antibody, but not with preimmune antibodies, led to almost-complete depletion of the 400K GIP complex, as determined by decoration of the endogenous GIP complex with an anti-Tom40 antibody following BN-PAGE analysis (Fig. 5; compare lanes 3 and 4 with lane 2).
These results demonstrate that the bulk of AAC precursor molecules accumulated in tim10-2 mitochondria are associated with the GIP complex. We expect that the antibody shift BN-PAGE analysis will be a useful tool for identifying the composition of protein complexes and particularly translocation intermediates, which often are present only in small quantities and detectable only by use of radioactively labeled proteins.
Accumulation of AAC at surface receptors in tim10-2 mitochondria.
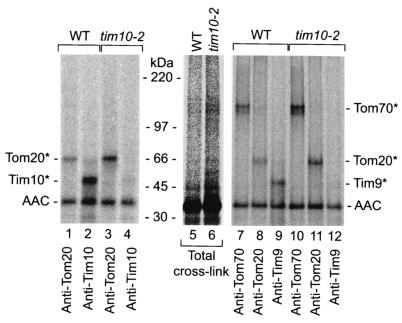
Next we wondered if the arrested AAC precursor was associated with import components situated before or after the GIP complex, i.e., the initial receptors Tom70 and Tom20 as well as the Tim9-Tim10 complex. Because neither Tom70 nor the Tim9-Tim10 complex stably associates with the GIP complex and Tom20 is only weakly associated, especially at the higher digitonin concentrations used for BN-PAGE analysis (12, 40, 42), we used cross-linking with the homobifunctional chemical cross-linking reagent EGS instead of antibody shift BN-PAGE. Upon incubation of radiolabeled AAC with wild-type or tim10-2 mitochondria in the absence of a membrane potential, EGS was added. The mitochondria were reisolated and washed extensively. Upon lysis of the mitochondrial membranes under denaturing conditions, proteins associated with the AAC precursor were identified by immunoprecipitation. In the first set, we compared the efficiency of cross-linking of the AAC precursor to Tom20 and Tim10 in wild-type and mutant mitochondria. Whereas wild-type mitochondria formed a strong cross-linked product between AAC and Tim10, this product was virtually absent in tim10-2 mitochondria (Fig. 6, lanes 2 and 4), yet unexpectedly, the cross-linking of AAC to Tom20 was significantly increased in tim10-2 mitochondria (Fig. 6, lane 3). Similarly, the AAC precursor was cross-linked to Tom20 in tim10-2 mitochondria in the presence of a membrane potential also (data not shown).
FIG. 6.
AAC accumulates at surface receptors in tim10-2 mitochondria. Radiolabeled AAC in reticulocyte lysate was incubated with yeast wild-type and tim10-2 mitochondria in the absence of a membrane potential (presence of valinomycin and oligomycin). Following cross-linking with EGS, the mitochondria were reisolated, washed, and subjected to immunoprecipitation under stringent conditions with the antibodies indicated. The samples were separated by SDS-PAGE and analyzed by digital autoradiography. “Total cross-link” represents 5% of the material used for immunoprecipitation. Asterisks indicate cross-linked products of AAC and specified proteins.
Since the lack of detection of an AAC-Tim10 cross-linked product in the mutant mitochondria could at least in part be due to the amino acid alterations in Tim10-2, we analyzed the second component of this intermembrane space TIM complex, Tim9, which is not affected by the mutation. In wild-type mitochondria, but not in tim10-2 mitochondria, the AAC precursor formed a cross-linked product with Tim9 in the absence of a membrane potential (Fig. 6, lanes 9 and 12), indicating that AAC is indeed unable to contact the Tim9-Tim10 complex in tim10-2 mitochondria. Moreover, tim10-2 mitochondria formed strongly increased amounts not only of the AAC-Tom20 cross-linked product, but also of an AAC-Tom70 cross-linked product, relative to wild-type mitochondria (Fig. 6, lane 11 versus lane 8 and lane 10 versus lane 7). The possibility that the increased binding of the AAC intermediate to Tom receptors in tim10-2 mitochondria was merely a backup of AAC due to a downstream blockage in the import pathway at the GIP complex can be excluded, since the experiments were performed with radiolabeled precursors which constitute a very small, nonsaturating amount of protein with respect to the number of total import sites available (37, 42, 44). These findings indicate that a block of AAC interaction with the Tim9-Tim10 complex in tim10-2 mitochondria leads to an arrest of the precursor in the TOM complex at a surprisingly early stage, where it is still in close proximity to the primary receptors Tom70 and Tom20.
