Abstract
Proteome science relies on bioinformatics tools to characterize proteins via their proteolytic peptides which are identified via characteristic mass spectra generated after their ions undergo fragmentation in the gas phase within the mass spectrometer. The resulting secondary ion mass spectra are compared with protein sequence databases in order to identify the amino acid sequence. Although these search tools (e.g. SEQUEST, Mascot, X!Tandem, Phenyx) are frequently successful, much is still not understood about the amino acid sequence patterns which promote/protect particular fragmentation pathways, and hence lead to the presence/absence of particular ions from different ion series. In order to advance this area, we have developed a database, PepSeeker (http://nwsr.smith.man.ac.uk/pepseeker), which captures this peptide identification and ion information from proteome experiments. The database currently contains >185 000 peptides and associated database search information. Users may query this resource to retrieve peptide, protein and spectral information based on protein or peptide information, including the amino acid sequence itself represented by regular expressions coupled with ion series information. We believe this database will be useful to proteome researchers wishing to understand gas phase peptide ion chemistry in order to improve peptide identification strategies. Questions can be addressed to j.selley@manchester.ac.uk.
INTRODUCTION
Proteomics is growing rapidly as a technique in functional genomics. Driven by advances in mass spectrometry and analytical chemistry, coupled with the expanding number of completely sequenced genomes, proteomics is becoming a widely exploited technology for characterizing the proteins found in living systems. There are a growing number of proteome databases appearing on the internet (1–5) along with maturing data standards in proteomics driven by the Proteome Standards Initiative (PSI) (6–8). Existing databases cover a wide-range of mass spectrometry-based proteomics data, including data stored on basic identifications of proteins and peptides, the samples studied, instruments used, and software search tools employed. Notable examples include the PeptideAtlas database developed by Aebersold and colleagues (1), the Global Proteome Machine (GPM) from Beavis and co-workers (2), the Open Proteomics Database (3), the PEDRo proteome repository developed locally in Manchester (4), and the PRIDE database at the EBI (5). This growing list of resources offers a range of approaches for the capture, storage and dissemination of proteomic experimental data and reflects the fact that proteomics has now come of age in the post-genomic era and is delivering large, complex datasets which are rich in information.
These advances in proteomics are supported by bioinformatics search tools which allow the mass spectra generated to be compared with the protein sequence databases in order to identify the protein. Typically, this is done by identifying the protein from peptides produced by hydrolysing the polypeptide chain with a proteolytic enzyme such as trypsin. The tryptic peptides are then separated and analysed in the mass spectrometer. Proteins can then be characterized either from the mass-to-charge values of the peptide ions themselves (known as Peptide Mass Fingerprinting), or increasingly by tandem mass spectrometry (MS) where the peptide ion is itself induced to fragment via energetic collision with a gas in the instrument (Peptide Fragment Fingerprinting). This latter technique is part of the popular MudPIT (Multidimensional Protein Identification Technology) approach originated from the Yates lab (9,10), where thousands of peptides are separated via liquid chromatography directly fed into a mass spectrometer, yielding many thousands of spectra for bioinformatic analysis. Search tools such as Mascot (11), SEQUEST (12), X!Tandem (13) and Phenyx (14) are then employed to determine the most probable matching peptide in a sequence database. This is dependent on the quality of the spectrum, the ions observed and many other factors. Despite several excellent studies [Wysocki and other refs, (15–17)], the fragmentation of peptide ions in the gas phase is still only partially understood, and these algorithms primarily exploit the differences in the mass-to-charge values of the ions in order to identify candidate peptides which match the experimental spectra. A more complete understanding of how different amino acid sequences promote or influence fragmentation pathways will lead to improvements in our ability to predict the relative presence/absence of particular peaks from different ion series in the tandem MS spectra. This in turn can be exploited in these software search tools to make better peptide identifications, both in terms of the number of peptides which can be identified and the overall confidence which can be placed in them. This is important since a large fraction of the spectra currently analysed do not lead to confident peptide identifications (18) and proteomics still does not offer a truly genome-wide coverage.
