Abstract
There are many components which govern the function of a protein within a cell. Here, we focus on the molecular recognition of small molecules and the prediction of common recognition by similarity between protein–ligand binding sites. SitesBase is an easily accessible database which is simple to use and holds information about structural similarities between known ligand binding sites found in the Protein Data Bank. These similarities are presented to the wider community enabling full analysis of molecular recognition and potentially protein structure–function relationships. SitesBase is accessible at http://www.bioinformatics.leeds.ac.uk/sb.
INTRODUCTION
Rapid advances in structural biology have resulted in a large increase in the number of protein structures deposited in the Protein Data Bank (1). New structural genomics initiatives have reinforced this growth and structures are now being solved prior to any knowledge of their functions. With this expansion has come the need to rapidly analyse protein structures and determine their functions. SitesBase is a new database of protein–ligand binding sites that contains pre-calculated all-against-all binding site similarities which can be used for rapid retrieval and examination of other binding sites with similar structure, independent of overall fold or sequence similarity from the existing PDB. The WWW interface provides a simple resource for structural and functional studies of ligand binding sites. Assessing binding site similarity is assisted with a statistical P-value measure, which gives the probability of obtaining a given score by random chance. The user can visualize multiple alignments and perform full structural superpositions. Links to other structural databases [PDBsum (2) and SCOP (3)] are also provided for additional structural examination. The resource can be automatically updated as new proteins become available independent of whether they have been structurally or functionally annotated.
Our approach differs from other functional site similarity searching methods (4–12) by using an all-atom representation of binding sites, and hence we lose no information regarding possible atomic level similarity and the possible retention or otherwise of hydrogen bonds, electrostatic or steric interactions with the ligand. This information may be particularly useful in detailed studies of comparative molecular recognition. This could include designing ligand specificity or understanding cross-reactivity in structure-based drug design. Additionally, we have chosen to focus on creating a database of pre-calculated protein–ligand binding site similarities allowing fast retrieval of results and providing the basis of a ligand binding site classification scheme independent of overall fold and sequence similarity.
OVERVIEW OF SITESBASE
SitesBase is held within a MySQL relational database allowing rapid retrieval of data and providing an easily maintainable resource which is updated monthly as more structures become available. The construction and contents of the database are described below.
Binding site identification
Ligands were automatically identified within 32 508 PDB (1) files (June 2005) by the presence of bound ligands (identified by keywords HETATM) and their atoms stored within the database. The database excludes protein/peptide ligands and treats Het-groups as individual ligand binding sites. Protein atoms within a 5 Å radius of any ligand atom define its binding site and these are also stored. This located over 125 000 binding sites. The database also holds the results of binding site comparisons (see below). We have removed ligands with fewer than six atoms which discards metal ions and small solvents such as sulfate and acetate ions. Commonly occurring solvents (e.g. glycerol, ethylene glycol, etc.) and post-translational modifications to amino acids are also removed leaving 33 168 binding sites. A full list of ‘discarded’ ligands is given in the Supplementary Data.
Binding site comparison
Geometric hashing was used to perform an all-against-all comparison of the 33 168 ligand binding sites. The method has been described in detail previously (13) and proceeds by identifying equivalent atom constellations between pairs of binding sites. Equivalent atoms must have the same element type (i.e. carbon, oxygen, nitrogen, etc.) and occur in similar relative spatial orientations. Similarity is measured by an atom–atom score (number of atoms comprising the largest possible matching constellation). In addition, an adjusted score is calculated to take into account the maximum possible match size (atom–atom score divided by the size of the smallest site). Each pair-wise atom–atom score is stored along with the adjusted score, root-mean-square deviation (RMSD), a list of the equivalenced atoms and a rotation/translation matrix to superimpose the sites. Restrictions are in place to prevent numerous random matches being stored in the database; hence, scores are only stored if the atom–atom score is ≥20 or the adjusted score is ≥0.3 or if there is any detectable sequence similarity between the sites (see seq-sim score calculations below).
