Abstract
Genomes that have been highly conserved following increases in ploidy (by duplication or hybridization) like Glycine max (soybean) present challenges during genome analysis. At http://soybeangenome.siu.edu the Soybean Genome Database (SoyGD) genome browser has, since 2002, integrated and served the publicly available soybean physical map, bacterial artificial chromosome (BAC) fingerprint database and genetic map associated genomic data. The browser shows both build 3 and build 4 contiguous sets of clones (contigs) of the soybean physical map. Build 4 consisted of 2854 contigs that encompassed 1.05 Gb and 404 high-quality DNA markers that anchored 742 contigs. Many DNA markers anchored sets of 2–8 different contigs. Each contig in the set represented a homologous region of related sequences. GBrowse was adapted to show sets of homologous contigs at all potential anchor points, spread laterally and prevented from overlapping. About 8064 minimum tiling path (MTP2) clones provided 13 473 BAC end sequences (BES) to decorate the physical map. Analyses of BES placed 2111 gene models, 40 marker anchors and 1053 new microsatellite markers on the map. Estimated sequence tag probes from 201 low-copy gene families located 613 paralogs. The genome browser portal showed each data type as a separate track. Tetraploid, octoploid, diploid and homologous regions are shown clearly in relation to an integrated genetic and physical map.
INTRODUCTION
Soybean (Glycine max L. Merr.) has a genome size of 1.1–1.15 Gb (1). The soybean genome is a partially diploidized tetraploid. The genome is the product of a diploid ancestor (n = 11), which underwent aneuploid loss (n = 10), polyploidization (2n = 20) and diploidization (n = 20) (2). Two genome duplications or hybridizations may have occurred (3–6). The duplicated regions have been segmented and reshuffled (7,8). Microsatellites markers can amplify single loci from genomic DNA (9), but only 35% of such markers identify single loci among bacterial artificial chromosome (BAC) pools (10–13). Similarly ∼35% of expressed sequence tag (EST) probes identify BACs from single loci (14). Therefore, both marker and EST analyses suggest that ∼35% of the soybean genome is extensively diploidized.
Highly conserved polyploid regions (>98% identity) of 150–500 kb have been inferred from regions where many BACs coalesce into contigs (8,10–13). Polyploid regions exist in either tetraploid or octoploid states. Contigs of polyploid regions contain 2- or 4-fold the BAC clone representation per unique band. Approximately 25% of the soybean genome is both polyploid and highly conserved.
The recent developments of more robust genetic map (9,15–17) and physical map (10–13) datasets can make soybean genomics more efficient (18,19). These complex sets of data have needed to be integrated and presented to researchers in a visually simple informative format. This short report describes representation of the physical map of soybean at the Soybean Genome Database (SoyGD) with recent improvements. SoyGD has served the integrated physical and genetic map of soybean since October 2002. Data at SoyGD were represented in a modified GBrowse (20) format at http://soybeangenome.siu.edu to allow clearer representation and annotation of polyploid and homologous duplicated regions.
DATABASE COMPONENTS
SoyGD is a relational database built on a Poweredge 2400 server provided by supplemental funding to NSF 98-72635. The database is housed at the Center for Excellence in Soybean Research Teaching and Outreach, Southern Illinois University, Carbondale. The web interface provides genome information in relation to the genetic map framework. The operating system (platform) used is Windows Server 2003. The database management system is Mysql 4.0. The scripting language is ActiveState Perl. The server is Apache Web Server. The standard Perl modules used are CGI 2.56, DBI and DBD::mysql.
Request from users are sent as queries to the SOYGB web server. In SOYGB the query (handled by the Apache Web Server) is validated. If valid, the query is passed to the scripting language (Perl). The Perl modules check the query and extract the appropriate data from the Mysql database. The results are sent to the client.
Representation of integrated genetic and physical map data
Genetic maps represent chromosomes as linkage groups with features (loci and markers) at centimorgan (cM) positions. Physical maps represent chromosomes as contiguous stretches of DNA sequence with features at base pair positions. SoyGD assumes that the 2512 cM genetic map of soybean encompasses its entire genome (1.1 Gb). The cM positions on the genetic map were converted to base pair positions on Gbrowse such that 1 cM is equivalent to 437 kb.
