Abstract
Maize is a staple crop of the grass family and also an excellent model for plant genetics. Owing to the large size and repetitiveness of its genome, we previously investigated two approaches to accelerate gene discovery and genome analysis in maize: methylation filtration and high C0t selection. These techniques allow the construction of gene-enriched genomic libraries by minimizing repeat sequences due to either their methylation status or their copy number, yielding a 7-fold enrichment in genic sequences relative to a random genomic library. Approximately 900 000 gene-enriched reads from maize were generated and clustered into Assembled Zea mays (AZM) sequences. Here we report the current AZM release, which consists of ∼298 Mb representing 243 807 sequence assemblies and singletons. In order to provide a repository of publicly available maize genomic sequences, we have created the TIGR Maize Database (http://maize.tigr.org). In this resource, we have assembled and annotated the AZMs and used available sequenced markers to anchor AZMs to maize chromosomes. We have constructed a maize repeat database and generated draft sequence assemblies of 287 maize bacterial artificial chromosome (BAC) clone sequences, which we annotated along with 172 additional publicly available BAC clones. All sequences, assemblies and annotations are available at the project website via web interfaces and FTP downloads.
INTRODUCTION
The availability of the highly accurate genome sequences of Arabidopsis and rice, along with the draft genome sequences that have been generated such as for the poplar genome (http://genome.jgi-psf.org/Poptr1/Poptr1.home.html), or in progress such as those of sorghum (http://www.jgi.doe.gov/sequencing/why/CSP2006/sorghum.html), a moss (http://www.jgi.doe.gov/sequencing/why/CSP2005/physcomitrella.html) and a spikemoss (http://www.jgi.doe.gov/sequencing/why/CSP2005/selaginella.html), has created the opportunity to perform comparative genomics studies among a broad variety of plants. All of the plant genomes previously selected for sequencing are relatively small. Large plant genomes pose a big challenge for genome sequencing owing to their high level of repetitiveness and frequent polyploidy. This sequencing limitation has so far excluded most economically important crops and key plant model systems from comprehensive genome analysis.
Maize (Zea mays), is the most important crop in the United States and one of the most important worldwide. It belongs to the cereal family, which includes other crop plants, such as rice, wheat, sorghum, barley and rye, and has been widely used as a classical model for genetics studies. Nevertheless, funding for sequencing its complete genome has become available only recently (http://www.nsf.gov/pubs/2004/nsf04614/nsf04614.htm). Its large size and frequent, highly conserved repetitive elements (1,2) are likely to generate assembly artifacts that are difficult to resolve, significantly increasing the efforts needed for completion. For such reasons, we explored alternative technologies for gene-targeted sequencing in maize as a prelude to generating the complete genome.
Two gene-enrichment technologies were tested in maize: methylation filtration (MF) and high-C0t selection (HC). MF takes advantage of the differential methylation of DNA in plants: genic sequences have been shown to be hypomethylated, whereas repetitive sequences are hypermethylated (3,4). An MF library consists of random, small insert genomic clones (5) constructed using an McrBC+ Escherichia coli host strain (6). The McrBC (modified cytosine restriction) system selectively propagates the hypomethylated sequences but not the methylated sequences. The HC approach is based on the differential reassociation kinetics of high- and low-copy-number DNA after denaturation. During the renaturation process, the repetitive sequences reanneal quickly while the low-copy-number sequences remain as single-stranded DNA for a longer time. This single-stranded, low-copy DNA can be separated from double-stranded high-copy-number sequences using hydroxyapatite chromatography. The second strand of the low-copy-number DNA is then synthesized in vitro so that the DNA can be cloned and sequenced (7,8). Both the MF and HC gene-enrichment techniques have proved successful in several plant species (7,9–12).