Binding site annotation
Each binding site is annotated with up to date SCOP (3) classifications (currently version 1.67) to facilitate analysis of the results, e.g. to separate trivial matches (family relatives with high sequence similarity) from more distant relatives. Structural classification assignments are not always straightforward because binding sites can occur between domains. The following method is used to list all domains contributing to the binding site: each atom within a binding site has an associated SCOP classification. If multiple domains are located within one binding site, they are ranked by the number of contributing atoms. The highest ranking class is termed primary. Others are listed in ranked order as secondary and tertiary classes. In most cases, primary class annotation is properly assigned and consistent with other family members; however, sometimes one of the other assigned classes may be more appropriate when a site is seen in context with other class relatives.
The probability (P-value) of achieving a given atom–atom score by random chance is calculated by comparison with the tail of the random extreme value distribution (EVD). The random EVD model is generated for each individual query site by selecting scores from the database and removing relatives, i.e. sites within the same primary or secondary SCOP superfamily as the query. Data which do not fit to an EVD are usually explained by retention of significant high scoring hits within the random model, e.g. where there are significant hits between proteins with different folds or when the query protein is unclassified in SCOP. In these cases, the P-value will be a conservative estimate of the probability for a given score. It is also important to note that observed P-values will change as new binding sites are incorporated into the databases, following the release of new PDB entries.
We also use the method of Stark et al. (10) to calculate the sequence similarity of local structural similarities. This gives a seq-sim score for matching binding site pairs based on a structure-based sequence similarity score. The seq-sim score is a measure of the significance of attaining a given RMSD for a given number of residues with matching sequence. Hence, low seq-sim score indicates high sequence similarity with important atoms in similar positions. The method uses residue abundances to calculate the significance of matching residue types. A match between two amino acids is identified only if certain atoms are found to be equivalent. For all residues, the Cα atoms must match in similar relative spatial orientations. Other residues (AFILPV) must match both the Cα and Cβ atoms. The following residues (DENQTCSRKHYMW) must additionally match a further functional atom (14).
Contents of SitesBase
Currently, (June 2005) SitesBase houses the local protein structure of 33 168 ligand binding sites annotated with SCOP codes where available (see Table 1).
Table 1.
A summary of the contents of SitesBase
| Number | |
|---|---|
| Summary of SitesBase (June 2005) | |
| Total number of PDB files (with ligand ≥6 atoms) | 12 898 |
| Total number of binding sites | 33 168 |
| Number of primary SCOP families | 1032 |
| Ligand binding sites by SCOP classification | |
| All α | 4172 |
| All β | 4166 |
| α/β | 10 763 |
| α+β | 4239 |
| Multidomain proteins (α and β) | 855 |
| Membrane and cell surface proteins/peptides | 1453 |
| Small proteins | 108 |
| Othera | 200 |
| As yet unclassified in SCOP | 7302 |
aCoiled-coil proteins, low resolutions structures, peptides and designed proteins.
