Abstract
RNA splicing is an essential step in gene expression, and is often variable, giving rise to multiple alternatively spliced mRNA and protein isoforms from a single gene locus. The design of effective databases to support experimental and computational investigations of alternative splicing (AS) is a significant challenge. In an effort to integrate accurate exon and splice site annotation with current knowledge about splicing regulatory elements and predicted AS events, and to link information about the splicing of orthologous genes in different species, we have developed the Hollywood system. This database was built upon genomic annotation of splicing patterns of known genes derived from spliced alignment of complementary DNAs (cDNAs) and expressed sequence tags, and links features such as splice site sequence and strength, exonic splicing enhancers and silencers, conserved and non-conserved patterns of splicing, and cDNA library information for inferred alternative exons. Hollywood was implemented as a relational database and currently contains comprehensive information for human and mouse. It is accompanied by a web query tool that allows searches for sets of exons with specific splicing characteristics or splicing regulatory element composition, or gives a graphical or sequence-level summary of splicing patterns for a specific gene. A streamlined graphical representation of gene splicing patterns is provided, and these patterns can alternatively be layered onto existing information in the UCSC Genome Browser. The database is accessible at http://hollywood.mit.edu.
INTRODUCTION
Gene expression is controlled at several levels, and in metazoan genomes, where the majority of protein-coding genes contain introns, the splicing of precursors to mRNAs (pre-mRNAs) constitutes a critical step for regulation of gene expression (1–3). RNA splicing occurs in the nucleus and is catalyzed by a large ribonucleoprotein (RNP) complex known as the spliceosome, which is composed of several small nuclear RNPs and over one hundred proteins (4). The processing of pre-mRNAs is often variable, giving rise to multiple alternatively spliced mRNAs, which may serve to produce distinct protein isoforms (5–7). Typical mammalian gene loci span tens of thousands of nucleotides (nt), with an average of nine exons/eight introns and the coding region typically spanning ∼1500 nt (8–10). In addition to the precise recognition of splice sites among many possible pseudo-sites, the removal of introns and the production of the correct message, the spliceosome must also produce tissue- and developmental stage-specific mRNA isoforms and integrate RNA splicing decisions with other steps in RNA processing, such as capping, cleavage and polyadenylation (11,12). Correct pre-mRNA splicing is generally required for cell viability. At least 15% of point mutations that cause genetic defects do so by altering splice site sequences (13), and the misregulation of alternative splicing (AS) is associated with a number of human diseases (14–17).
Alternative pre-mRNA splicing is estimated to affect more than half of actively transcribed human genes (18), and the systematic identification of AS events is important for the fundamental understanding of the regulation of gene expression in development, differentiation and human disease (19,20). A number of AS databases have been constructed, based on either searches of the scientific literature (21–23) or automated large-scale comparisons of transcript and genomic sequences (see below). The latter approach is made possible by the availability of large repositories of complementary DNA (cDNA) sequences and expressed sequence tags (ESTs), derived from different tissues or cell lines. Available data enable large-scale computational analysis of AS in human and mouse, and a few other organisms, with an average of >200 transcripts available for each annotated human gene (24). Transcript-based AS databases include: the Alternative Splicing Database Project, ASAP (25), the Alternative Splicing Database, ASD (26), the Extended Alternatively Spliced EST Database, EASED (27), SPLICEINFO (28) and ECGENE (29), to name a few (18,30) (http://hollywood.mit.edu/db/). Bioinformatics studies relying on such databases have proven useful in revealing differences in AS patterns between tissues (31–33), in identifying conserved AS events in orthologous genes (34–38), and for describing disease-associated AS (39). However, the AS events recorded differ significantly between different databases, owing to differences in primary sequence data used, in the algorithms used to generate spliced alignments, and in the stringency of alignment quality filtering.
More recently, splicing-sensitive microarrays have been designed and used for the detailed analysis of tissue-specific and other types of AS (40–45), and a cross-linking/immunoprecipitation strategy has been introduced for the systematic identification of RNAs bound by a given splicing factor (46). These newly developed methods set a direction toward increasingly parallel experimental analysis of splicing regulation, and AS databases will become increasingly important in both experimental design and data analysis for these types of functional genomic approaches.