Physical map data are based on contigs built by the FingerPrinted Contig (FPC) program (11,12). Custom Perl scripts were written to convert data from .FPC format into a generic flat file (GFF) format. GFF files can be loaded into databases suitable for GBrowse access. Build versions 3 and 4 can be viewed at the SoyGD website (http://soybeangenome.siu.edu) with each as a separate database. Results are also summarized in tables that include information on chromosome coordinates, anchored clones, overlapping clones, contigs, duplicated contigs and QTL in the region. The code is open source and can be downloaded from the SoyGD website.
Classification of features
A classification scheme was developed to identify types of features and manipulate them as sets in the algorithms (Figure 1). Briefly, gamma features were primary groups, similar to a marker anchoring a BAC that anchors a contig. A beta feature would be associated with a gamma feature. For example, a beta BAC is predicted to be in the same contig as a gamma BAC by fingerprints. A beta contig is predicted to overlap with a gamma contig by shared BAC clones or contig end band overlaps. Alpha features contain only one of the features; normally a marker anchor without an anchored BAC or contig. The classification scheme was attached to contig labels.
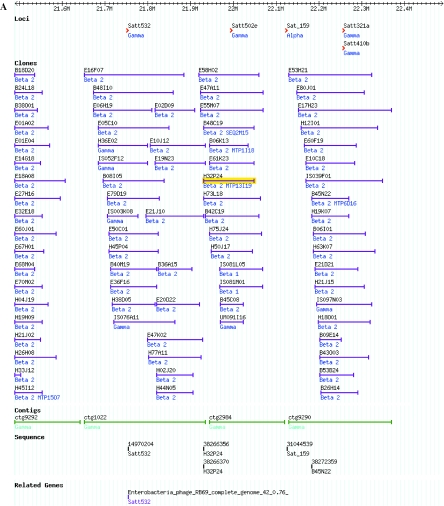
Figure 1.
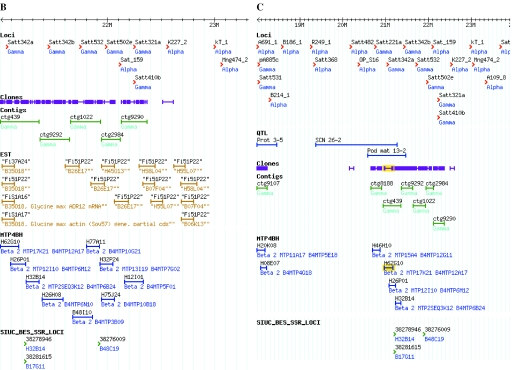
Representation in SoyGD by GBrowse of a portion of the soybean genome from linkage group D1A+Q showing duplicated regions, multiple markers and multiple contigs per marker. (A) An 1 Mb region of build 4 SoyGD image with contigs, clones, markers sequences and related genes is shown. Contigs are shown as blue bars. Some of the BAC clones in the contigs are shown as green bars, some were cut off for brevity in this figure. Markers are shown as red arrowheads. BES and marker sequences are shown as magenta bars. Gene annotations are shown as purple lines (the five most probable at P < 10−5 are listed; only one is shown here). (B) Part of a 4 Mb view encompassing the same region, note that clones coalesce to single bars at this resolution to aid clone density estimates. Sequences and related genes are de-selected and thus hidden but ESTs, MTP4BH and BES-SSRs are selected in this view. EST hybridizing clones are shown as golden bars. MTP4BH BAC clones are shown as blue bars. BES derived microsatellite markers are shown as green arrowheads (BES-SSR). (C) A part of a 10 Mb view encompassing the same region. QTL are shown as red bars and can be viewed in relationship to other resources. QTL views in a 4–10 Mb window are most useful because the map accuracy for QTL of ∼10–25 cM is common. All sequences and features with entries at GenBank are linked to their gene index numbers by mouse over. Views ∼10 Mb can be arranged by contacting the administrator. In views above 10 Mb, sequence and related genes coalesce to single bars and are useful for gene density estimates and correlations with chromosome karyotpe.
Representation of homologous regions
Heuristic algorithms were devised to spread out features that were assigned to the same cM and bp locations. The algorithms also used heuristics to assign probable locations for features in duplicated areas of the genome. When loci anchored contiguous sets of overlapping clones (contigs) on more than one marker linkage group, the contigs were placed in all of the locations, and sublabeled ‘a’, ‘b’, ‘c’, etc. A Perl script was written to allow separation of contigs that anchor to the same marker.