This requirement for better search tools in tandem MS for automatic peptide identification is important both for searches against known protein sequence databases, and the more challenging de novo search problem. To facilitate these efforts, we have designed and implemented a database system, PepSeeker, to capture and store this information, and allow users to query it to help mine rules and explore fragmentation patterns observed in peptide sequences studied in the mass spectrometer. This has been in part motivated by a local project seeking to mine the data using machine learning methods to discover rules to model peptide spectra including the relative peak heights of the fragment ions. To this end, our PepSeeker database contains both peptide identifications and the associated fragment ion details used to identify that amino acid sequence. It is intended to complement the more holistic proteome databases, with the primary focus on the identification itself allied to the peptide sequence data, coupled to the underlying ion series. To this extent, PepSeeker supports novel searches not available via other databases and tools, where spectra and specific ion information can be retrieved with respect to amino acid patterns.
DATA CAPTURE STRATEGY
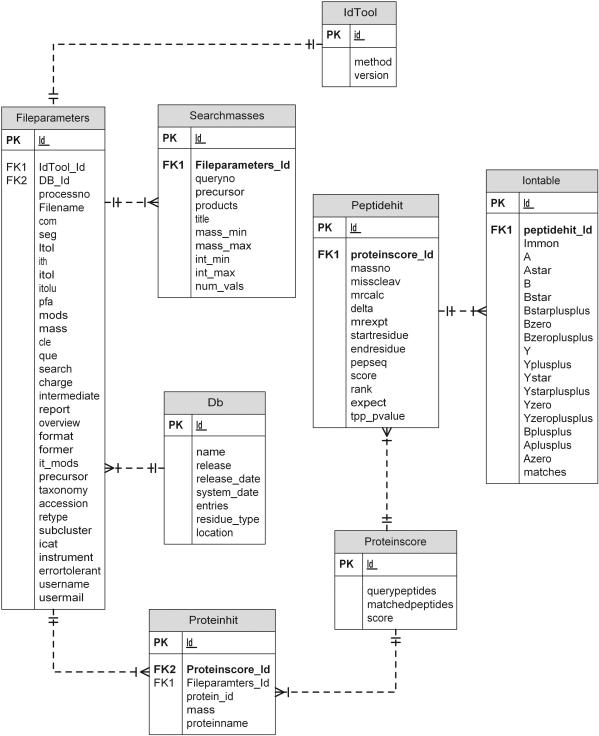
The current implementation of PepSeeker has been developed using a MySQL platform with a simple schema designed to capture data obtained primarily from a local Mascot-based proteomics pipeline. This strategy enables data to be captured from a range of instruments and vendors. The database schema is shown in Figure 1, which shows the data captured at present, including basic file searching parameters relating to the spectra, instrument and database, as well as protein and peptide hit information from the identifications. Rather than base this around the PEDRo or PSI XML schema currently under development, we elected to design a simplified model targeted directly at the identification stage of the proteomics pipeline, and do not capture all the information associated with the mzData/mzXML standards concerning full spectra and processing details or comprehensive instrument parameters. This is largely equivalent to the mzIdent/analysisXML standard currently under development by the PSI group (6). Results from the Mascot searches are parsed and loaded into the database directly from Mascot ‘.dat’ format flatfiles, although we are able to parse and accept other formats such as Sequest's ‘.dta’ and ‘.out’ files in a semi-automated fashion. A web-based submission form is under development to accept all formats. This provides us with an interim solution whilst the mzIdent/analysisXML matures as a standard, since it is expected that all search engines will be tailored to deliver this as output in the near future. Data is captured based on simple filtering criteria concerning the user/email/database, so that only the desired data is captured from groups willing to share data. The following data tables are used: SearchMasses (containing precursor and product ions, their intensities and charges), Fileparameters (containing information about file, instrument and software settings), Proteinhit (containing information about all of the top protein hits), Proteinscore (containing protein score and information on number of peptide queries matched), Peptidehit (containing information about matched peptides) and IonTable (containing information on ions matching peptide ion fragments). Currently, the database contains peptide identifications from species including Saccharomyces cerevisiae, Schizosaccharomyces pombe, Escherichia coli, Plasmodium falciparum, mouse and human. Statistics relating to the number of peptides in the database are shown in Table 1.