Accumulation of AAC at the TOM complex depends on the essential Tim9-Tim10 complex but not on the Tim8-Tim13 complex.
Besides the essential Tim9-Tim10 complex, the mitochondrial intermembrane space contains a second complex of small Tim proteins, the Tim8-Tim13 complex (27, 34, 36, 49). This complex is not essential for cell viability and has been reported to play a role in the import of some precursor proteins such as the precursor of Tim23, in particular when special import conditions such as dissipation of the membrane potential were used (10, 27, 36, 45). Since the AACGIP intermediate described here represents a new tool for analyzing the stages of AAC import into mitochondria, we asked if a possible involvement of the Tim8-Tim13 complex in AAC transport across the outer membrane has been overlooked so far and may be discernible at the level of the AACGIP translocation intermediate. We constructed a mutant yeast strain in which the complete ORFs of both TIM8 and TIM13 were deleted. Import of AAC into these mitochondria was analyzed in the absence or in the presence of a membrane potential and compared to import into wild-type and tim10-2 mitochondria. No evidence for increased formation of the AACGIP intermediate in tim8Δ tim13Δ mitochondria compared to that in wild-type mitochondria was observed (Fig. 7, lanes 3 and 6 versus lanes 1 and 4). These results support the view that the Tim8-Tim13 complex is not involved in the transfer of AAC through the TOM machinery.
DISCUSSION
We have defined two steps in the translocation of carrier proteins through the GIP of the mitochondrial outer membrane and thus can explain the various and seemingly conflicting results that have been reported on the role of the essential Tim9-Tim10 complex of the intermembrane space in the import of carrier proteins. The initial insertion of the AAC precursor protein into the TOM/GIP complex is independent of the Tim9-Tim10 complex, in agreement with the observation that purified outer-membrane vesicles devoid of soluble intermembrane-space Tim proteins can efficiently accumulate AAC in the GIP complex (40). The completion of AAC translocation through the GIP, however, requires a functional Tim9-Tim10 complex. The completion of translocation involves the release of AAC precursor molecules from the two surface receptors Tom70 and Tom20. This finding explains why AAC, which was accumulated in mitochondria defective in Tim9 or Tim10, was still accessible to proteases added to the isolated mitochondria (1, 14, 26, 56).
The finding that a functional Tim9-Tim10 complex is needed for release of AAC from import receptors is quite surprising but is consistent with the view that carrier proteins are imported in nonlinear chain conformations and thus may require additional assistance to effectively transfer to the intermembrane space of mitochondria. Whereas presequence-containing preproteins typically contain a single amino-terminal targeting signal which directs their import by sequential binding to Tom and Tim proteins (31, 40, 46), interaction of AAC with the receptor Tom70 is mediated by the multiple internal signals of the precursor, which cooperate in recruiting several receptor molecules. The subsequent stepwise transfer of AAC to Tom proteins acting later in its import pathway occurs while some targeting signals are still in contact with Tom70 (61). This model can now be extended to the translocation of AAC across the outer membrane, suggesting a transmembrane cooperation between surface receptors and Tim9-Tim10 in AAC transfer. Only upon stable interaction of part of an AAC precursor molecule with the Tim9-Tim10 complex in the intermembrane space is the precursor released from the Tom proteins on the mitochondrial surface and thus able to complete the translocation through the GIP complex.