THE INTERFACE
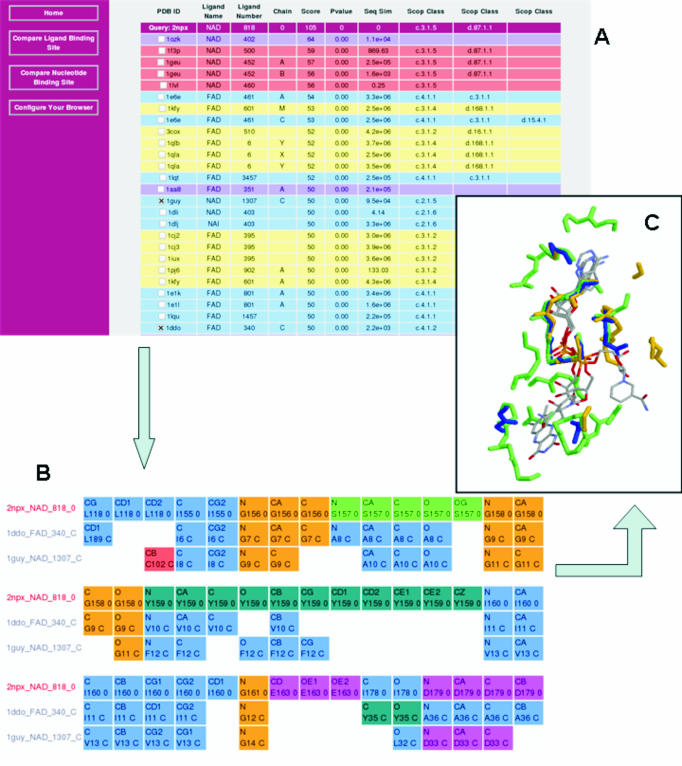
The SitesBase interface provides three ways to search for a query site: (i) a PDB code, e.g. 1hdx, (ii) a ligand three-letter code, corresponding to the Residue name in PDB format, e.g. NAD and (iii) a keyword or phrase, e.g. alcohol dehydrogenase. Once a site is selected, the query can be submitted to search for all similar binding sites. This retrieval is rapid because all binding site comparisons are stored within the database. Similarity scores to a selected query binding site can be retrieved using the web interface presenting all binding sites and their relatives in a fast and informative way while hiding technical SQL (Structured Query Language) statements from the user. Binding site similarities are returned in a list ranked by decreasing atom–atom score. Each hit is displayed with a score, P-value and seq-sim score and is coloured according to its structural relationship to the query in the current SCOP database (Figure 1A). There are also hypertext links to both PDBsum (2) and SCOP (3) for further structural and functional information. Check-box selection of matching sites can be submitted to rapidly retrieve a multiple alignment of the binding site atoms to the query site (Figure 1B) and a PDB format file containing the superimposed binding sites (Figure 1C).
Figure 1.
Collection of screen shots from SitesBase showing results of a search (A) with query 2npx (NADH peroxidase). This binding site has two SCOP classes attributed. The primary SCOP class is c.3.1.5 the NAD/FAD reductase family with an NAD/FAD binding domain fold. The SCOP and PDB codes are linked to the SCOP and PDBsum databases, respectively. Four of the listed hits are family relatives of the query (pink), ten are superfamily relatives (yellow) and ten are fold relatives (blue). There are also two high scoring hits which are yet to be classified in the SCOP database (purple). Note: in this illustration some high scoring hits have been removed in order to show more distantly related proteins. (B) Gives a multiple alignment of two selected hits with the same SCOP class (i.e. with different folds) as the query and clearly shows the sequence conserved GXGXXG dinucleotide binding motifs. (C) Illustrates these three protein sites (2npx, blue; 1guy, yellow; and 1ddo, green) superimposed in 3D.
Binding site similarity
Binding site similarity is assessed by the number of matching atoms (atom–atom score) and by P-value. While family and superfamily relatives often have similar functions this is not always the case and significant differences in ligand recognition can occur. Therefore, it is useful to analyse atomic level detail of ligand recognition given the existence of fold or even sequence similarity. Additionally, site similarity may provide evidence of common ligand recognition or function when there is little or no similarity in the overall fold of a protein. Figure 1 gives examples of binding site similarities in the absence of fold similarity. Here, the NAD binding site within NADH peroxidase (2npx) was compared with the entries in SitesBase. It is depicted with two other binding sites superimposed. In each case, the proteins have different overall folds to the query. Similarity can be seen around the glycine-rich dinucleotide binding motif (15), suggesting common modes of molecular recognition. It is interesting that D-amino acid oxidase (1ddo) binds a different but related molecule (FAD) with similarity only around the common AMP part of the ligands showing that the method is useful in locating both full and partial binding site matches. This information can be important in determining possible side effects of drugs where a drug could bind to multiple targets.
SUMMARY AND FUTURE WORK
Currently, the database is useful for studies of protein structure–function relationships and to generate protein structure-based alignments for ligand-based 3D pharmacophore generation. Additionally, the database can be used to corroborate functional similarity given sequence or fold similarity. The database can be used to produce an all-against-all map of similarity giving a global view of binding site similarity. This will form a basis for binding site structural classification in the future. We also plan to allow users to upload files to search for similarities to known binding sites within SitesBase. This will prove useful for the study of newly determined proteins of unknown function or help in identifying alternate or even new ligand binding sites in existing proteins.