Figure 1.
PepSeeker database scheme, showing the relationship between tables.
Table 1.
Pepseeker database statistics
| Viewable spectra | 1 397 159 |
| Proteins | 49 537 |
| Peptides (total) | 186 873 |
| Unique peptides | 47 732 |
| Average peptide length | 11.6 amino acids |
| Range of peptide lengths | 3–66 amino acids |
PepSeeker INTERFACE
The interface is built using Perl CGI and DBI to interact with and query the MySQL database, providing a variety of entry points depending on a particular researcher's search parameters. The first entry point is via the ‘Filename’ search form. This supports searches via user or even the search title for the search, and is aimed mainly at contributors to the dataset allowing researchers to track their experiments and results.
The ‘Protein’ search form relates to the putative parent proteins of identified peptides, allowing queries to retrieve identified peptides from proteins based on keywords, protein mass ranges, taxonomies and/or specific flatfile databases that were searched. We capture version information for publicly available databanks such as Swiss-Prot, Uniprot, MSDB and allow non-standard databases to be downloaded separately via ftp. Given the inherent problems associated with unambiguously assigning peptides to specific proteins, we simply capture and store all reported matches listed in Mascot output. This is deliberate, so that all putative protein–peptide relationships are captured bearing in mind that the focus of this resource is very much on the peptide identifications rather than the protein identifications.
The ‘Peptide’ search form is useful to look for specific amino acid sequence patterns, occurring in isolation or coupled with other patterns. Searches support regular expressions allowing quite complex queries, which may be coupled with restriction by protein accession/identifier and quality control on the peptide confidence (via expectation values or Mascot ion scores). The supported regular expression pattern matching is explained in the online help, and the matching pattern is highlighted in the output from a query.
The ‘Ions (simple)’ search provides a means to search for specific ion types identified by the search engines associated with particular amino acids and locations within a peptide sequence. As in the peptide search, this can be anchored to a specific protein. A more advanced search can also be performed, where many specific ions can be associated with specific positions in a peptide sequence, each subtended by a selected amino acid. In addition, the C-terminal amino acid can be specified (any, arginine or lysine) as the differing basicity of their sidechains can produce differing fragmentation patterns. The ion type query forms [both simple sequence search ‘Ions (simple)’ and advanced ion search ‘Ions (advanced)’] are particularly important to this project, to provide users with a means to examine the presence or absence of given ion types for given peptide sequence patterns which may relate to specific ion fragmentation pathways, or be useful to examine spectra which exemplify given trends identified by machine learning.
