Abstract
The high similarity of tunicates and vertebrates during their development coupled with the transparency of tunicate larvae, their well-studied cell lineages and the availability of simple and efficient transgenesis methods makes of this subphylum an ideal system for the investigation of vertebrate physiological and developmental processes. Recently, the sequencing of two different Ciona genomes has lead to the identification of numerous genes. In order to better understand the regulation of these genes, a database was created containing information on regulation of tunicate genes collected from literature. It includes for instance information regarding the minimal promoter length, the transcription factors involved and their binding sites, as well as the localization of the gene expression. Additionally, binding sites for characterized transcription factors were predicted based on published in vitro recognition sites. Comparison of the promoters of homologous genes in different species is also provided to allow identification of conserved cis elements. At the time of writing, information about 184 promoters, containing 73 identified binding sites and >2000 newly predicted binding sites is available. This database is accessible at http://dbtgr.hgc.jp.
INTRODUCTION
Tunicates, which include larvaceans, thaliaceans and sedentary ascidians, or sea squirts, such as Ciona intestinalis and Ciona savignyi, are lower chordates and share basic gene repertoires and many characteristics, both developmental and physiological, with vertebrates (1–3). Although the adult ascidian shows no resemblance to vertebrate animals, its fertilized egg develops within a day into a tadpole-like larva consisting of ∼2600 cells which shares a basic body-plan with vertebrates (2). The well-established cell lineage (4,5) and the transparency of the larva allow the visualization of the spatial and temporal gene expression pattern in detail during development, making tunicates an efficient tool to elucidate the genetic regulatory systems underlying the developmental and physiological processes of vertebrates. Furthermore, simple electroporation methods permit the simultaneous transformation of several hundred synchronously developing embryos (6) and transient transgenesis has been applied successfully for efficient expression of exogenous genes (6–8), thus providing researchers simple and reliable tools to carry out large-scale investigations of the different tunicate gene expression networks.
Although the regulation of specific genes has been investigated for several years (9), the recent availability of the draft genome of C.intestinalis and C.savignyi, coupled with the results of systematic in situ hybridization experiments, offers new possibilities with regards to the identification of cis-elements involved in both the spatial and temporal gene regulation (10).
DBTGR, the DataBase of Tunicate Gene Regulation, was constructed in order to provide a comprehensive access to published information regarding regulation of gene expression in tunicates. It further offers information on putative binding sites for the identified transcription factors. Alignments of orthologous promoter sequences are also provided for comparative analysis of regulatory elements. Finally, user-defined motif searches can be carried out on the complete set of available promoter sequences.
The web pages provided by DBTGR are generated by PHP scripts from the information stored in a MySQL database. The database itself consists of several cross-linked tables containing data about the promoters, the transcription factors and the binding sites, as well as the relation between them. The consensus and weight matrix searches are performed by external C programs and their results cached to provide fast access to frequent queries.
OVERVIEW
DBTGR mainly consists of a collection of experimentally characterized promoters for which the location of gene expression is given along with the transcription factor driving it. In addition, when available, the position of known binding sites of transcription factors as well as other regulatory elements are given for each promoter.
Information about the gene product and the localization of its expression during the development of the organism is given both as a graphical overview and a detailed text (Figure 1). The graphical overview represents a larva consisting of six domains: the epidermis, the nervous system, the endoderm, the brain, the notochord and the muscles. Each domain in which expression is found is filled with a different color, thus providing an easy way to identify promoters targeting expression to specific areas. The text entry gives detailed information about the localization of the gene expression by enumerating the different tissues and cells where it was experimentally observed.
Figure 1.
Graphical representation of gene expression. The schematic larva is separated in six domains, which are highlighted with different colors depending on the localization of gene expression.
Promoter regions critical for gene expression, as well as the size of the minimal functional promoter are given provided they were determined during the promoter characterization. Similarly, information regarding the recognition sequences of the involved transcription factors is given when available.
A list of the identified binding sites, separated in two categories to distinguish between binding sites for which experiments have shown involvement in gene expression and predicted binding sites, is then presented, indicating among others the transcription factor involved, the position of the binding site and the strand on which it was identified. Selected binding sites can easily be added or removed from the sequence displayed at the bottom of the page by ticking the corresponding checkbox and reloading the page via the provided button.
Cross-references to the corresponding gene location in both version 1.00 [available from the US Department of Energy Joint Genome Institute (JGI) http://www.jgi.doe.gov] and version 1.95 [available from Ensembl http://www.ensembl.org (11)] of the draft genome of C.intestinalis is also given, and links are provided to the respective genome browsers and gene entry pages. Furthermore, when predicted promoters for homologous genes could be obtained from the C.savignyi genome, a link to the alignment of both promoter sequences where conserved regions are highlighted is provided.