How can the different conclusions of Murphy et al. (43) and our study be reconciled? Murphy et al. (43) used a yeast strain with the the tim10-1 mutant gene in combination with a suppressor allele of TIM9 and a deletion of TIM8. In this strain, the steady-state level of Tim22 was decreased. In the isolated mutant mitochondria, no accumulation or association of the AAC precursor with the TOM complex was observed, and only the insertion of AAC into the inner membrane was impaired (43). Taking the various reports together, it is now evident that the essential Tim subunits of the intermembrane space play critical roles at three stages during the import of AAC. (i) They are required for the release of the AAC precursor from the import receptors and movement through the GIP. (ii) The Tim9-Tim10 complex functions in a chaperone-like manner, guiding the hydrophobic AAC precursor through the aqueous intermembrane space (8, 39). The release of AAC from the GIP complex occurs independently of the Tim8-Tim13 complex, in agreement with the detailed analysis of Curran et al. (8), which showed the specificity of the Tim9-Tim10 complex for carrier proteins. (iii) The Tim9-Tim10 complex associates with Tim12 and is thus also involved in the insertion of AAC into the inner membrane via the TIM22 complex (1, 26, 28, 43, 56). The differences between our study and that of Murphy et al. (43) with respect to the translocation of AAC across the outer membrane are most likely due to the different mutant forms of tim10 employed. The new tim10-2 allele isolated here revealed a strong phenotype, impairing the first stage of involvement of Tim9-Tim10, while the mutant strain used by Murphy et al. (43) affected the later stage of AAC import. The tim10-2 mutant allele thus provides the first experimental possibility of accumulating the authentic precursor of AAC in the GIP complex of the outer membrane. This demonstrates directly that authentic AAC is translocated via the GIP complex.
Acknowledgments
We thank R. Jensen for the antiserum against Tim17; C. Koehler, K. Tokatlidis, and G. Schatz for the antiserum against Tim12; and T. Prinz for experimental advice.
This work was supported by the Deutsche Forschungsgemeinschaft, the Sonderforschungsbereich 388 Freiburg, and the Fonds der Chemischen Industrie/BMBF. K.N.T. was supported by a long-term fellowship from the Alexander von Humboldt-Stiftung.
REFERENCES
- 1.Adam, A., M. Endres, C. Sirrenberg, F. Lottspeich, W. Neupert, and M. Brunner. 1999. Tim9, a new component of the TIM22·54 translocase in mitochondria. EMBO J. 18:313-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alconada, A., F. Gärtner, A. Hönlinger, M. Kübrich, and N. Pfanner. 1995. Mitochondrial receptor complex from Neurospora crassa and Saccharomyces cerevisiae. Methods Enzymol. 260:263-286. [DOI] [PubMed] [Google Scholar]
- 3.Baker, K. P., A. Schaniel, D. Vestweber, and G. Schatz. 1990. A yeast mitochondrial outer membrane protein essential for protein import and cell viability. Nature 348:605-609. [DOI] [PubMed] [Google Scholar]
- 4.Bauer, M. F., S. Hofmann, W. Neupert, and M. Brunner. 2000. Protein translocation into mitochondria: the role of TIM complexes. Trends Cell Biol. 10:25-31. [DOI] [PubMed] [Google Scholar]
- 5.Bömer, U., A. C. Maarse, F. Martin, A. Geissler, A. Merlin, B. Schönfisch, M. Meijer, N. Pfanner, and J. Rassow. 1998. Separation of structural and dynamic functions of the mitochondrial translocase: Tim44 is crucial for the inner membrane import sites in translocation of tightly folded domains, but not of loosely folded preproteins. EMBO J. 17:4226-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonneaud, N., O. Ozier-Kalogeropoulos, G. Y. Li, M. Labouesse, L. Minvielle-Sebastia, and F. Lacroute. 1991. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors Yeast 7:609-615. [DOI] [PubMed]
- 7.Burke, D., D. Dawson, and T. Stearns. 2000. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual, 2000 ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N. Y.