Representation of polyploid regions
Polyploid regions were annotated based on clones per unique band. Contigs in the 8000 series were inferred to be tetraploid and contigs in the 9000 series were inferred to be octoploid. SoyGD users should examine the inference of polyploidy critically as some diploid regions of the genome may also be overrepresented in the fingerprinted BAC clone dataset.
Features not assigned to linkage groups or chromosomes
GBrowse represents the data in SoyGD in relation to the composite genetic map (9). Contigs that could not be assigned to a position in the genetic map were assigned to Queue and listed numerically. The map will be updated regularly, as Queued contigs are anchored, as new improved FPC builds are made and as revised genetic and new QTL maps are made available (Figure 1).
DATABASE CONTENTS
The data represented in Gbrowse at SoyGD were modified from Wu et al. (12) by the addition of satellite markers anchored to BACs made from soybean cultivar ‘Forrest’ DNA (Table 1) (11,21). Genetic and physical distances were based on the March 2003 release of the soybean composite genetic map (www.soybase.org) that showed 2512 cM. The soybean physical map build 3 used the FPC file described by Wu et al. (12). Build 4 used the FPC file developed by Shultz (11). Both build 3 and build 4 FPC files are freely available and can be downloaded from http://bioinformatics.siu.edu (Table 2).
Table 1.
Microsatellite markers were used to anchor BAC clones of three genotypes comprising the physical map builds 3 and 4 in SoyGD (the 86 RFLP markers used are not listed in the table)
| Genotype(s) | Marker numbers |
|---|---|
| Williams only | 67 |
| Faribault only | 45 |
| Forrest only | 66 |
| Faribault and Williams 82 | 80 |
| Faribault and Forrest | 11 |
| Williams 82 and Forrest | 12 |
| Faribault, Williams 82 and Forrest | 32 |
| Total integrated | 318 |
Table 2.
Progress in the soybean physical map (builds 2–4) shown in SoyGD
| Automated | Manual edit | Manual edit | |
|---|---|---|---|
| Build 2 | Build 3 | Build 4 | |
| September 2001 | August 2002 | October 2003 | |
| Total (Repeat) | |||
| BAC clones in FPC database | 81 024 | 78 001 | 78 001 |
| BACs used in contig assembly | 75 568 | 76 749a | 72 942b |
| Number of singletons | 5884 | 3702 | 27 810 |
| Marker anchored singletons | 0 | 0 | 120 |
| Clones in contigs (fold genome) | 69 684 | 73 047 | 45 130 |
| Fold genome in contigs | 8.7 | 9.1 | 5.6 |
| Number of contigs | 5597 | 2905 | 2854 (646) |
| Anchoring markers | 0 | 385 | 404 |
| Anchored contigs | 0 | 781 | 742 (181) |
| Q contigs | n/a | 1040 | 0 |
| Contigs that contained | |||
| >25 clones | 220 | 921 | 477 (268) |
| 10–25 clones | 3038 | 920 | 1458 (433) |
| 3–9 clones | 1845 | 850 | 820 (0) |
| 2 clones | 385 | 216 | 99 (0) |
| Unique bands within contigs | 396 843 | 345 457 | 258 240 (54 560)c |
| Length of the contigs (Gb) | 1.667d | 1.451d | 1.037 (0.258) |
aAdjusted for 1252 ‘NoFp’ 0 band clones only (includes all clones with ≥1 band).
bAdjusted for all 0–4 band clones, contamination-suspect clones and most high band number (>65) clones.
cBased on 4.5 kb per unique band, for 2854 contigs containing ∼68 unique bands in 15 clones, 264 duplicated region contigs containing ∼68 unique bands in 30 clones, 15 840 unique bands, and 406 highly repeated region contigs containing ∼68 unique bands in 60 clones, 48 720 unique bands.
dBased on 4.0 kb per unique band.
BAC anchors with microsatellite markers
Microsatellite and restriction fragment length polymorphism (RFLP) DNA markers provided 404 useful genetic map anchors for the physical map contigs. There were 86 reliable RFLP markers and 318 reliable microsatellite markers (Table 1). There were marker anchors to fingerprinted clones from five BAC libraries made from three soybean cultivars (13,21–23). BAC clones made from cultivar Forrest DNA (21) have B (BamHI), H (HindIII) or E (EcoRI) as the prefixes that identifies which restriction enzyme was used to construct the library. BAC clones made from cultivar Williams 82 DNA (with EcoRI) (22) have IS as their prefix. BAC clones made from cultivar Faribault (with HindIII) (23) have UM as their prefix (Figure 1).