The promoter sequence presented at the bottom of the page consists of at most the 3000 bases upstream of the protein-coding region and was originally obtained from the JGI version 1.00 genome draft. When possible, the corresponding sequence in the JGI version 2.00 genome draft was extracted by BLAST search (12). Although only the latest available promoter sequence is given as an online image, when present, both sequence versions are available as FASTA files for download. In the online image, binding sites are shown as arrows located above or under the nucleotide sequence depending on the strand on which they are located. Their color corresponds to that of the previously listed binding site description, and a question mark is added next to predicted binding sites. The information available in the binding sites list can also be obtained by moving the mouse over the arrow representing it.
Both the listed and the image binding sites are linked to a motif-specific page providing a list of all the genes where that motif was found, the sequence of the corresponding binding sites with the bases matching the consensus sequence being highlighted, and both a position-specific weight matrix and a consensus sequence for that motif computed based on the binding sites for which experimental evidences of their involvement in regulation are present. Similarly, the transcription factors listed are linked to a page providing information on the genes they regulate and the binding site they recognize.
FEATURES
In order to provide the user with information about promoter regions conserved between several species, predicted promoter sequences for C.savignyi genes were extracted from the C.savignyi genome draft based on the whole genome alignment made with the JGI version 1.00 C.intestinalis genome draft available from VISTA (13). As for C.intestinalis promoter sequences, the C.savignyi sequences are available as FASTA files for download, and as online images displaying predicted binding sites.
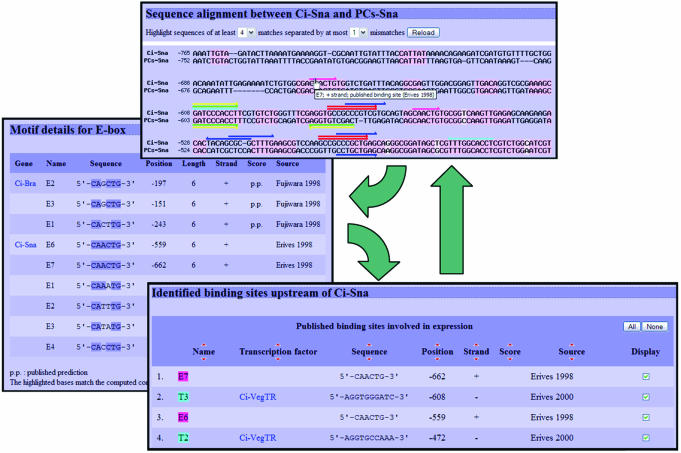
Furthermore, the latest version of the C.intestinalis promoter sequences were aligned using ClustalW (14) with the predicted C.savignyi promoter sequences. Links to these alignments are available from the detailed promoter pages. Each alignment is shown as a dynamically generated online image with the regions conserved in both promoter sequences highlighted (Figure 2). The possibility to modify the extent of the highlighted conserved regions is provided via the adjustment of two parameters: the minimum size of a conserved block and the maximum length of the non-conserved region between two such blocks. In addition to the highlighted conserved regions, the binding sites identified in either sequences are also shown, so that a correlation between the predicted or proven binding sites and sequence conservation can easily be visualized.
Figure 2.
Alignment of the C.intestinalis and C.savignyi snail promoter sequences. The two promoter sequences were aligned with ClustalW. Arrows above the alignment represent binding sites identified in the upper sequence, while arrows under the alignment refer to binding sites in the lower sequence. The direction of the arrows indicates on which strand the specific binding site was found. Binding sites with the same color are bound by the same transcription factor. The two overlapping red arrows indicate that this specific site was both reported in publications and predicted by consensus searches, while overlapping arrows with different colors identify a binding site recognized by two or more transcription factors. Clicking on an arrow leads to a motif-specific page listing all occurrences of that motif, the concerned promoters being linked back to their detailed page. There, all identified binding sites are given, with the possibility to toggle their display on the single promoter sequence, or to obtain the pre-aligned sequences again.
Searches for particular motifs can be performed in the complete dataset by using either a consensus sequence or a position-specific weight matrix. The extent of the returned results can be modified by adjusting the number of mismatches allowed in the case of a consensus search, and the cutoff threshold in the case of a weight matrix search. As alternative to user-defined matrices, the provided JASPAR weight matrices (15) can also be used.