To aid in computational and large-scale experimental studies of AS, we developed Hollywood, a comparative relational database of AS. Hollywood integrates accurate exon and splice site annotation derived from spliced alignments of transcripts to genomic sequences with current knowledge about splicing regulatory elements and predicted AS events, and links information about the splicing of orthologous genes in different species to facilitate comparative analyses. A compact representation of the splicing pattern of any desired gene is provided, and sets of alternative or constitutive exons can be obtained using complex queries for features such as splice site strength, type of AS event, tissue expression, splicing regulatory element content or conservation of the AS event between human and mouse.
Hollywood DATABASE
The design and implementation of Hollywood followed certain guiding principles: (i) all exon and isoform data should derive from high-quality spliced alignment of transcripts to genome sequences; (ii) AS events should be identified without requiring designation of an arbitrary ‘reference’ transcript; (iii) current knowledge about splice sites, splicing regulatory elements and predicted AS events should be incorporated into the database to allow efficient searches; (iv) the database should be integrated with other widely-used databases and genome browsers when possible; and (v) two main types of queries should be supported—queries for splicing information about a particular gene or genes and queries for sets of exons with particular properties. Examples of the output format for each of these types of queries are shown in Figure 1A for a gene query for the human fragile X mental retardation syndrome-related (FXR1) gene, and in Figure 1B for an exon query yielding exon 16 of the FXR1 gene.
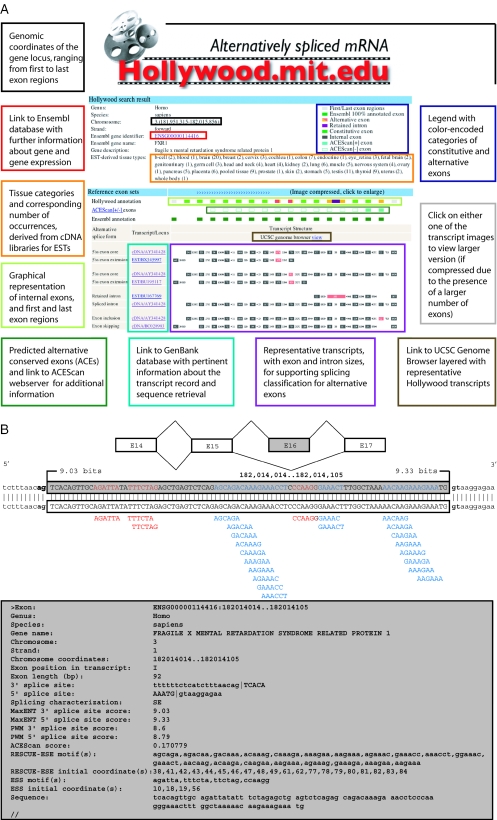
Figure 1.
(A) Screen shot of the Hollywood graphical interface summarizing splicing patterns for a single gene. The top of the interface summarizes species, locus, Ensembl-linked gene number and name, gene description, and EST-derived tissue types with corresponding number of occurrences. The annotation performed by Hollywood is complemented by ACEScan-predicted splice-conservation and exons that were annotated by Ensembl. Color-coded boxes are used to link display features with explanatory information. For each alternative exon splice type, Hollywood displays GenBank-linked accession numbers and primary transcript structures of representative pairs of transcripts, which can be used to identify the alternative exons, and layers each structure onto the UCSC Genome Browser. (B) Hollywood exon record for skipped exon E16 identified in the human fragile X mental retardation syndrome-related (FXR1) gene. FXR1 is an autosomal homolog of the FXR gene and encodes an RNA-binding protein. Figure 1 shows data for exon E16, by searching Hollywood with ‘FXR1’ for gene name and ‘Skipping’ for exon type, with a schematic representation of features at top and the standard text output below. FXR1 is shown with two isoforms that alternatively skip/include E16. Skipping of E16 results in a shift of the reading-frame that is predicted to alter and shorten the C-terminus of the FXR1 protein. E16 is an ACEScan[+] exon with a score of ∼0.2, and hence the orthologous exon of the mouse FXR1 is predicted to undergo exon skipping. This exon is perfectly conserved in sequence and contains two clusters of RESCUE-ESE hexamers. Transcripts aligned to the locus of FXR1 show E16 included in more than a dozen transcripts (two cDNAs AY341428 and HSU25165, and >10 ESTs) and excluded in ∼30 other transcripts (cDNA BC028983 and other ESTs), and ESTs suggest that FXR1 is expressed in many tissues.