Relatively few markers were monogenetic when amplified from BAC pools (Table 3). Contigs with two markers from different groups were tested for clone contamination. If no contamination was detected they were either shown at both locations or placed in Queue for additional analysis. Forty-five microsatellite markers used in Ref. (12) were removed from SoyGD because fingerprints suggested well-to-well contamination of the BACs (10). The anchors are available on request.
Table 3.
Frequency of duplicated regions detected by anchors in SoyGD
| State | Anchors | Clones |
|---|---|---|
| One anchor 1 contig | 124 | 124 |
| One anchor 2 contigs | 110 | 220 |
| One anchor 3 contigs | 45 | 135 |
| One anchor 4 contigs | 31 | 124 |
| One anchor 5 contigs | 17 | 85 |
| One anchor 6 contigs | 4 | 24 |
| One anchor 7 contigs | 2 | 14 |
| One anchor 8 contigs | 2 | 16 |
| Total | 363a | 762 |
aIncludes 318 replicated high quality microsatellites and 45 potentially contaminated anchor markers.
Contig classification by ploidy in build 4
The build 4 map has 45 130 clones in 2854 contigs (Table 2). Contigs were numbered in series to aid users in their understanding of the representation in GBrowse (Figure 1). In the contigs (ctg), ctg1–2208 were inferred to be diploidized regions. The BAC clone depth per unique band was 6-fold or less. The clone depth was similar to the 6-fold haploid genome redundancy of the fingerprinted BAC libraries used to construct build 4. Contigs in the ctg8000 series were inferred to contain clones from two homologous regions because BAC clone depth was close to 12-fold. Contigs in the ctg9000 series were inferred to contain clones from four homologous regions because BAC clone depth was close to 24-fold. The total number of unique bands in all contigs was 194 907 or ∼0.78 Gb, 77% of the soybean genome but almost 100% of the total genome because conserved duplicated regions need be sampled only once.
Minimum tile path
Three different minimum tile path (MTP) clone sets were created from the contigs. Build 3 provided contigs for MTP2 development and clones from the HindIII/BamHI and EcoRI BAC libraries were included. Build 4 provided the contigs for MTP4 development. Separate clone lists were generated based on coverage of all contigs using only HindIII and BamHI BAC clones for MTP4BH. Clone lists for MTP4E used only the EcoRI BAC clone subset. The separation of the libraries in MTP4 assists users since MTP4BH clones were made with pCLCD04541 (AF184978), a 27.6 kb BIBAC vector, useful for plant transformation whereas MTP4E clones were made in pECBAC1, a small vector (7.5 kb), so that MTP4E clones will be useful for genome sequencing. In the SoyGD, clones from the MTPs are annotated with the suffix MTP2 or MTP4 below the clone (Figure 1).
Anchoring contigs with composite map microsatellite markers
Among 1,025 composite map markers available, 318 were judged high quality anchors in replicated pool decovolution experiments. There were 742 contigs anchored with 404 replicated RFLP and microsatellite markers in build 4 (Table 3). Most of the remaining 707 non replicated marker anchors are available from SIUC as clone or pool adresses on request. A significant portion (∼65%) of the soybean genome in contigs was located on the existing soybean genetic map. However, many contigs were located to more than one position because they contained homologs of the markers employed.
BAC end sequences (BESs)
About 16 128 forward and reverse sequence reads (GenBank accession nos CG812653–CG826126) were attempted from 8064 MTP2 clones. There are 13 473 BES reads. Mean read length was 736 bp (24). The total amount of sequence generated from MTP2 were 9.9 Mb (∼1%) of the soybean genome. There are 5555 paired reads. Approximately 15% of sequences contained (2111) gene-like sequences at E-values of 10−5 when they were compared with GenBank accession no. in May 2005 (24).