Using MF and HC, we have sequenced 895 731 gene-enriched sequence reads from maize (9). We have assembled these gene-enriched reads based on sequence similarity to generate the Assembled Zea mays (AZM) sequences aiming to reconstruct the genic regions. The maize MF and HC sequences have also been assembled and analyzed by other groups including the Maize Assembled Genomic Islands database (http://www.plantgenomics.iastate.edu/maize) and the Plant Genome Database (http://www.plantgdb.org/prj/GSSAssembly/zeamays). We have also investigated the feasibility of generating bacterial artificial chromosome (BAC) assemblies by sequencing, assembling and annotating 287 maize BAC clones to a medium (5- to 7.5-fold) sequence coverage. In addition, we have annotated 172 maize BAC assemblies downloaded from the public domain. BAC annotation was performed using a series of automated processes. We have performed a number of analyses on the AZMs and BAC assemblies, which include anchoring AZMs to the genetic map, annotation for the gene content of the AZMs and BAC assemblies and construction of a maize repeat database, including both known and novel repeats. All data are accessible from the TIGR Maize Database website, a centralized and comprehensive resource of maize genomic assemblies and annotation (http://maize.tigr.org).
RESULTS
Assembly of gene-enriched sequences
MF and HC libraries with an average insert size of ∼1.5 kb were constructed along with a whole-genome shotgun (unfiltered or UF) library as a control. The AZMs were built every 4 months. In the latest release (AZM 4.0), a total of 450 166 MF, 445 565 HC and 50 877 UF reads were generated after vector trimming and removal of low-quality sequences. Using comparable datasets of MF, HC and UF reads, the levels of gene-enrichment were ∼7-fold (13). The sequence reads include a combination of paired-end and single-end reads with an average edit length of 751 bp. For each data release, four individual assembly builds were carried out: (i) MF and HC reads combined, (ii) MF reads only, (iii) HC reads only and (iv) UF reads only. In each assembly build, the input reads were first repeat-masked to avoid assembly artifacts from highly similar repeat sequences. The repeat-masked sequences were clustered based on the identity of the overlapping ends (>40 bp) and mate-pair information. The sequence reads grouped into each cluster were assembled with the TIGR assembler