Hollywood incorporates current knowledge about splice sites and splicing regulatory elements. It uses both a standard position-specific weight matrix model as well as a sophisticated maximum entropy-based model for the quantification of 3′ and 5′ splice site (3′ss and 5′ss) strength; the latter has been shown to more accurately distinguish authentic and pseudo splice sites (47,48). In addition to classical 3′ss and 5′ss motifs, it is now well established that other cis-regulatory elements including exonic splicing enhancers (ESEs) and silencers (ESSs) play common and important roles in exon and splice site choice (49). Hollywood annotates exons with sets of candidate ESE and ESS elements that have been identified in recent computational and experimental screens (50,51). The database also incorporates information about ‘alternative-conserved exons’ (ACEs)—orthologous exon pairs whose alternative splicing is conserved between human and mouse—from two sources: ∼450 exons with transcript evidence of AS in both species are annotated, as well as ∼2000 candidate ACEs predicted by the ACEScan algorithm (35).
Primary data
In building the Hollywood system, five major data sources were used, all of which are publicly available: (i) Ensembl gene chromosomal locations and gene identifiers (52), corresponding to genome assemblies from GoldenPath version hg16 of the human and version mm3 of the mouse genome (http://genome.ucsc.edu); (ii) transcript sequences from GenBank release 139.0, including the repositories gbpri, gbrod and gbhtc; (iii) EST sequences from dbEST, release 01122004, totaling ∼5.4 million human and ∼4.5 million mouse ESTs [dbEST records were grouped into one of about 40 human or mouse primary tissue types according to their cDNA library information, as described previously (32)]; (iv) mammalian interspersed repeat sequences from the RepBase repository (53); and (v) sets of ESEs and ESSs from the RESCUE-ESE and FAS-hex2 datasets, respectively (50,51).
Exon and feature annotation
Genomic sequences were extracted spanning an Ensembl gene from the start to the end of the annotation, plus an additional 5000 nt upstream and downstream of the start and end, respectively; these sequences are referred to as gene ‘slices’. The set of slices for all Ensembl genes was obtained from EnsMart (54). Use of the Ensembl gene annotation to define gene slices enables use of standard gene names and identifiers and enables linking to external databases. However, beyond the definition of slice boundaries, Ensembl annotation is not explicitly used in Hollywood: all exon/intron annotation and splicing information derives directly from transcript alignments. For convenience, Hollywood generally uses gene slice-based coordinates, which are converted to global chromosomal coordinates as needed.
Large-scale spliced alignments of transcript sequences to genomic DNA are conducted using the genome annotation system Genoa (http://genes.mit.edu/genoa), which will be described in greater detail elsewhere. Briefly, Genoa detects statistically significant blocks of identity between repeat-filtered cDNA sequences and gene slices, then conducts spliced alignments of best-matched cDNAs to corresponding gene loci using the algorithm mRNAvsGen. To avoid problems attendant to automated annotation of genomic regions, which are subject to frequent rearrangement such as immunoglobulin loci, cDNAs from certain classes of immune-related genes are optionally excluded by Genoa. Statistically significant matches are then identified between EST sequences and aligned repeat-filtered cDNAs, and best-matched ESTs are aligned to the corresponding gene slices using the Sim4 algorithm (55). Genoa was applied with stringent alignment criteria, requiring a sequence identity above 93% for cDNA alignments. For ESTs, the first and last aligned segments were required to be at least 30 nt long, with a sequence identity of at least 90%, and the entire alignment was required to have a sequence identity of at least 90%, over at least 90% of EST nucleotides. Using ∼22 200 human and 25 000 mouse gene slices, these alignments criteria were passed by ∼79 000 out of 115 000 human cDNAs for ∼19 300 gene slices, and by roughly one-fifth out of 5.4 million human ESTs, highlighting the stringency of the applied quality filter. The same alignment criteria were passed by only ∼27 000 out of 102 000 mouse cDNAs for ∼13 500 gene slices, while roughly one-fifth out of 4.1 million mouse ESTs met these criteria. Genoa aligned 2–4% of ESTs and ∼1% of cDNAs to multiple loci on different chromosomes.