New microsatellite markers in BESs
Common soybean microsatellite-like repeats (ATT)n, (AT)n and (CA)n were found in 8% of the BESs. The MTP2- and BES-derived markers are shown on a separate track from loci and labeled BES-SSR (Figure 1B). Genetic map analysis showed 72% of the newly designed markers were polymorphic in eight common cultivars and that the markers could be located in gaps in the maps built using existing soybean recombinant inbred line populations (11,16) (www.intl-pag.org/13/abstracts/PAG13_P141.html). There were only about 1000 microsatellite markers in the soybean composite map in 2004 (9). The release of the 1053 BES-SSR markers to SoyGD by SIUC makes available more than 2000 microsatellite markers for soybean. Since the second set of a thousand microsatellite markers were derived from an MTP of BAC clones the BES-SSR appeared to be evenly distributed, with about one marker every 500–1000 kb (1–3 cM). Gaps exist in all genetic maps of soybean (9) and the gaps will be filled by some of the BES-SSR markers. Furthermore, when a BES-SSR marker has been placed on a linkage group of a genetic map, the BAC and contig that contain the marker become located on the same linkage group in SoyGD as gamma features.
Composite map DNA markers in BESs
The 42 BESs contained sequences highly homologous (over 80–341 bp with E-values between 10−30 and 10−300) to 30 previously sequenced genetic markers (10 RFLPs and 20 microsatellites) of the composite genetic map (9). The 30 markers represent ∼2% of the unique markers with sequences reported in GenBank (May 2005). Contigs that contain BAC clones with BES highly homologous to markers were placed onto the appropriate linkage group as gamma features. Development of a paired BES library of the remaining 37 000 build 4 clones in contigs will provide SoyGD a further 200–300 marker anchors by in silico detection at a cost much lower than the gel-based methods of marker amplicon detection in BACs.
Paralog integration and candidate gene identification
EST hybridization probes were able to map the relative positions of 613 candidate paralogs of 201 gene families (14). Among the probes were 108 ESTs that had homology to genes of known function. The BACs that hybridized to ESTs are shown as a separate track encompassing a whole BAC clone. Comparison of the inferred paralog positions from both EST hybridization and BES derived gene models with the position of a QTL (Figure 1C) can be used to identify potential candidate genes that might underlie the QTL.
GBrowse representation of duplicate features
Duplicate genomic regions were identified in two forms (Figure 1). In build 4, a region is marked duplicated on the marker track of GBrowse if the contig contains about 12 or 24 clones per unique band (i.e. it is in the 8000 or 9000 series). For example Satt321a and Satt410b anchored a single contig (ctg9290) that contained 22 clones but encompassed just 205 kb. All the 22 clones share the central 39 of 82 unique bands in the contig, so this central region of the contig was inferred to be octoploid. The contig ends are expected to merge with other contigs at four different genomic positions. The types of contigs in the 8000 or 9000 series might be split into two or four homologous regions with higher resolution fingerprint or DNA sequence data.
The second form of duplicate genomic region was identified by single genetic markers that anchored two contigs (within such contigs, the composite clones could not be merged by an editor). Examples include Satt342 that anchored two contigs, ctg439 and ctg9292, and Satt402 that anchored two contigs, ctg3787 and ctg9040. Each contig may either represent the loci marked by Satt342 and Satt402 on linkage group D1A+Q or a homologous region from some other place of the genome. In this case, all anchored contigs are shown at each location. Algorithms to represent the data were developed at SoyGD that placed contigs anchored by a common marker adjacent to one another (Figure 1) rather than stacking contigs and mixing their composite clones and anchors as GBrowse would normally do (20). The algorithm developed at SoyGD spreads these contigs out to the left and right, without allowing them to overlap. In most cases two contigs identified by a single marker are adjacent. For example, Satt342a anchored a diploid contig, ctg439. SoyGD showed it next to Satt342b that anchors an octoploid contig, ctg9292 (Figure 1B and C). In some cases contigs may become separated by contigs anchored by other markers (e.g. Satt402a and 402b are split by NP008_1).
CONCLUSIONS
Assembling a physical map for a polyploid or extensively duplicated genome presented special challenges for database visualization. Novel bioinformatic approaches were used to create a set of contigs that present soybean genome structure to users in a manner that informs whether the region is likely to be octoploid, tetraploid or diploid. Homologous regions to specific loci could be identified through representation of marker amplicons. The 2854 contigs represented here will provide a platform for further map resolution by additional research.
Marker amplicons and homologous regions
Multiple marker amplicons (Table 3) were the most difficult problem to contend with both when the genetic markers were integrated into the physical map and when the integrated map was presented to users. However, a more complete catalog of the homologous regions identified by the marker amplicons can significantly enhance the soybean genome map (4,11).