- 8.Curran, S. P., D. Leuenberger, W. Oppliger, and C. M. Koehler. 2002. The Tim9p-Tim10p complex binds to the transmembrane domains of the ADP/ATP carrier. EMBO J. 21:942-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daum, G., P. C. Böhni, and G. Schatz. 1982. Import of proteins into mitochondria: cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J. Biol. Chem. 257:13028-13033. [PubMed] [Google Scholar]
- 10.Davis, A. J., N. B. Sepuri, J. Holder, A. E. Johnson, and R. E. Jensen. 2000. Two intermembrane space TIM complexes interact with different domains of Tim23p during its import into mitochondria. J. Cell Biol. 150:1271-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dekker, P. J. T., F. Martin, A. C. Maarse, U. Bömer, H. Müller, B. Guiard, M. Meijer, J. Rassow, and N. Pfanner. 1997. The Tim core complex defines the number of mitochondrial translocation contact sites and can hold arrested preproteins in the absence of matrix Hsp70-Tim44. EMBO J. 16:5408-5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dekker, P. J. T., M. T. Ryan, J. Brix, H. Müller, A. Hönlinger, and N. Pfanner. 1998. Preprotein translocase of the outer mitochondrial membrane: molecular dissection and assembly of the general import pore complex. Mol. Cell. Biol. 18:6515-6524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dietmeier, K., A. Hönlinger, U. Bömer, P. J. T. Dekker, C. Eckerskorn, F. Lottspeich, M. Kübrich, and N. Pfanner. 1997. Tom5 functionally links mitochondrial preprotein receptors to the general import pore. Nature 388:195-200. [DOI] [PubMed] [Google Scholar]
- 14.Endres, M., W. Neupert, and M. Brunner. 1999. Transport of the ADP/ATP carrier of mitochondria from the TOM complex to the TIM22·54 complex. EMBO J. 18:3214-3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gärtner, F., W. Voos, A. Querol, B. R. Miller, E. A. Craig, M. G. Cumsky, and N. Pfanner. 1995. Mitochondrial import of subunit Va of cytochrome c oxidase characterised with yeast mutants: independence from receptors, but requirement for matrix hsp70 translocase function. J. Biol. Chem. 270:3788-3795. [DOI] [PubMed] [Google Scholar]
- 16.Guthrie, C., and G. R. Fink (ed.). 1991. Methods in enzymology, vol. 194. Guide to yeast genetics and molecular biology. Academic Press, San Diego, Calif. [PubMed]
- 17.Harlow, E., and D. Lane. 1999. Using antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N. Y.
- 18.Hartl, F. U., J. Ostermann, B. Guiard, and W. Neupert. 1987. Successive translocation into and out of the mitochondrial matrix: targeting of proteins to the intermembrane space by a bipartite signal peptide. Cell 51:1027-1037. [DOI] [PubMed] [Google Scholar]
- 19.Haucke, V., M. Horst, G. Schatz, and T. Lithgow. 1996. The Mas20p and Mas70p subunits of the protein import receptor of yeast mitochondria interact via the tetratricopeptide repeat motif in Mas20p: evidence for a single hetero-oligomeric receptor. EMBO J. 15:1231-1237. [PMC free article] [PubMed] [Google Scholar]
- 20.Hill, J. E., A. M. Myers, T. J. Koerner, and A. Tzagoloff. 1986. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast 2:163-167. [DOI] [PubMed] [Google Scholar]
- 21.Hill, K., K. Model, M. T. Ryan, K. Dietmeier, F. Martin, R. Wagner, and N. Pfanner. 1998. Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins. Nature 395:516-521. [DOI] [PubMed] [Google Scholar]
- 22.Ito, K., T. Date, and W. Wickner. 1980. Synthesis, assembly into the cytoplasmic membrane, and proteolytic processing of the precursor of coliphage M13 coat protein. J. Biol. Chem. 255:2123-2130. [PubMed] [Google Scholar]
- 23.Jensen, R. E., and A. E. Johnson. 2001. Opening the door to mitochondrial protein import. Nat. Struct. Biol. 8:1008-1010. [DOI] [PubMed] [Google Scholar]