EXAMPLE DATABASE QUERIES
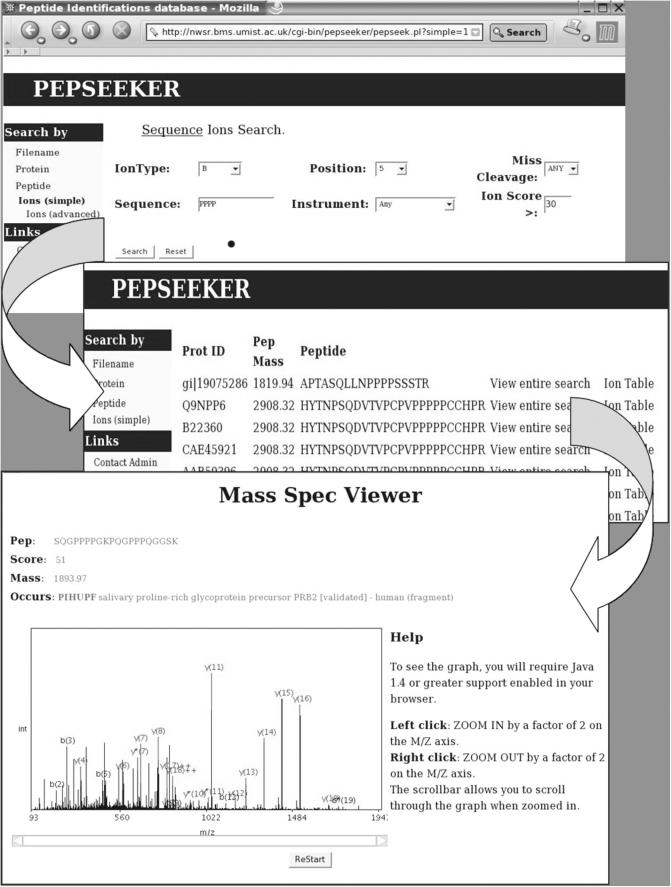
To illustrate the utility of this resource, an example query is shown in Figure 2. This relates to a Peptide search for a specific fragmentation pattern. Most peptide ion fragmentation yields ions in one of two ion series, a b-series and a y-series resulting from fragmentation at the peptide bond. Proline residues are well known to promote fragmentation in the gas phase, and hence for example, we can query the database to show all examples of a proline cleavage where it has generated a y5 ion. We can query the database for examples where there is more than one proline present in the peptide and use the information there to compare with results from machine learning experiments, or indeed to discover unusual or novel patterns which may be investigated further by designing a series of peptides for further study.
Figure 2.
Screen-shot of the PepSeeker front-end, showing an example of the navigation from a simple ion search to a list of the matching peptides through to a graphical representation of the spectra and associated ion information. An example PepSeeker query is shown searching for all peptides within the database containing the sequence PPPP. The first window is the query entry, the second window is the output and the third window displays the spectrum and table associated to the peptide SQGPPPPGKPQGPPPQGGSK.
The example in Figure 2 show how you can move around the database from the initial query. Here, a search is conducted in the ‘Ions (simple)’ search query form which reveals a set of peptides matching the selected regular expression (‘PPPP’), which reports back all peptides in the database containing this pattern. A further click on the ‘Ion table’ link brings up a simple peptide secondary ion spectra in which the user can view the ions present in the spectrum along with an ion table showing all possible ions with those actually present highlighted. The user can then zoom in on the spectrum, to see if any putative ions of low level intensity which were left unassigned by the search tool.
DATA SUBMISSION AND FUTURE DIRECTIONS
The database is publicly available via the following URL (http://nwsr.smith.man.ac.uk/pepseeker). Our principal aim is to make peptide identification data available to the community to provide datasets for groups developing tools for peptide identification. In addition, we provide a means to search the data for characteristic ion patterns linked to amino acid sequence. Currently, contributors wishing to contribute data can send us Mascot ‘.dat’ files or similar formats from other vendors (‘.dta’ and ‘.out’ files from Sequest), but we plan to operate an upload site in the near future. Likewise, we plan to support PSI-compliant XML formats such as mzIdent/analysisXML when they become available, permitting both upload and download in such a format. Groups wishing to contribute data should contact J.Selley@manchester.ac.uk.