Investigations of whether common marker anchors to BACs from the three cultivars could identify different contigs provided no evidence for cultivar differences in genome architecture (Table 1 and Figure 1) (25). For example, contig ctg1022 (Figure 1A) contained an anchor between Satt532 and the BAC clone H36E02 from cultivar Forrest; and between Satt532 and the BAC clones ISO52F12, ISO76A11 and ISO03K08 made from cultivar ‘Williams 82’. Whereas, neighboring contig ctg9290 contained two anchors, Satt321 and Satt401, from different genetic map positions (linkage groups D2 and H) on a single clone ISO97N03.
Showing all homologous contigs at each possible location was necessary to prevent users from focusing on single contigs within homologous sets. Spreading the contigs was preferred to stacking the contigs because no merge was implied. Users are encouraged to analyze all contigs sharing a homologous relationship. A more complete catalog of homologus regions will allow the soybean community to reinvestigate QTL bearing regions and discover homologous, duplicated QTL within the genome.
Physical maps of polyploid regions
The inference that 23% of the contigs contained coalesced polyploid regions was made based on the distribution of clone number per unique band. Similarly, genome duplication was shown to result in contigs containing two or more homologous genomic regions that encompassed DNA marker amplicons. Contigs inferred to contain four duplicated regions were twice as frequent as those containing just two. Genome representation that clearly identified potentially polyploid contigs was a key factor in improving map quality and SoyGD user satisfaction from 2002 to 2005. The partitioning of euchromatin and heterochromatin was not directly measured, but contig distribution, potential EST paralog distribution and BES gene model distribution suggest the polyploid genome regions are gene rich and unlikely to be heterochromatic (14,19).
Informatics
Before SoyGD, access to the physical map was not standardized and required users to learn new interfaces from site-to-site. During SoyGD development it became clear that some researchers needed to operate FPC locally to query the data, as their projects required that contig merges be made. However, a large portion of the community does not need this type of access. The majority of users have benefited from the standardized interface provided by GBrowse. To further assist users, the way in which GBrowse showed duplications was improved over earlier attempts (26). However, it is not possible for any researcher to fingerprint any clone and add it to the dataset. This still requires the cooperation of multiple research entities and is not yet perfectly bi-directional.
Future initiatives
SoyGD will represent in future the new build 5 for the physical map that is in progress based on the 76 749 fingerprinted clones available to the public. New QTL data will be incorporated from the newly released Essex by Forrest RIL population (26). Gene expression data (27) may be added to the gene models represented in SoyGD. All of the EST placed in SoyGD to date have been used for expression analysis (28) and are related to responses of roots to biotic stresses. Increasing the amount of data from expression analysis will strengthen the usefulness of SoyGD for the identification of candidates for genes underlying QTL and other genetic loci.
Direct query of the data in SoyGD with users own data is envisioned and will be made possible over the Internet. Layering on the map fingerprint data from the Williams 82 cultivar is envisioned when the data are released. The new SoyGD GBrowse GFF files will be transferred to NCGR for use in their Legume Information System (http://www.comparative-legumes.org) to update the C-Map and show the homologous regions at that site.
Soybean researchers will be provided with continued electronic access to BAC clones encompassing regions likely to contain genes and QTL of agronomic importance. The genome browser interface will be improved using as yet unpublished data in several ways: by showing sequences and gene predictions from 37 000 BESs in addition to those in Ref. (24); by showing the locations of additional microsatellite anchors, SNP anchors and BES-SSR anchors; by showing many well-annotated contiguous sequenced regions of more than 100 kb (19); and by showing the locations of about 2000 resistance gene analog positive clones (nucleotide-binding leucine rich repeat paralogs), 700 receptor-like kinases and 512 additional low-copy gene family paralogs (14). In addition to adding soybean data, the adoption of GBrowse tools that allow comparisons of synteny among genomes will be a priority (29).
Acknowledgments
The authors thank Dr Q. Tao for assistance with fingerprinting. We thank Chester Langin for his work on GBrowse from 2004 to 2005. We thank LIS for their C-Map representation of our GBrowse data. This research was funded in part by a grant from the NSF 9872635, ISPOB 98-122-02 and 02-127-03 and USB 2228-5228. The SoyGD material was based upon work supported by the National Science Foundation under Grant No. 9872635. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation, ISPOB or USB. The continued support of SIUC, College of Agriculture and Office of the Vice Chancellor for Research to M.J.I. and D.A.L. is appreciated. Funding to pay the Open Access publication charges for this article was provided by USB 6228.