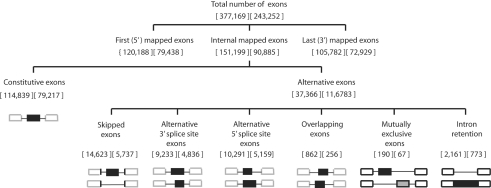
The annotation of exons as constitutive or alternative is made by the program runHollywood, which implements a set of computational rules to identify splice types of alternative exons (to be described elsewhere). Hollywood annotates constitutive exons, skipped exons, mutually exclusive exons, alternative 3′ss and/or 5′ss exons and retained introns. By default, every exon is labeled as ‘constitutive’ and this label remains unless specific criteria are met for annotation as another alternative. Figure 2 shows for the human and mouse genome the numbers of annotated constitutive and alternative exons, together with a pictorial representation of the criteria required for identifying each of these alternative exon types. This annotation is not restricted to one splice type per exon, but allows for exons to be included in multiple categories, e.g. an exon may exhibit both skipping and alternative 5′ss usage.
Figure 2.
Tree representation of numbers of human (left) and mouse (right) alternative and constitutive exons in Hollywood. All exons are supported by spliced alignments of transcript sequences with minimal acceptor and donor splice sites AG/ and /GT or /GC, respectively. Splice sites required to identify constitutive and alternative exons are marked bolded in black. Hollywood branches its annotation as follows: (i) on the first level, it distinguishes between first, internal and last exons; (ii) on the next level, it distinguishes between constitutive exons, with constant 3′ss and 5′ss, and alternative exons, with varying 3′ and/or 5′ss; (iii) on the last level, alternative exons are annotated as skipped, alternative 3′ss, alternative 5′ss, overlapping, or mutually exclusive exons. In addition, introns that are retained in mature mRNA are annotated as intron retention events. Alternative exons may undergo multiple splice variations and can belong to multiple branches.
Data model and implementation as a relational database
The Hollywood system consists of a generalized alignment parser framework, a relational database and a web interface. Hollywood defines a relational data model that distinguishes three primary tables—‘exon’, ‘gene’ and ‘transcript’—such that updated or new information can be represented in a structure consistent with existing data. A generalized parsing module simplifies the process of incorporating information, which is provided in flat file format. The parser currently inserts data into the PostgreSQL relational database management system, but could in principle support various database back ends. The structured query language (SQL) provides the ability to perform a wide range of powerful queries. Hollywood can also be queried through a web interface (Figure 3), with which users can build queries without knowledge of SQL. This interface allows users to retrieve sets of exons or transcripts that satisfy constraints defined on any number of supported features. Data are output in flat file or XML-based formats for downstream bioinformatic analysis.
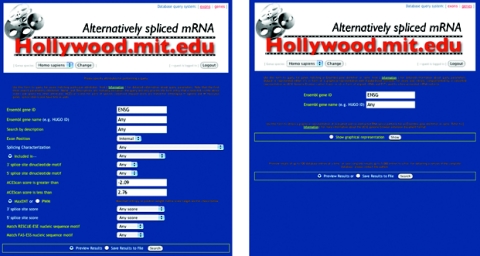
Figure 3.