to generate a consensus sequence using a modified pipeline designed to assemble expressed transcript sequences (14). Sequence reads that could not be clustered or assembled were collected as singletons. The output sequences, including assemblies and singletons, from each build were referred to as AZMs. Individual assemblies can be accessed through a web-based AZM report as shown in Figure 1.
Figure 1.
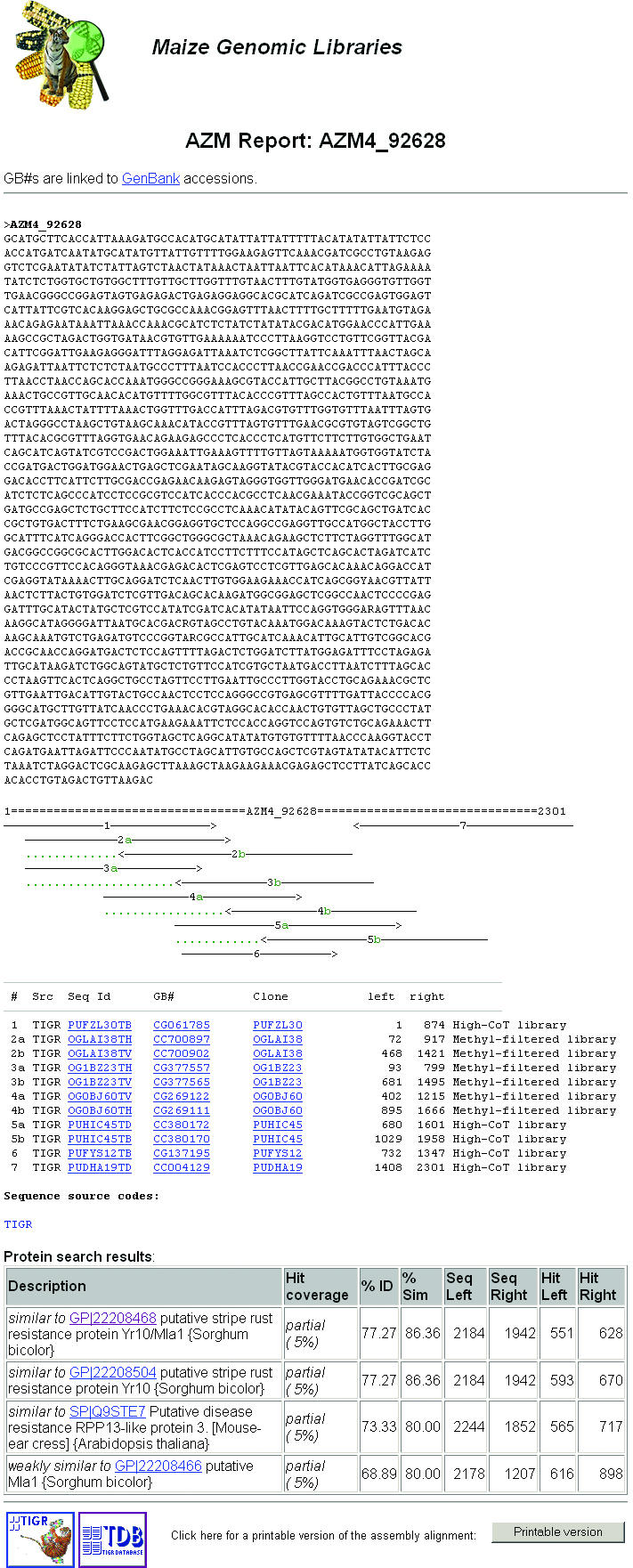
An AZM assembly report showing the assembled consensus sequence, a graphical layout of the component reads and related protein hits.
A total of 243 807 AZMs spanning 298 Mb were generated in the AZM 4.0 MF and HC combined build from 895 731 input reads (673 Mb). The average assembly length is 1.2 kb and the average number of component reads in each assembly is 5.5 (Table 1). Using a curated set of maize gene models, it was estimated that the coverage of the genic fraction of the genome by the AZMs is ∼2-fold (W.B. Barbazuk, unpublished data).
Table 1.
A summary for the maize genomic assembly release 4.0 (AZM4)
| Before assembly | |
| Gene-rich sequence reads | 895 731 |
| After assembly | |
| Number of assemblies generated | 144 999 |
| Average number of reads per assembly | 5.5 |
| Average length of assembly (bp) | 1624 |
| Maximum length of assembly (bp) | 16 340 |
| Singleton sequences | 98 808 |
| Assemblies and singletons (AZM4) | 243 807 |
| % Repeat-masked | 30 |
| % GC-content | 46 |
Anchoring the AZM sequences to chromosomes
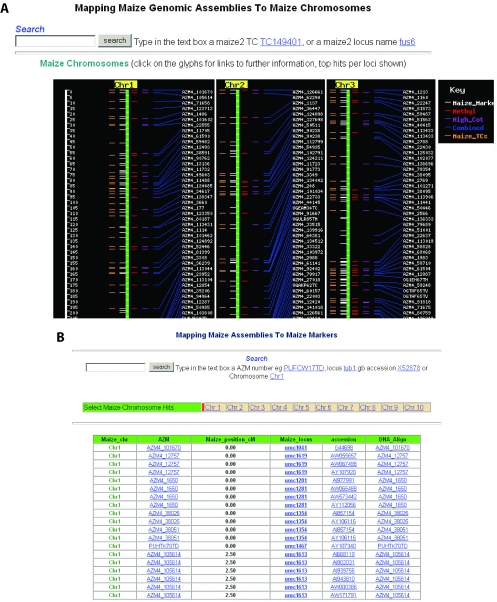
Unlike fingerprinted BAC assemblies, AZMs are not linked to the physical or genetic maps. To place the AZMs onto the maize chromosome arms, we have anchored the AZMs to the genetic map using the ‘IBM2 neighbors’ collection of sequence-based genetic markers (http://www.maizemap.org/resources.htm). An in silico alignment of AZMs to 2530 maize genetic loci was carried out using BLAT searches (15). Only alignments of at least 95% identity along at least 20% of the marker sequence were reported. A total of 1767 genetic loci (70% of all available markers) were aligned to the AZMs. A chromosome-based genome viewer was set up in our website to provide a global display of the distribution of AZMs and genetic markers along individual chromosomes (Figure 2A). The AZM mapping information can also be selected by chromosome, AZM accession, genetic locus or genetic marker accession (Figure 2B).
Figure 2.
