Abstract
Guanine-rich nucleic acids are known to form highly stable G-quadruplex structures, also known as G-quartets. Recently, there has been a tremendous amount of interest in studying G-quadruplexes owing to the realization of their biological importance. G-rich sequences (GRSs) capable of forming G-quadruplexes are found in the vicinity of polyadenylation regions and are involved in regulating 3′ end processing of mammalian pre-mRNAs. G-rich motifs are also known to play an important role in alternative, tissue-specific splicing by interacting with hnRNP H protein subfamily. Whether quadruplex structure directly plays a role in regulating RNA processing events requires further investigation. To date there has not been a comprehensive effort to study G-quadruplexes near RNA processing sites. We have applied a computational approach to map putative Quadruplex forming GRSs within the transcribed regions of a large number of alternatively processed human and mouse gene sequences that were obtained as fully annotated entries from GenBank and RefSeq. We have used the computed data to build the GRSDB database that provides a unique avenue for studying G-quadruplexes in the context of RNA processing sites. GRSDB website offers visual comparison of G-quadruplex distribution patterns among all the alternative RNA products of a gene with the help of dynamic graphics. At present, GRSDB contains data from 1310 human and mouse genes, of which 1188 are alternatively processed. It has a total of 379 223 predicted G-quadruplexes, of which 54 252 are near RNA processing sites. GRSDB is a good resource for researchers interested in investigating the functional relevance of G-quadruplexes, especially in the context of alternative RNA processing. It can be accessed at http://bioinformatics.ramapo.edu/grsdb/.
INTRODUCTION
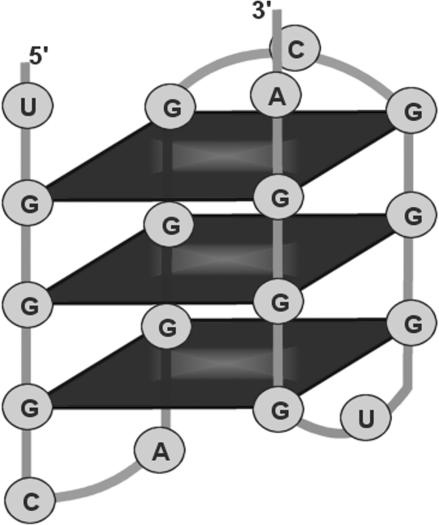
Guanine-rich nucleic acids are known to form higher order structures. Their ability to form highly stable quadruplex structures was discovered more than four decades ago (1). The G-quadruplex structure, also known as a G-quartet, is composed of stacked G-tetrads, which are square co-planar arrays of four guanine bases each. Cyclic Hoogsteen hydrogen bonding between the four guanines within each tetrad renders a high level of stability to the quadruplex (Figure 1). Although structures with three or more G-tetrads are considered to be more stable, many nucleotide sequences are known to form quadruplexes with two G-tetrads (2,3). G-quadruplexes may be formed by repeated folding of a single nucleic acid molecule (unimolecular G-quadruplex) or by interaction of two or four strands. The former is more likely to be encountered in physiological conditions (4,5). (The present work focuses only on the unimolecular quadruplexes.) Formation of G-quadruplexes in vivo is facilitated by proteins (6). Some proteins are also implicated in resolving the G-quadruplex structure (7,8).
Figure 1.
QGRS: 5′-GGGCAGGGCAGGUGGGA-3′. Predicted intramolecular G-quadruplex formed by a GRS.
G-quadruplex sequence motifs have been reported in telomeric, promoter and other regions of mammalian genomes. Formation of a G-quadruplex in the promoter region has been associated with transcription regulation of the c-myc oncogene and is being considered as a potential target for therapeutic purposes (9,10). Owing to the realization of their biological importance, recently, there has been a tremendous amount of interest in studying G-quadruplexes. This is evident from a surge in the published literature. [for reviews see (8,11)].