Web interface offering two feature-selection forms for searches for sets of exons with specific splicing characteristics or splicing regulatory element composition (left), or for the splicing picture for a specific gene (right). Each feature is linked to the online documentation with explanatory information about feature utilization, values or nomenclature. Ensembl gene identifiers are most reliable for querying, as gene names are often not standardized, and response time is typically within seconds, depending on the complexity of the query.
A primary design goal was to ease the process of importing new data, and to this end Hollywood utilizes Perl packages that hide database implementation details. The parser interacts with the relational database backend, and the data model is optimized for efficient storage and data retrieval. Proper normalization techniques are employed in Hollywood, which facilitate the removal of redundant information and contribute to the logical organization of the data model, and the primary exon, gene and transcript data were decoupled in order to minimize the number of tables that have to be reloaded when a single input file is updated. For instance, for an update of gene names/descriptions one only needs to reload the corresponding table, and without further dependencies, the rest of the database remains unaffected.
Hollywood web interface and example applications
The Hollywood system is accessible at http://hollywood.mit.edu. Human and mouse gene slices, comprising roughly one-third of the human or mouse genome, are available for download and organized by chromosomes. Gene locus-based (local) coordinates are provided in 5′–3′ direction corresponding to the transcriptional orientation of the gene in each slice; corresponding chromosome-based (global) coordinates are also provided.
Retrieval of exon sequences and features
A feature-selection form allows the user of Hollywood to extract sets of constitutive and/or alternative exons, either for a single gene, specified by its Ensembl gene identifier or name, or for genes that share similar descriptions (e.g. kinases). The user can select exon features such as internal, constitutive or alternative (e.g. skipped), inclusion in transcripts derived from a particular tissue, conservation or presence of a particular ESE hexamer (e.g. GAAGAA). After the user has selected the features and submitted the form, exon records that meet the criteria are retrieved from the database. Figure 1A shows the data structure of such a record for exon E16 of the FXR1 gene, which undergoes tissue-specific AS (32,56).
As an example, a query with feature selection ‘hnRNP’ for gene description, ‘Skipped exon’ for splice type, and ‘Internal’ for exon position retrieved ∼40 human exons, including skipped exons in genes encoding hnRNP A1, hnRNP C and hnRNP R. A second query for exons with features ‘Internal’, ‘Skipped exon’, ‘Testis’ for tissue type and ‘TTCCTT’ for ESS sequence element retrieved ∼160 skipped human exons, which included the sperm-specific thioredoxin 2 (Sptrx) gene and the gene encoding the spermatogenesis cell apoptosis-related protein 1. A third query for exons with features ‘Internal’, ‘Constitutive exon’ and positive ACEScan scores (orthologous exon pairs predicted to be alternatively spliced in both human and mouse). The Hollywood system retrieved ∼1400 ACEs, including exon 5 of the tissue-specific RNA-binding protein NOVA-1, which has recently been identified as a skipped exon that is auto-regulated by NOVA-1 (57). As a final example, Hollywood was queried first with the feature selection ‘Skipped exon’ for splice type, ‘Brain’ and ‘GAAGAA’ for ESE sequence element, and was then queried with ‘GGTAAG’ for ESS sequence element, keeping the remaining features as selected previously. It retrieved ∼470 exon records for the first query, and ∼50 exon records for the latter query. These examples anticipate some of the types of analysis of alternative exons and functional sequence elements that can be conducted using Hollywood.
Viewing AS patterns
In addition to specific sets of exons, queried for by employing the feature selection form, Hollywood allows the user to display a summary of the splicing information contained in the database for one gene at a time. The user simply queries for a specific gene by name or by Ensembl identifier, and selects the display option. Alternatively, one may obtain a sequence-based representation of the display, and download all transcripts mapped to the gene locus.