Conflict of interest statement. None declared.
REFERENCES
- 1.Arumuganathan K., Earle E.D. Estimation of nuclear DNA content of plants by flow cytometry. Plant Mol. Biol. Rep. 1991;9:229–241. [Google Scholar]
- 2.Singh R.J., Hymowitz T. The genomic relationship between Glycine max L. Merr. and G. soja Sieb. and Zucc. as revealed by pachytene chromosome analysis. Theor. Appl. Genet. 1988;76:705–711. doi: 10.1007/BF00303516. [DOI] [PubMed] [Google Scholar]
- 3.Blanc G., Wolfe K.H. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell. 2004;16:1667–1678. doi: 10.1105/tpc.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shoemaker R.C., Polzin K., Labate J., Specht J., Brummer E.C., Olson T., Young N.D., Concibido V., Wilcox J., Tamulonis J.P., et al. Genome duplication in soybean (Glycine subgenus soja) Genetics. 1996;144:329–338. doi: 10.1093/genetics/144.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shoemaker R., Keim P., Vodkin L., Retzel E., Clifton S.W., Waterston R., Smoller D., Coryell V., Khanna A., Erpelding J., et al. A compilation of soybean ESTs: generation and analysis. Genome. 2002;45:329–338. doi: 10.1139/g01-150. [DOI] [PubMed] [Google Scholar]
- 6.Tian A.G., Wang J., Cui P., Han Y.J., Xu H., Cong L.J., Huang X.G., Wang X.L., Jiao Y.Z., Wang B.J., et al. Characterization of soybean genomic features by analysis of its expressed sequence tags. Theor. Appl. Genet. 2004;108:903–913. doi: 10.1007/s00122-003-1499-2. [DOI] [PubMed] [Google Scholar]
- 7.Grant D., Cregan P., Shoemaker R.C. Genome organization in dicots: genome duplication in Arabidopsis and synteny between soybean and Arabidopsis. Proc. Natl Acad. Sci. USA. 2000;97:4168–4173. doi: 10.1073/pnas.070430597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan H.H., Mudge J., Kim D.J., Larsen D., Shoemaker R.C., Cook D.R., Young N.D. Estimates of conserved microsynteny among the genomes of Glycine max, Medicago truncatula and Arabidopsis thaliana. Theor. Appl. Genet. 2003;106:1256–1265. doi: 10.1007/s00122-002-1183-y. [DOI] [PubMed] [Google Scholar]
- 9.Song Q.J., Marek L.F., Shoemaker R.C., Lark K.G., Concibido V.C., Delannay X., Specht J.E., Cregan P.B. A new integrated genetic linkage map of the soybean. Theor. Appl. Genet. 2004;109:122–128. doi: 10.1007/s00122-004-1602-3. [DOI] [PubMed] [Google Scholar]
- 10.Shultz J., Meksem K., Lightfoot D.A. Evaluating physical maps by clone location comparison. Genome Lett. 2003a;2:99–107. [Google Scholar]
- 11.Shultz J. 2005. Bioinformatic approaches to improving the efficiency of physical map builds; p. 247. PhD thesis, SIUC, Carbondale, IL. [Google Scholar]
- 12.Wu C., Sun S., Nimmakayala P., Santos F.A., Meksem K., Springman R., Ding K., Lightfoot D.A., Zhang H.B. A BAC- and BIBAC-based physical map of the soybean genome. Genome Res. 2004;14:319–326. doi: 10.1101/gr.1405004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu C.C., Nimmakayala P., Santos F.A., Springman R., Scheuring C., Meksem K., Lightfoot D.A., Zhang H.B. Construction and characterization of a soybean bacterial artificial chromosome library and use of multiple complementary libraries for genome physical mapping. Theor. Appl. Genet. 2004;109:1041–1050. doi: 10.1007/s00122-004-1712-y. [DOI] [PubMed] [Google Scholar]
- 14.Shopinski K., Iqbal M.J., Afzal J., Ahsan R., Shultz J., Lightfoot D.A. Integration of 144 ESTs with the soybean physical map. Agron. Abstr. 2003;102:388. [Google Scholar]
- 15.Zhu Y.L., Song Q.J., Hyten D.L., Van Tassell C.P., Matukumalli L.K., Grimm D.R., Hyatt S.M., Fickus E.W., Young N.D., Cregan P.B. Single-nucleotide polymorphisms in soybean. Genetics. 2003;163:1123–1134. doi: 10.1093/genetics/163.3.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kazi S. Minimum tile derived microsatellite Markers improve the getic and physical map of the soybean genome. 2005. p. 189. MS thesis, MBMB, SIUC, Carbondale, IL62901.