- 24.Kerscher, O., J. Holder, M. Srinivasan, R. S. Leung, and R. E. Jensen. 1997. The Tim54p-Tim22p complex mediates insertion of proteins into the mitochondrial inner membrane. J. Cell Biol. 139:1663-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kerscher, O., N. B. Sepuri, and R. E. Jensen. 2000. Tim18p is a new component of the Tim54p-Tim22p translocon in the mitochondrial inner membrane. Mol. Biol. Cell 11:103-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koehler, C. M., E. Jarosch, K. Tokatlidis, K. Schmid, R. J. Schweyen, and G. Schatz. 1998. Import of mitochondrial carriers mediated by essential proteins of the intermembrane space. Science 279:369-373. [DOI] [PubMed] [Google Scholar]
- 27.Koehler, C. M., D. Leuenberger, S. Merchant, A. Renold, T. Junne, and G. Schatz. 1999. Human deafness dystonia syndrome is a mitochondrial disease. Proc. Natl. Acad. Sci. USA 96:2141-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koehler, C. M., S. Merchant, W. Oppliger, K. Schmid, E. Jarosch, L. Dolfini, T. Junne, G. Schatz, and K. Tokatlidis. 1998. Tim9p, an essential partner subunit of Tim10p for the import of mitochondrial carrier proteins. EMBO J. 17:6477-6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koehler, C. M., S. Merchant, and G. Schatz. 1999. How membrane proteins travel across the mitochondrial intermembrane space. Trends Biochem. Sci. 24:428-432. [DOI] [PubMed] [Google Scholar]
- 30.Koehler, C. M., M. P. Murphy, N. A. Bally, D. Leuenberger, W. Oppliger, L. Dolfini, T. Junne, G. Schatz, and E. Or. 2000. Tim18p, a new subunit of the TIM22 complex that mediates insertion of imported proteins into the yeast mitochondrial inner membrane. Mol. Cell. Biol. 20:1187-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Komiya, T., S. Rospert, C. Koehler, R. Looser, G. Schatz, and K. Mihara. 1998. Interaction of mitochondrial targeting signals with acidic receptor domains along the protein import pathway: evidence for the ′acid chain' hypothesis. EMBO J. 17:3886-3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kovermann, P., K. N. Truscott, B. Guiard, P. Rehling, N. B. Sepuri, H. Müller, R. E. Jensen, R. Wagner, and N. Pfanner. 2002. Tim22, the essential core of the mitochondrial protein insertion complex, forms a voltage-activated and signal gated channel. Mol. Cell 9:363-373. [DOI] [PubMed] [Google Scholar]
- 33.Künkele, K. P., S. Heins, M. Dembowski, F. E. Nargang, R. Benz, M. Thieffry, J. Walz, R. Lill, S. Nussberger, and W. Neupert. 1998. The preprotein translocation channel of the outer membrane of mitochondria. Cell 93:1009-1019. [DOI] [PubMed] [Google Scholar]
- 34.Kurz, M., H. Martin, J. Rassow, N. Pfanner, and M. T. Ryan. 1999. Biogenesis of Tim proteins of the mitochondrial carrier import pathway: differential targeting mechanisms and crossing over with the main import pathway. Mol. Biol. Cell 10:2461-2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 36.Leuenberger, D., N. A. Bally, G. Schatz, and C. M. Koehler. 1999. Different import pathways through the mitochondrial intermembrane space for inner membrane proteins. EMBO J. 18:4816-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lim, J. H., F. Martin, B. Guiard, N. Pfanner, and W. Voos. 2001. The mitochondrial Hsp70-dependent import system actively unfolds preproteins and shortens the lag phase of translocation. EMBO J. 20:941-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 39.Luciano, P., S. Vial, M. A. S. Vergnolle, S. D. Dyall, D. R. Robinson, and K. Tokatlidis. 2001. Functional reconstitution of the import of the yeast ADP/ATP carrier mediated by the TIM10 complex. EMBO J. 20:4099-4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meisinger, C., M. T. Ryan, K. Hill, K. Model, J. H. Lim, A. Sickmann, H. Müller, H. E. Meyer, R. Wagner, and N. Pfanner. 2001. Protein import channel of the outer mitochondrial membrane: a highly stable Tom40-Tom22 core structure differentially interacts with preproteins, small Tom proteins, and import receptors. Mol. Cell. Biol. 21:2337-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moczko, M., U. Bömer, M. Kübrich, N. Zufall, A. Hönlinger, and N. Pfanner. 1997. The intermembrane space domain of mitochondrial Tom22 functions as a trans binding site for preproteins with N-terminal targeting sequences. Mol. Cell. Biol. 17:6574-6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Model, K., T. Prinz, T. Ruiz, M. Radermacher, T. Krimmer, W. Kühlbrandt, N. Pfanner, and C. Meisinger. 2002. Protein translocase of the outer mitochondrial membrane: role of import receptors in the structural organization of the TOM complex. J. Mol. Biol. 316:657-666. [DOI] [PubMed] [Google Scholar]