After a gene is selected, a diagram illustrating splicing patterns is computed in real-time and displayed. Such a display for the human FXR1 gene is shown in Figure 1A, supported by legends. The Hollywood annotation is presented as a set of evaluated reference exons, each of unit size and color-coded as first, internal or last exon, with internal exons further annotated according to their splicing properties as constitutive or alternative. The Hollywood annotation shown derives from exons supported by spliced alignments with minimal consensus 3′ss and 5′ss sequences (AG/ and /GT or /GC, respectively). For clarity, the first and last segments of EST alignments (which generally correspond to incomplete portions of exons) are not displayed. Similarly, first and last exons are clustered and represented as first/last ‘exon regions’ to simplify the diagrams and focus attention on splicing-related (rather than transcription- or polyadenylation-related) information. Hollywood does not display all obtained spliced alignments (which often number in the hundreds or more), but provides a streamlined representation, displaying pairs of ‘representative transcripts’, which support the inference of each AS event.
Finally, the Hollywood display can conveniently be layered onto the UCSC Genome Browser, by using custom tracks that are provided as links on the display, for detailed comparisons with other existing annotations and inspection of multi-species sequence conservation.
Hollywood MAINTENANCE AND DEVELOPMENT
The first-phase development of Hollywood has matured such that the public release 1.0 can serve as a central resource for data and graphical display of patterns of alternative pre-mRNA splicing. Updates, improvements and further developments will be ongoing, and are currently envisioned as extensions for comparative genomics, perhaps with layering onto the Ensembl Genome Browser, integration of external annotations and incorporation of splicing-specific microarray data with links to exon records.
Acknowledgments
The authors thank L. P. Lim and R.-F. Yeh for previous work that contributed to Hollywood, W.G. Fairbrother and Z. Wang for contributing the datasets of RESCUE-ESE and FAS-ESS sequence elements, respectively, G. Yeo for contributing the maximum entropy model of splice sites and ACEScan predictions, and U. Ohler for stimulating discussions. This work was supported by grants from the NSF and the NIH (C.B.B.). Funding to pay the Open Access publication charges for this article was provided by NSF and NIH funds.
Conflict of interest statement. None declared.
REFERENCES
- 1.Black D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 2.Lopez A.J. Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu. Rev. Genet. 1998;32:279–305. doi: 10.1146/annurev.genet.32.1.279. [DOI] [PubMed] [Google Scholar]
- 3.Maniatis T., Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- 4.Jurica M.S., Moore M.J. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 5.Cartegni L., Chew S.L., Krainer A.R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nature Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 6.Graveley B.R. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 7.Grabowski P.J., Black D.L. Alternative RNA splicing in the nervous system. Prog. Neurobiol. 2001;65:289–308. doi: 10.1016/s0301-0082(01)00007-7. [DOI] [PubMed] [Google Scholar]
- 8.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody J., Baldwin K., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 9.Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 10.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 11.Kornblihtt A.R. Promoter usage and alternative splicing. Curr. Opin. Cell. Biol. 2005;17:262–268. doi: 10.1016/j.ceb.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 12.Proudfoot N. Ending the message is not so simple. Cell. 1996;87:779–781. doi: 10.1016/s0092-8674(00)81982-0. [DOI] [PubMed] [Google Scholar]
- 13.Krawczak M., Reiss J., Cooper D.N. The mutational spectrum of signle base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum. Genet. 1992;90:41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- 14.Faustino N.A., Cooper T.A. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 15.Dredge B.K., Polydorides A.D., Darnell R.B. The splice of life: alternative splicing and neurological disease. Nature Rev. Neurosci. 2001;2:43–50. doi: 10.1038/35049061. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Blanco M.A., Baraniak A.P., Lasda E.L. Alternative splicing in disease and therapy. Nat. Biotechnol. 2004;22:535–546. doi: 10.1038/nbt964. [DOI] [PubMed] [Google Scholar]