In silico alignments of AZMs to maize chromosomes through sequenced genetic markers. (A) A section of a genome view displaying AZMs anchored to individual chromosomes through alignments to genetic markers. (B) Mapping AZM assemblies to genetic markers. In both displays, the AZM to marker mappings can be selected by chromosome, AZM accessions, genetic loci or genetic marker accessions.
Annotation of the AZM sequences
Since the AZMs are enriched in the genic regions of the maize genome, they represent a good resource for gene discovery and identification. To identify potential coding regions in the AZMs, we have set up an automated high-throughput gene annotation process based on alignments to protein and expressed transcript databases. Using BLAT, the AZMs were searched against a non-redundant protein database, and the TIGR plant gene index databases which consist of assembled cDNAs and expressed sequence tags referred to as the tentative consensus (TC) sequences from 22 plant species (http://www.tigr.org/tdb/tgi/plant.shtml). The criteria used for the protein and gene index searches were the best hit and the five best hits, respectively. Approximately 32 000 AZMs had a significant alignment to the protein database or TC database, thus, at least 13% of the AZMs contain potential coding sequences.
A web display has been set up for accessing the protein and TC alignments to the AZMs. The search results can be selected based on AZM accession, protein accession, TC accession and key word searches through the description of the protein or TC record.
Sequencing, assembly and annotation of maize BAC sequences
To gain further insight into the gene-rich regions of the maize genome, we have sequenced and generated phase I draft assemblies for a total of 287 gene-rich BACs. Sixty-five of the BAC clones correspond to a region in chromosome 1, which is duplicated in chromosome 9. Another 160 BAC clones were selected from gene-rich regions based on the presence of mapped cDNAs, and 62 additional BAC clones were selected by the maize community. The BAC clones were sequenced to ∼5- to 7.5-fold coverage. The reads were assembled using the sequence assembler Arachne (16). Owing to the repetitive content of the BAC sequences and the current sequence coverage, the BAC assemblies were created as unordered, unoriented phase I draft assemblies for submission to GenBank.
Annotation of the BAC assemblies was performed using a set of automated procedures, which process sequences and search results using a Sybase relational database (17). The annotation processes generate gene models from ab initio gene finders and also search the BAC sequences against nucleic acid and protein databases. First, the BAC sequences were processed by three gene finders, including FGENESH (18), GeneMark.hmm (19) and Genscan (20). The gene finders identify potential coding regions and generate predicted gene models. Second, the BAC sequences were searched against a non-redundant protein database and the TIGR plant gene index databases using the search tools from the AAT package, which performs optimal pairwise alignments to define the exon–intron boundaries in the query sequences using available protein and cDNA data (21). The current working gene models were generated using FGENESH outputs. The putative function of the gene product derived from a FGENESH prediction was assigned automatically based on the best match from the protein database search. All annotation data including predicted gene models, gene matches to the protein database and the plant gene index databases, and annotation reports of the working models are displayed through a suite of web pages from the open source MANATEE project (http://manatee.sourceforge.net).
Using the above approach, we have so far annotated 282 BAC clones (52 Mb) generated from our sequencing project and 167 maize BAC clones (25 Mb) collected from GenBank. Based on these two collections of 449 BAC sequences, our analysis showed that the average gene density in these BACs is 1 gene per 53 kb, the average gene size from the start to stop codons including introns is 3.2 kb, the average exon length is 236 bp, the average number of exons per gene is 5, and that the average length of the coding region is 1.2 kb.