- 17.Kassem M.A., Meksem K., Iqbal M.J., Wood A.J., Lightfoot D.A. Definition of Soybean Genomic Regions That Control Seed Phytoestrogen Amounts. J. Biomed. Biotechnol. 2004;1:52–60. doi: 10.1155/S1110724304304018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ashfield T., Bocian A., Held D., Henk A.D., Marek L.F., Danesh D., Penuela S., Meksem K., Lightfoot D.A., Young N.D., et al. Genetic and physical localization of the soybean Rpg1-b disease resistance gene reveals a complex locus containing several tightly linked families of NBS-LRR genes. Mol. Plant Microbe. Interact. 2003;16:817–826. doi: 10.1094/MPMI.2003.16.9.817. [DOI] [PubMed] [Google Scholar]
- 19.Triwitayakorn K., Njiti V.N., Iqbal M.J., Yaegashi S., Town C., Lightfoot D.A. Genomic analysis of a region encompassing QRfs1 and QRfs2: genes that underlie soybean resistance to sudden death syndrome. Genome. 2005;48:125–138. doi: 10.1139/g04-103. [DOI] [PubMed] [Google Scholar]
- 20.Stein L.D., Mungall C., Shu S., Caudy M., Mangone M., Day A., Nickerson E.J., Stajich E., Harris T.W., Arva A., et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meksem K., Zobrist K., Ruben E., Hyten D., Quanzhou T., Zhang H.B., amd Lightfoot D.A. Two large-insert soybean genomic libraries constructed in a binary vector: applications in chromosome walking and genome wide physical mapping. Theor. Appl. Genet. 2000;101:747–755. [Google Scholar]
- 22.Marek L.F., Shoemaker R.C. BAC contig development by fingerprint analysis in soybean. Genome. 1997;40:420–427. doi: 10.1139/g97-056. [DOI] [PubMed] [Google Scholar]
- 23.Danesh D., Penuela S., Mudge J., Denny R.L., Nordstrom H., Martinez J.P., Young N.D. A bacterial artificial chromosome library for soybean and identification of clones near a major cyst nematode resistance gene. Theor. Appl. Genet. 1998;96:196–202. [Google Scholar]
- 24.Shultz J., Meksem K., Shetty J., Town C.D., Koo H., Potter J., Wakefield K., Zhang H.B., Wu C., Lightfoot D.A. End sequencing of BACs comprising a provisional tiling path from a fingerprint physical map of soybean (Glycine max) cultivar Forrest. 2003. GenBank accession nos CG812653– CG826126 (13 473 sequences)
- 25.Keim P., Beavis W., Schupp J., Freestone R. Evaluation of soybean RFLP marker diversity in adapted germplasm. Theor. Appl. Genet. 1992;85:205–212. doi: 10.1007/BF00222861. [DOI] [PubMed] [Google Scholar]
- 26.Cheung J., Wilson M.D., Zhang J., Khaja R., MacDonald J.R., Heng H.H., Koop B.F., Scherer S.W. Recent segmental and gene duplications in the mouse genome. Genome Biol. 2003;4:R47. doi: 10.1186/gb-2003-4-8-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lightfoot D.A., Njiti V.N., Gibson P.T., Kassem M.A., Iqbal J.M., Meksem K. Registration of Essex × Forrest recombinant inbred line (RIL) mapping population. Crop Sci. 2005;45:1678–1681. [Google Scholar]
- 28.Iqbal M.J., Yaegashi S., Ahsan R., Shopinski K.L., Lightfoot D.A. Root response to Fusarium solani f. sp. glycines: temporal accumulation of transcripts in partially resistant and susceptible soybean. Theor. Appl. Genet. 2005;110:1429–1438. doi: 10.1007/s00122-005-1969-9. [DOI] [PubMed] [Google Scholar]
- 29.Pan X., Stein L., Brendel V. SynBrowse: a synteny browser for comparative sequence analysis. Bioinformatics. 2005;21:3461–3468. doi: 10.1093/bioinformatics/bti555. [DOI] [PubMed] [Google Scholar]