- 17.Caceres J.F., Kornblihtt A.R. Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet. 2002;18:186–193. doi: 10.1016/s0168-9525(01)02626-9. [DOI] [PubMed] [Google Scholar]
- 18.Modrek B., Lee C. A genomic view of alternative splicing. Nature Genet. 2002;30:13–19. [Google Scholar]
- 19.Black D.L. Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell. 2000;103:367–370. doi: 10.1016/s0092-8674(00)00128-8. [DOI] [PubMed] [Google Scholar]
- 20.Pagani F., Baralle F.E. Genomic variants in exons and introns: identifying the splicing spoilers. Nature Rev. Genet. 2004;5:389–396. doi: 10.1038/nrg1327. [DOI] [PubMed] [Google Scholar]
- 21.Stamm S., Zhu J., Nakai K., Stoilov P., Stoss O., Zhang M.Q. An alternative-exon database and its statistical analysis. DNA Cell Biol. 2000;19:739–756. doi: 10.1089/104454900750058107. [DOI] [PubMed] [Google Scholar]
- 22.Shah P., Jensen L.J., Boue S., Bork P. Extraction of transcript diversity from scientific literature. PLoS Comput. Biol. 2005;1:67–72. doi: 10.1371/journal.pcbi.0010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng C.L., Nair T.M., Gribskov M., Kwon Y.S., Li H.R., Fu X.D. A database designed to computationally aid an experimental approach to alternative splicing. Pac. Symp. Biocomput. 2004:78–88. doi: 10.1142/9789812704856_0008. [DOI] [PubMed] [Google Scholar]
- 24.Boguski M.S. The turning point in genome research. Trends Biochem. Sci. 1995;20:295–296. doi: 10.1016/s0968-0004(00)89051-9. [DOI] [PubMed] [Google Scholar]
- 25.Lee C., Atanelov L., Modrek B., Xing Y. ASAP: the Alternative Splicing Annotation Project. Nucleic Acids Res. 2003;31:101–105. doi: 10.1093/nar/gkg029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thanaraj T.A., Stamm S., Clark F., Riethoven J.J., Le Texier V., Muilu J. ASD: the Alternative Splicing Database. Nucleic Acids Res. 2004;32:D64–D69. doi: 10.1093/nar/gkh030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pospisil H., Herrmann A., Bortfeldt R.H., Reich J.G. EASED: Extended Alternatively Spliced EST Database. Nucleic Acids Res. 2004;32:D70–D74. doi: 10.1093/nar/gkh136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang H.D., Horng J.T., Lin F.M., Chang Y.C., Huang C.C. SpliceInfo: an information repository for mRNA alternative splicing in human genome. Nucleic Acids Res. 2005;33:D80–D85. doi: 10.1093/nar/gki129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim P., Kim N., Lee Y., Kim B., Shin Y., Lee S. ECgene: genome annotation for alternative splicing. Nucleic Acids Res. 2005;33:D75–D79. doi: 10.1093/nar/gki118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lareau L.F., Green R.E., Bhatnagar R.S., Brenner S.E. The evolving roles of alternative splicing. Curr. Opin. Struct. Biol. 2004;14:273–282. doi: 10.1016/j.sbi.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 31.Xu Q., Modrek B., Lee C. Genome-wide detection of tissue-specific alternative splicing in the human transcriptome. Nucleic Acids Res. 2002;30:3754–3766. doi: 10.1093/nar/gkf492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yeo G., Holste D., Kreiman G., Burge C.B. Variation in alternative splicing across human tissues. Genome Biol. 2004;5:R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taneri B., Snyder B., Novoradovsky A., Gaasterland T. Alternative splicing of mouse transcription factors affects their DNA-binding domain architecture and is tissue specific. Genome Biol. 2004;5:R75. doi: 10.1186/gb-2004-5-10-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sorek R., Shemesh R., Cohen Y., Basechess O., Ast G., Shamir R. A non-EST-based method for exon-skipping prediction. Genome Res. 2004;14:1617–1623. doi: 10.1101/gr.2572604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeo G.W., Van Nostrand E., Holste D., Poggio T., Burge C.B. Identification and analysis of alternative splicing events conserved in human and mouse. Proc. Natl Acad. Sci. USA. 2005;102:2850–2855. doi: 10.1073/pnas.0409742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohler U., Shomron N., Burge C.B. Recognition of unknown conserved alternatively spliced exons. PLoS Comput. Biol. 2005;1:e15. doi: 10.1371/journal.pcbi.0010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Modrek B., Lee C.J. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nature Genet. 2003;34:177–180. doi: 10.1038/ng1159. [DOI] [PubMed] [Google Scholar]