Although initially most of the studies focused on G-quadruplexes in the DNA, lately there have been many efforts to study G-quadruplex forming RNA (12–16). In fact, G- rich sequences capable of forming G-quadruplexes in the RNA have been implicated in a variety of important biological activities, such as mRNA turnover (6), Fragile X Mental Retardation Protein (FMRP) binding (14), translation initiation (15) as well as repression (16).
We have previously shown that a conserved auxiliary G-rich sequence (GRS) found near the polyadenylation regions can mediate efficient 3′ end processing of mammalian pre-mRNAs (17,18) by interacting with DSEF1/hnRNP H/H′ protein (19). However, hnRNP F has been shown to be a negative regulator of 3′ end processing (20). Regulated polyadenylation is an important component of differential gene expression. More than 50% of human and 32% of mouse genes are known to have alternative polyadenylation (21). An interplay among GRS-binding proteins, hnRNP H/H′ and F, helps in regulating alternative polyadenylation of immunoglobulin pre-mRNA (20) which, combined with alternative splicing, plays an important role in mouse B lymphocyte development (22).
In addition to differential gene expression, alternative splicing affects disease processes (23) and is a major source of protein diversity. More than two-thirds of human genes are thought to undergo alternative splicing (24). Members of the hnRNP H protein subfamily, that bind G-rich motifs, are known to be involved in alternative, tissue-specific, regulated splicing events (25–27). GRS motifs that are present near splice sites act as splicing regulators by interacting with hnRNP H (28). For example, binding of hnRNP H and F to G-rich tracts near 5′ splice site favors production of alternative pro-apoptotic Bcl-xs product (29). The regulatory G-rich motifs may be capable of forming quadruplex structures. Whether quadruplex structure directly plays a role in regulating RNA processing events requires investigation.
The majority of the mammalian poly(A) region GRS sequences that we had surveyed in our previous studies (18,19) are capable of forming unimolecular G-quadruplexes. Our preliminary analysis of ∼100 alternatively processed human transcripts has also revealed the presence of quadruplex forming sequences near alternative splice sites (30). However, a more detailed investigation into the distribution of G-quadruplex sequences near RNA processing sites requires a systematic large-scale analysis of mammalian genes. Although, there have been two recent surveys of quadruplexes in the human genome (31,32), to date there has not been a comprehensive effort to study G-quadruplexes near RNA processing sites.
We have used a computational approach (30) to map putative G-quadruplex forming sequences within the transcribed regions of a large number of alternatively processed human and mouse genes. The fully annotated genomic nucleotide sequences are obtained from NCBI-based GenBank and RefSeq for computational analysis. Based on our analysis of alternatively spliced and alternatively polyadenylated human and mouse genes, we have built the GRSDB database. GRSDB provides a unique avenue for studying G-quadruplex forming sequences in the context of RNA processing sites. In addition to providing data on composition and locations of mapped quadruplexes relative to the processing sites in the pre-mRNA sequence, GRSDB offers simultaneous visual comparison of G-quadruplex distribution patterns among all the alternative RNA products of individual genes with the help of dynamically generated graphics.
Researchers interested in investigating the functional relevance of G-quadruplex structure, in particular its role in regulating the gene expression by alternative processing, will find GRSDB to be of great value. It allows a comprehensive large-scale analysis as well as detailed studies in individual genes. GRSDB is also a good resource for performing large-scale analysis of G-quadruplex sequence composition, including study of loops, in the transcribed regions.
METHODS
Quadruplex forming GRS
The basic unit of study in GRSDB is the putative G-quadruplex that we have called QGRS (Quadruplex forming GRS). These sequences follow the motif GxNaGxNbGxNcGx. Here Gx refers to the group of guanines (which we will refer to as a G-group) that form a complex of x stacked G-tetrads. In the individual gene entries stored in GRSDB, x is generally 2, 3 or 4. The intervening arbitrary bases, Na, Nb and Nc, are called gaps or loops.