The maize repeat database
Approximately 60–80% of the maize genome consists of retrotransposons and other repetitive sequences. We have created a maize repeat database to aid in the identification and classification of the maize repeat elements. The repeat database is composed of three components: the cereal plant repeat sequences from GenBank, the maize repeats identified by similarity search and the novel maize repeats identified using a de novo repeat finder. A collection of cereal repeats obtained from GenBank was downloaded from the TIGR plant repeat database [http://www.tigr.org/tdb/e2k1/plant.repeats; (22)], which also includes a set of manually annotated transposable elements from maize (P. SanMiguel, unpublished data). The cereal repeat collection contains four categories: transposable elements, centromeric and telomeric repeats, ribosomal repeats and unclassified repeats. This dataset was included as the first component of the maize repeat database and was used as a reference set to search against maize genomic sequences, including AZMs, BAC assemblies and BAC end sequences, using BLAST searches and a cutoff threshold of at least 80% identity over a string longer than 100 bp. Subregions of the maize genomic sequences that satisfied the criteria were included as the second component of the maize repeat database. RECON is a de novo repeat finder that groups together similar elements from the input sequences using single-linkage clustering (23). The maize genomic sequences, which showed no significant hits to the GenBank cereal repeats were used as the input dataset for de novo repeat discovery using RECON. Repeat families with five or more elements were collected as the third component for the maize repeat database. In the current release, the maize repeat database contains 28 249 sequences (22 Mb), of which 74% are transposable elements, 1% are centromeric and telomeric repeats, 3% are ribosomal repeats and 22% are unclassified repeats.
CONCLUSIONS
We have created the TIGR Maize Database with the goal to set up a central repository of maize genomic assemblies and annotation. We have generated 298 Mb of AZM assemblies and 52 Mb of BAC assemblies, representing most of the maize gene space and 14% of the whole genome. We are in the process of assembling 97 000 additional MF reads generated by Cold Spring Harbor Laboratory (11) into the AZMs. The AZM assemblies can be accessed through a web-based AZM report which displays the assembled consensus sequence, a layout graph of the component reads and related protein hits. For AZMs that have been anchored to the genetic map via genetic markers, they can be accessed through a genome viewer or selected through a web display. Predicted gene models on the BAC assemblies can be accessed through the MANATEE annotation report, which displays putative functions of the gene product, gene structure, protein domains and related protein sequences.
Both the component reads and AZM and BAC assemblies are available as ftp downloads. The AZM assemblies and the maize repeat database are available for BLAST searches using a blast server at the maize project website (http://tigrblast.tigr.org/tgi_maize/index.cgi).
While the maize genome sequencing effort is underway, BAC clones from a minimal tiling will be sequenced. The AZMs could be aligned to the sequenced BAC clones, thereby helping to complete some BAC sequences and also helping to anchor them to the physical map. The maize genomic assemblies we generated can facilitate large-scale genomic annotation and analyses, such as gene duplications, classification of paralogous genes and comparative genomics, to further our understanding of the maize genome.
Acknowledgments
The authors would like to acknowledge the participants in the Consortium for Maize Genomics project (Robert Citek, Muhammad Budiman, Andrew Nunberg, Joseph Bedell, Nathan Lakey and Karel Schubert; http://maize.danforthcenter.org) for their valuable contributions, and Robin Buell for helpful suggestions on the manuscript. This work was supported by National Science Foundation award DBI-0221536. Funding to pay the Open Access publication charges for this article was provided by The Institute for Genomic Research.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hake S., Walbot V. The genome of Zea mays, its organization and homology to related grasses. Chromosoma. 1980;79:251–270. [Google Scholar]
- 2.SanMiguel P., Bennetzen J.L. Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotransposons. Ann. Bot. 1998;82:37–44. [Google Scholar]