- 38.Thanaraj T.A., Clark F., Muilu J. Conservation of human alternative splice events in mouse. Nucleic Acids Res. 2003;31:2544–2552. doi: 10.1093/nar/gkg355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu Q., Lee C. Discovery of novel splice forms and functional analysis of cancer-specific alternative splicing in human expressed sequences. Nucleic Acids Res. 2003;31:5635–5643. doi: 10.1093/nar/gkg786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson J.M., Castle J., Garrett-Engele P., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 41.Hu G.K., Madore S.J., Moldover B., Jatkoe T., Balaban D., Thomas J., Wang Y. Predicting splice variant from DNA chip expression data. Genome Res. 2001;11:1237–1245. doi: 10.1101/gr.165501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clark T.A., Sugnet C.W., Ares M., Jr Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science. 2002;296:907–910. doi: 10.1126/science.1069415. [DOI] [PubMed] [Google Scholar]
- 43.Neves G., Zucker J., Daly M., Chess A. Stochastic yet biased expression of multiple Dscam splice variants by individual cells. Nature Genet. 2004;36:240–246. doi: 10.1038/ng1299. [DOI] [PubMed] [Google Scholar]
- 44.Pan Q., Shai O., Misquitta C., Zhang W., Saltzman A.L., Mohammad N., Babak T., Siu H., Hughes T.R., Morris Q.D., et al. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol. Cell. 2004;16:929–941. doi: 10.1016/j.molcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 45.Blanchette M., Labourier E., Green R.E., Brenner S.E., Rio D.C. Genome-wide analysis reveals an unexpected function for the Drosophila splicing factor U2AF50 in the nuclear export of intronless mRNAs. Mol. Cell. 2004;14:775–786. doi: 10.1016/j.molcel.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 46.Ule J., Jensen K.B., Ruggiu M., Mele A., Ule A., Darnell R.B. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 47.Yeo G., Burge C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 48.Eng L., Coutinho G., Nahas S., Yeo G., Tanouye R., Babaei M., Dork T., Burge C., Gatti R.A. Nonclassical splicing mutations in the coding and noncoding regions of the ATM Gene: maximum entropy estimates of splice junction strengths. Hum. Mutat. 2004;23:67–76. doi: 10.1002/humu.10295. [DOI] [PubMed] [Google Scholar]
- 49.Smith C.W., Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 2000;25:381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- 50.Fairbrother W.G., Yeh R.F., Sharp P.A., Burge C.B. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 51.Wang Z., Rolish M.E., Yeo G., Tung V., Mawson M., Burge C.B. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 52.Birney E., Andrews D., Bevan P., Caccamo M., Cameron G., Chen Y., Clarke L., Coates G., Cox T., Cuff J., et al. Ensembl 2004. Nucleic Acids Res. 2004;32:D468–D470. doi: 10.1093/nar/gkh038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jurka J. Repbase update: a database and an electronic journal of repetitive elements. Trends Genet. 2000;16:418–420. doi: 10.1016/s0168-9525(00)02093-x. [DOI] [PubMed] [Google Scholar]
- 54.Kasprzyk A., Keefe D., Smedley D., London D., Spooner W., Melsopp C., Hammond M., Rocca-Serra P., Cox T., Birney E. EnsMart: a generic system for fast and flexible access to biological data. Genome Res. 2004;14:160–169. doi: 10.1101/gr.1645104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Florea L., Hartzell G., Zhang Z., Rubin G.M., Miller W. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kirkpatrick L.L., McIlwain K.A., Nelson D.L. Alternative splicing in the murine and human FXR1 genes. Genomics. 1999;59:193–202. doi: 10.1006/geno.1999.5868. [DOI] [PubMed] [Google Scholar]
- 57.Ule J., Ule A., Spencer J., Williams A., Hu J.S., Cline M., Wang H., Clark T., Fraser C., Ruggiu M., et al. Nova regulates brain-specific splicing to shape the synapse. Nature Genet. 2005;37:844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]