Two sequences are said to be overlapping if their positions in the nucleotide sequence do overlap. The default action of GRSDB is to display non-overlapping sequences, but the user can display all QGRS.
Structure of GRSDB
GRSDB is a relational database built using MySQL. The GRSDB website can be accessed at http://bioinformatics.ramapo.edu/grsdb/. This database primarily stores information about putative G-quadruplex sequences (QGRS) for genes that are alternatively processed (either alternatively spliced or alternatively polyadenylated). GRSDB is structured to facilitate queries about alternatively processed genes and to display information on the G-quadruplex sequences contained in the transcribed regions of the gene and their locations relative to RNA processing sites. Table 1 shows the types of objects found in the database.
Table 1.
Structure of GRSDB
| Data type | Comments |
|---|---|
| Gene | Includes gene name, gene location, whether or not gene is alternatively spliced or alternatively polyadenylated |
| Product | Includes RNA product name, and numbers of introns, exons and poly(A) signals |
| Intron/exon map | Maps start and stop positions of introns and exons |
| Poly(A) map | Maps start and stop position of poly(A) signal |
| QGRS | Includes actual sequences, their position and G-scores |
GRSDB is populated using an auxiliary program, QGRS-Mapper, that is based on previously published methods (30) and was developed using BioPerl. Once appropriate genes have been identified, this program links to GenBank or RefSeq, downloads the corresponding genomic nucleotide sequence entry of the gene, and parses the entry for product, intron, exon, poly(A) and related information. The program then processes the nucleotide sequence to find all QGRS and map their location within the gene and their distance from relevant RNA processing sites.
QGRS scoring
A scoring method is applied to each QGRS. The computed score, called a G-score (30), is formulated to reward sequences with smaller, more even gaps between the G-groups in addition to larger G-group size, thereby favoring the arrangement that is more likely to form a unimolecular complex. This choice of scoring system is in agreement with the existing literature on loop structures in G-quadruplexes (31–35). In particular, the data gathered in this research points to loop sizes tending to be small and preferentially equal or nearly equal.
Interfaces
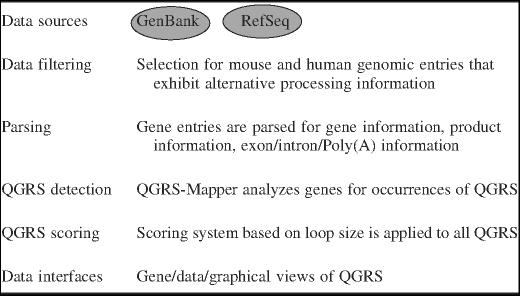
The data flow for GRSDB is summarized in Table 2. After the gene information is downloaded from NCBI, parsed, processed for QGRS, and scored, it is then uploaded into GRSDB. At this point the database is ready for user queries. There are three different interfaces provided for viewing database contents: the gene view, the data view and the graphical view.
Table 2.
Data flow of GRSDB
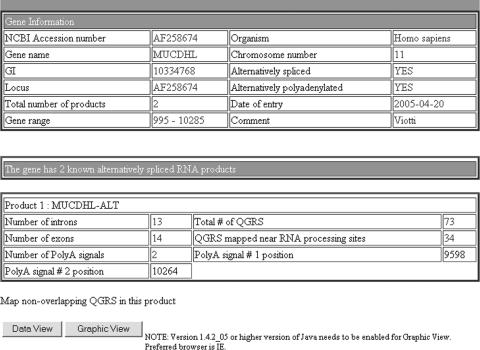
Database users are given a variety of options in formulating a query, including searching for genes that are alternatively spliced or alternatively polyadenylated. Once a query has been entered, a table is displayed of all genes satisfying the query. Information for individual genes is displayed in a table as shown in Figure 2 for the particular gene MUCDHL. This is what we call the gene view. One can see that MUCDHL is both alternatively spliced and alternatively polyadenylated.