Acknowledgments
The authors wish to thank Alexander Stark and Robert Russell for their help with implementing the seq-sim score calculations and the BBSRC for financial support (grant B18760). Funding to pay the Open Access publication charges for this article was provided by the University of Leeds and JISC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laskowski R.A., Hutchinson E.G., Michie A.D., Wallace A.C., Jones M.L., Thornton J.M. PDBsum: a Web-based database of summaries and analyses of all PDB structures. Trends Biochem. Sci. 1997;22:488–490. doi: 10.1016/s0968-0004(97)01140-7. [DOI] [PubMed] [Google Scholar]
- 3.Murzin A.G., Brenner S.E., Hubbard T., Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 4.Artymiuk P.J., Poirrette A.R., Grindley H.M., Rice D.W., Willett P. A graph-theoretic approach to the identification of three-dimensional patterns of amino acid side-chains in protein structures. J. Mol. Biol. 1994;243:327–344. doi: 10.1006/jmbi.1994.1657. [DOI] [PubMed] [Google Scholar]
- 5.Binkowski T.A., Adamian L., Liang J. Inferring functional relationships of proteins from local sequence and spatial surface patterns. J. Mol. Biol. 2003;332:505–526. doi: 10.1016/s0022-2836(03)00882-9. [DOI] [PubMed] [Google Scholar]
- 6.Kinoshita K., Furui J., Nakamura H. Identification of protein functions from a molecular surface database, eF-site. J. Struct. Funct. Genomics. 2002;2:9–22. doi: 10.1023/a:1011318527094. [DOI] [PubMed] [Google Scholar]
- 7.Kleywegt G.J. Recognition of spatial motifs in protein structures. J. Mol. Biol. 1999;285:1887–1897. doi: 10.1006/jmbi.1998.2393. [DOI] [PubMed] [Google Scholar]
- 8.Schmitt S., Kuhn D., Klebe G. A new method to detect related function among proteins independent of sequence and fold homology. J. Mol. Biol. 2002;323:387–406. doi: 10.1016/s0022-2836(02)00811-2. [DOI] [PubMed] [Google Scholar]
- 9.Shulman-Peleg A., Nussinov R., Wolfson H.J. Recognition of functional sites in protein structures. J. Mol. Biol. 2004;339:607–633. doi: 10.1016/j.jmb.2004.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stark A., Sunyaev S., Russell R.B. A model for statistical significance of local similarities in structure. J. Mol. Biol. 2003;326:1307–1316. doi: 10.1016/s0022-2836(03)00045-7. [DOI] [PubMed] [Google Scholar]
- 11.Wallace A.C., Borkakoti N., Thornton J.M. TESS: a geometric hashing algorithm for deriving 3D coordinate templates for searching structural databases. Application to enzyme active sites. Protein Sci. 1997;6:2308–2323. doi: 10.1002/pro.5560061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ivanisenko V.A., Pintus S.S., Grigorovich D.A., Kolchanov N.A. PDBSite: a database of the 3D structure of protein functional sites. Nucleic Acids Res. 2005;33:D183–D187. doi: 10.1093/nar/gki105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brakoulias A., Jackson R.M. Towards a structural classification of phosphate binding sites in protein-nucleotide complexes: an automated all-against-all structural comparison using geometric matching. Proteins. 2004;56:250–260. doi: 10.1002/prot.20123. [DOI] [PubMed] [Google Scholar]
- 14.Russell R.B. Detection of protein three-dimensional side-chain patterns: new examples of convergent evolution. J. Mol. Biol. 1998;279:1211–1227. doi: 10.1006/jmbi.1998.1844. [DOI] [PubMed] [Google Scholar]
- 15.Wierenga R.K., de Maeyer M.C.H., Hol W.G.J. The interaction of pyrophosphate moeities with a-helices in dinucleotide binding proteins. Biochemistry. 1985;24:1346–1357. [Google Scholar]