Acknowledgments
We acknowledge all the groups donating data, including Kathleen Carroll, Sarah Hart, Josip Lovric and Paul Sims. T.M., S.J.H. and S.J.G. thank BBSRC for support via the 3GP programme (G17608), J.S. and S.J.H. via BBSRC grant (G17520), J.A.L. and K.W.L. for BBSRC (EGM17685) support and to J.A.S., S.J.H. and S.J.G. for BBSRC support on the ISPIDER project (BBSB17204). Funding to pay the Open Access publication charges for this article was provided by JISC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Desiere F., Deutsch E.W., Nesvizhskii A.I., Mallick P., King N.L., Eng J.K., Aderem A., Boyle R., Brunner E., Donohoe S., et al. Integration with the human genome of peptide sequences obtained by high-throughput mass spectrometry. Genome Biol. 2005;6:R9. doi: 10.1186/gb-2004-6-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Craig R., Cortens J.P., Beavis R.C. An open source system for analyzing, validating and storing protein identification data. J. Proteome Res. 2004;3:1234–1242. doi: 10.1021/pr049882h. [DOI] [PubMed] [Google Scholar]
- 3.Prince J.T., Carlson M.W., Wang R., Lu P., Marcotte E.M. The need for a public proteomics repository (commentary) Nat. Biotechnol. 2004;22:471–472. doi: 10.1038/nbt0404-471. [DOI] [PubMed] [Google Scholar]
- 4.Garwood K., McLaughlin T., Garwood C., Joens S., Morrison N., Taylor C.F., Carroll K., Evans C., Whetton A.D., Hart S., et al. PEDRo: a database for storing, searching and disseminating experimental proteomics data. BMC Genomics. 2004;5:68–79. doi: 10.1186/1471-2164-5-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martens L., Hermjakob H., Jones P., Taylor C., Gevaert K., Vandekerckhove J., Apweiler R. PRIDE: The PRoteomics IDEntifications database. Proteomics. 2005;5:3537–3545. doi: 10.1002/pmic.200401303. [DOI] [PubMed] [Google Scholar]
- 6.Orchard S., Kersey P., Hermjakob H., Apweiler R. The HUPO Proteomics Standards Initiative meeting: towards common standards for exchanging proteomics data. Comput. Funct. Genomics. 2003;4:16–19. doi: 10.1002/cfg.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor C.F., Paton N.W., Garwood K.L., Kirby P.D., Stead D.A., Yin Z., Deutsch E.W., Selway L., Walker J., Riba-Garcia I., et al. A systematic approach to modeling, capturing, and disseminating proteomics experimental data. Nat. Biotechnol. 2003;21:1–8. doi: 10.1038/nbt0303-247. [DOI] [PubMed] [Google Scholar]
- 8.Hermjakob H., Montecchi-Palazzi L., Bader G., Wojcik J., Salwinski L., Ceol A., Moore S., Orchard S., Sarkans U., von Mering C., et al. The HUPO PSI molecular interaction format—a community standard for the representation of protein interaction data. Nat. Biotechnol. 2004;22:177–183. doi: 10.1038/nbt926. [DOI] [PubMed] [Google Scholar]
- 9.Washburn M.P., Wolters D., Yates J.R. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 10.Paoletti A.C., Zybailov B., Washburn M.P. Principles and applications of multidimensional protein identification technology. Expert Rev. Proteomics. 2004;1:275–282. doi: 10.1586/14789450.1.3.275. [DOI] [PubMed] [Google Scholar]
- 11.Perkins D., Pappin D., Creasy D., Cottrell J. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 12.Eng J.K., McCormack A.L., Yates J.R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 13.Robertson C., Beavis R.C. Tandem: matching proteins with mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 14.Coling J., Masselot A., Giron M., Dessingy T., Magnin J. OLAV: towards high-throughput tandem mass spectrometry data identification. Proteomics. 2003;3:1454–1463. doi: 10.1002/pmic.200300485. [DOI] [PubMed] [Google Scholar]
- 15.Huang Y.Y., Wysocki V.H., Ji L., Triscari J.M., Smith R.D., Pasa-Tolic L., Anderson G.A., Lipton M.S. Data mining of 30 000 peptide dissociation spectra: How cleavage varies with charge. Abstracts Am. Chem. Soc. 2004;227 128-PHYS part 2. [Google Scholar]
- 16.Tabb D.L., Huang Y.Y., Wysocki V.H., Yates J.R. Influence of basic residue content on fragment ion peak intensities in low-energy—collision induced dissociation spectra of peptides. Anal. Chem. 2004;76:1243–1248. doi: 10.1021/ac0351163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breci L.A., Tabb D.L., Yates J.R., Wysocki V.H. Cleavage N-terminal to praline: Analysis of a database of peptide mass spectra. Anal. Chem. 2003;75:1963–1971. doi: 10.1021/ac026359i. [DOI] [PubMed] [Google Scholar]
- 18.Bern M., Goldberg D., McDonald W.H., Yates J.R., III Automatic quality assessment of peptide tandem mass spectra. Bioinformatics. 2004;20:i49–i54. doi: 10.1093/bioinformatics/bth947. [DOI] [PubMed] [Google Scholar]