- 43.Murphy, M. P., D. Leuenberger, S. P. Curran, W. Oppliger, and C. M. Koehler. 2001. The essential function of the small Tim proteins in the TIM22 import pathway does not depend on formation of the soluble 70-kilodalton complex. Mol. Cell. Biol. 21:6132-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palmisano, A., V. Zara, A. Hönlinger, A. Vozza, P. J. T. Dekker, N. Pfanner, and F. Palmieri. 1998. Targeting and assembly of the oxoglutarate carrier: general principles for biogenesis of carrier proteins of the mitochondrial inner membrane. Biochem. J. 333:151-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paschen, S. A., U. Rothbauer, K. Kaldi, M. F. Bauer, W. Neupert, and M. Brunner. 2000. The role of the TIM8-13 complex in the import of Tim23 into mitochondria. EMBO J. 19:6392-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pfanner, N., and A. Geissler. 2001. Versatility of the mitochondrial protein import machinery. Nat. Rev. Mol. Cell Biol. 2:339-349. [DOI] [PubMed] [Google Scholar]
- 47.Pfanner, N., and W. Neupert. 1987. Distinct steps in the import of ADP/ATP carrier into mitochondria. J. Biol. Chem. 262:7528-7536. [PubMed] [Google Scholar]
- 48.Pfanner, N., M. Tropschug, and W. Neupert. 1987. Mitochondrial protein import: nucleoside triphosphates are involved in conferring import-competence to precursors. Cell 49:815-823. [DOI] [PubMed] [Google Scholar]
- 49.Roesch, K., S. P. Curran, L. Tranebjaerg, and C. M. Koehler. 2002. Human deafness dystonia syndrome is caused by a defect in assembly of the DDP1/TIMM8a-TIMM13 complex. Hum. Mol. Genet. 11:477-486. [DOI] [PubMed] [Google Scholar]
- 50.Ryan, M. T., H. Müller, and N. Pfanner. 1999. Functional staging of ADP/ATP carrier translocation across the outer mitochondrial membrane. J. Biol. Chem. 274:20619-20627. [DOI] [PubMed] [Google Scholar]
- 51.Ryan, M. T., W. Voos, and N. Pfanner. 2001. Assaying protein import into mitochondria. Methods Cell Biol. 65:189-215. [DOI] [PubMed] [Google Scholar]
- 52.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulphate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 53.Schägger, H., and G. von Jagow. 1991. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199:223-231. [DOI] [PubMed] [Google Scholar]
- 54.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sirrenberg, C., M. F. Bauer, B. Guiard, W. Neupert, and M. Brunner. 1996. Import of carrier proteins into the mitochondrial inner membrane mediated by Tim22. Nature 384:582-585. [DOI] [PubMed] [Google Scholar]
- 56.Sirrenberg, C., M. Endres, H. Fölsch, R. A. Stuart, W. Neupert, and M. Brunner. 1998. Carrier protein import into mitochondria mediated by the intermembrane proteins Tim10/Mrs11 and Tim12/Mrs5. Nature 391:912-915. [DOI] [PubMed] [Google Scholar]
- 57.Söllner, T., J. Rassow, and N. Pfanner. 1991. Analysis of mitochondrial protein import using translocation intermediates and specific antibodies. Methods Cell Biol. 34:345-358. [DOI] [PubMed] [Google Scholar]
- 58.Truscott, K. N., N. Pfanner, and W. Voos. 2001. Transport of proteins into mitochondria. Rev. Physiol. Biochem. Pharmacol. 143:81-136. [DOI] [PubMed] [Google Scholar]
- 59.van Wilpe, S., M. T. Ryan, K. Hill, A. C. Maarse, C. Meisinger, J. Brix, P. J. T. Dekker, M. Moczko, R. Wagner, M. Meijer, B. Guiard, A. Hönlinger, and N. Pfanner. 1999. Tom22 is a multifunctional organizer of the mitochondrial preprotein translocase. Nature 401:485-489. [DOI] [PubMed] [Google Scholar]
- 60.Vieira, J., and J. Messing. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259-268. [DOI] [PubMed] [Google Scholar]
- 61.Wiedemann, N., N. Pfanner, and M. T. Ryan. 2001. The three modules of ADP/ATP carrier cooperate in receptor recruitment and translocation into mitochondria. EMBO J. 20:951-960. [DOI] [PMC free article] [PubMed] [Google Scholar]