- 3.Rabinowicz P.D., Schutz K., Dedhia N., Yordan C., Parnell L.D., Stein L., McCombie W.R., Martienssen R.A. Differential methylation of genes and retrotransposons facilitates shotgun sequencing of the maize genome. Nature Genet. 1999;23:305–308. doi: 10.1038/15479. [DOI] [PubMed] [Google Scholar]
- 4.Bennetzen J.L., Schrick K., Springer P.S., Brown W.E., SanMiguel P. Active maize genes are unmodified and flanked by diverse classes of modified, highly repetitive DNA. Genome. 1994;37:565–576. doi: 10.1139/g94-081. [DOI] [PubMed] [Google Scholar]
- 5.Rabinowicz P.D. Constructing gene-enriched plant genomic libraries using methylation filtration technology. Methods Mol. Biol. 2003;236:21–36. doi: 10.1385/1-59259-413-1:21. [DOI] [PubMed] [Google Scholar]
- 6.Raleigh E.A., Wilson G. Escherichia coli K-12 restricts DNA containing 5-methylcytosine. Proc. Natl Acad. Sci. USA. 1986;83:9070–9074. doi: 10.1073/pnas.83.23.9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan Y., SanMiguel P.J., Bennetzen J.L. High-Cot sequence analysis of the maize genome [Erratum (2003) Plant J., 36, 430.] Plant J. 2003;34:249–255. doi: 10.1046/j.1365-313x.2003.01716.x. [DOI] [PubMed] [Google Scholar]
- 8.Barbazuk W.B., Bedell J.A., Rabinowicz P.D. Reduced representation sequencing: a success in maize and a promise for other plant genomes. Bioessays. 2005;27:839–848. doi: 10.1002/bies.20262. [DOI] [PubMed] [Google Scholar]
- 9.Whitelaw C.A., Barbazuk W.B., Pertea G., Chan A.P., Cheung F., Lee Y., Zheng L., van Heeringen S., Karamycheva S., Bennetzen J.L., et al. Enrichment of gene-coding sequences in maize by genome filtration. Science. 2003;302:2118–2120. doi: 10.1126/science.1090047. [DOI] [PubMed] [Google Scholar]
- 10.Rabinowicz P.D., Citek R., Budiman M.A., Nunberg A., Bedell J.A., Lakey N., O'Shaughnessy A.L., Nascimento L.U., McCombie W.R., Martienssen R.A. Differential methylation of genes and repeats in land plants. Genome Res. 2005;15:1431–40. doi: 10.1101/gr.4100405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palmer L.E., Rabinowicz P.D., O'Shaughnessy A.L., Balija V.S., Nascimento L.U., Dike S., de la Bastide M., Martienssen R.A., McCombie W.R. Maize genome sequencing by methylation filtration. Science. 2003;302:2115–2117. doi: 10.1126/science.1091265. [DOI] [PubMed] [Google Scholar]
- 12.Bedell J.A., Budiman M.A., Nunberg A., Citek R.W., Robbins D., Jones J., Flick E., Rholfing T., Fries J., Bradford K., et al. Sorghum genome sequencing by methylation filtration. PLoS Biol. 2005;3:e13. doi: 10.1371/journal.pbio.0030013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Springer N.M., Xu X., Barbazuk W.B. Utility of different gene enrichment approaches toward identifying and sequencing the maize gene space. Plant Physiol. 2004;136:3023–3033. doi: 10.1104/pp.104.043323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pertea G., Huang X., Liang F., Antonescu V., Sultana R., Karamycheva S., Lee Y., White J., Cheung F., Parvizi B., et al. TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics. 2003;19:651–652. doi: 10.1093/bioinformatics/btg034. [DOI] [PubMed] [Google Scholar]
- 15.Kent W.J. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Batzoglou S., Jaffe D.B., Stanley K., Butler J., Gnerre S., Mauceli E., Berger B., Mesirov J.P., Lander E.S. ARACHNE: a whole-genome shotgun assembler. Genome Res. 2002;12:177–189. doi: 10.1101/gr.208902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haas B.J., Wortman J.R., Ronning C.M., Hannick L.I., Smith R.K., Jr, Maiti R., Chan A.P., Yu C., Farzad M., Wu D., et al. Complete reannotation of the Arabidopsis genome: methods, tools, protocols and the final release. BMC Biol. 2005;3:7. doi: 10.1186/1741-7007-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salamov A.A., Solovyev V.V. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 2000;10:516–522. doi: 10.1101/gr.10.4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lukashin A.V., Borodovsky M. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burge C., Karlin S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 21.Huang X., Adams M.D., Zhou H., Kerlavage A.R. A tool for analyzing and annotating genomic sequences. Genomics. 1997;46:37–45. doi: 10.1006/geno.1997.4984. [DOI] [PubMed] [Google Scholar]
- 22.Ouyang S., Buell C.R. The TIGR Plant Repeat Databases: a collective resource for the identification of repetitive sequences in plants. Nucleic Acids Res. 2004;32:D360–D363. doi: 10.1093/nar/gkh099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bao Z., Eddy S.R. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 2002;12:1269–1276. doi: 10.1101/gr.88502. [DOI] [PMC free article] [PubMed] [Google Scholar]