The C.intestinalis promoter entries are linked to the JGI and Ensembl genome browsers and gene information pages, as well as to the corresponding ANISEED and Ghost entries.
FUTURE PROSPECTS
The available promoter sequences will be updated when new genome versions are released using a similar method as described here for the extraction of the JGI version 2.0 sequences.
Further improvements such as the inclusion of information on the regulation of Halocynthia roretzi, C.savignyi and Oikopleura dioica genes extracted from the literature are planned. In addition, contributions from researchers are welcomed, either under the form of links to recently published work containing relevant data, or by direct submission of newly characterized promoter sequences and their features. Furthermore, recent discussions showed an interest for centralized contact information regarding the various promoter constructs available from the different groups of the tunicate community. This information could similarly be added to DBTGR by extraction from the literature or direct submission from the community.
Close cooperation with the recently formed ‘Model Organism Database Working Group’ of the tunicate community will ensure that DBTGR grows in accordance with and in response to its needs, as well as its integration with the several other complementary databases provided by the community.
Acknowledgments
Computation time was provided by the Super Computer System, Human Genome Center, Institute of Medical Science, The University of Tokyo. The JGI sequence data were produced by the US Department of Energy Joint Genome Institute http://www.jgi.doe.gov/. The authors would like to thank the Japan Society for the Promotion of Science for the support they provided to N.S. This work was supported by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (No. 17310114). Funding to pay the Open Access publication charges for this article was provided by the Japan Society for the Promotion of Science.
Conflict of interest statement. None declared.
REFERENCES
- 1.Dehal P., Satou Y., Campbell R.K., Chapman J., Degnan B., De Tomaso A., Davidson B., Di Gregorio A., Gelpke M., Goodstein D.M., et al. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- 2.Satoh N. The ascidian tadpole larva: comparative molecular development and genomics. Nature Rev. Genet. 2003;4:285–295. doi: 10.1038/nrg1042. [DOI] [PubMed] [Google Scholar]
- 3.Seo H.C., Edvardsen R.B., Maeland A.D., Bjordal M., Jensen M.F., Hansen A., Flaat M., Weissenbach J., Lehrach H., Wincker P., et al. Hox cluster disintegration with persistent anteroposterior order of expression in Oikopleura dioica. Nature. 2004;431:67–71. doi: 10.1038/nature02709. [DOI] [PubMed] [Google Scholar]
- 4.Conklin E.G. The organization and cell lineage of the ascidian egg. J. Acad. Natl Sci. 1905;13:1–119. [Google Scholar]
- 5.Nishida H. Cell lineage analysis in ascidian embryos by intracellular injection of a tracer enzyme. III. Up to the tissue restricted stage. Dev. Biol. 1987;121:526–541. doi: 10.1016/0012-1606(87)90188-6. [DOI] [PubMed] [Google Scholar]
- 6.Corbo J.C., Levine M., Zeller R.W. Characterization of a notochord-specific enhancer from the Brachyury promoter region of the ascidian, Ciona intestinalis. Development. 1997;124:589–602. doi: 10.1242/dev.124.3.589. [DOI] [PubMed] [Google Scholar]
- 7.Hikosaka A., Kusakabe T., Satoh N., Makabe K.W. Introduction and expression of recombinant genes in ascidian embryos. Dev. Growth Differ. 1992;34:627–634. doi: 10.1111/j.1440-169X.1992.tb00031.x. [DOI] [PubMed] [Google Scholar]
- 8.Zeller R.W. Generation and use of transgenic ascidian embryos. Methods Cell Biol. 2004;74:713–730. doi: 10.1016/s0091-679x(04)74029-7. [DOI] [PubMed] [Google Scholar]
- 9.Corbo J.C., Di Gregorio A., Levine M. The ascidian as a model organism in developmental and evolutionary biology. Cell. 2001;106:535–538. doi: 10.1016/s0092-8674(01)00481-0. [DOI] [PubMed] [Google Scholar]
- 10.Kusakabe T. Decoding cis-regulatory systems in ascidians. Zoolog. Sci. 2005;22:129–146. doi: 10.2108/zsj.22.129. [DOI] [PubMed] [Google Scholar]
- 11.Hubbard T., Andrews D., Caccamo M., Cameron G., Chen Y., Clamp M., Clarke L., Coates G., Cox T., Cunningham F., et al. Ensembl 2005. Nucleic Acids Res. 2005;33:D447–D453. doi: 10.1093/nar/gki138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frazer K.A., Pachter L., Poliakov A., Rubin E.M., Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sandelin A., Alkema W., Engstrom P., Wasserman W.W., Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]