Figure 2.
MUCDHL—Gene View. The first table in this view gives basic information about the gene being analyzed, including the alternatively spliced or polydenylated status of the gene. The second table provides information about the first alternatively spliced RNA product of the gene, including numbers of exons/introns/poly(A) signals. Also displayed are the total number of QGRS in the product and near RNA processing sites (i.e. within 120 nt of a site). Located below each product table are buttons allowing the user to view the QGRS information for that product in two ways; the data view and the graphic view, shown in Figures 3 and 4. Not shown are the second product table and the button placed at the bottom of the page allowing the user to analyze all RNA products simultaneously. To view the entire screen shot, see the Supplementary Data.
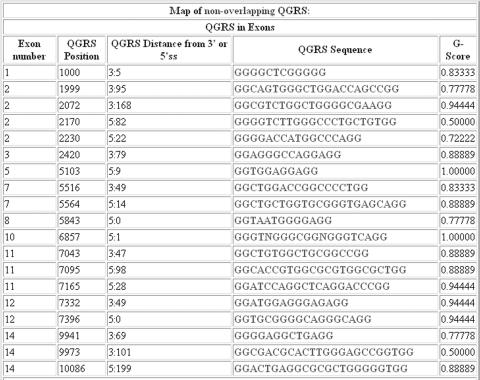
At this point the user can choose to analyze one of the products or all products simultaneously. There are two types of analysis possible, the data view or graphical view. Figure 3 represents the data view analysis for Product 1 of MUCDHL, showing all non-overlapping QGRS. The table shows the location of each QGRS, its distance from the nearest splice site, and its G-score.
Figure 3.
MUCDHL—Data View. A table of the mapping data for the non-overlapping QGRS in the product (refer to the text for explanation of non-overlapping) is shown. The table provides the location of QGRS in exons, the distance from 3′ and 5′ splice sites, the actual sequence, and its G-score (refer to the text for explanation). Not shown are the tables for QGRS mapping data in introns and in poly(A) regions. To view the entire screen shot, see Supplementary Data.
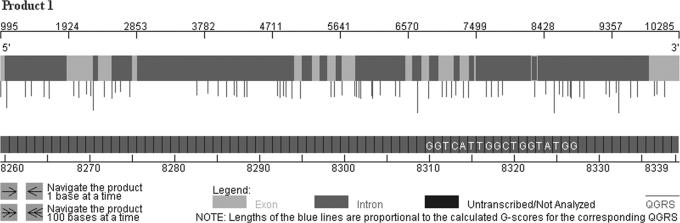
Alternatively, the user can select the graphical view of any product (or again, all products together), which is shown in Figure 4. A visual model of the product is displayed, showing the location of exons, introns and untranscribed regions. Further, the location of each QGRS is indicated by a vertical line. The length of the line is proportional to the G-score for the sequence.
Figure 4.
MUCDHL gene—Graphic View. A visual representation for RNA Product 1 of this gene. The upper graph shows the location of exons/introns in the product, along with a scale to locate their positions. The QGRS in the product are indicated by the vertical bars, whose length is proportional to the G-scores of the QGRS. The lower graph represents a zoom-in of the RNA product 1 displaying the QGRS at position 8310. The arrows at the bottom left are used to navigate the RNA product with the interactive-zoom tool. It is possible to visually compare both the alternatively spliced products, for example, to identify differential association of QGRS with alternative sites. See Supplementary Data for product comparison.
CONCLUSIONS
GRSDB provides curated information on composition and distribution of putative QGRSs in the transcribed regions of alternatively processed human and mouse genes. The data are based on the analysis of fully annotated GenBank/RefSeq human and mouse genomic nucleotide entries that exhibit alternative processing information. Although the NCBI databases contain a large number of mRNA sequence records, at present the number of genomic entries that will provide information needed for our studies is limited.
At present, our database contains information obtained from 1310 human and mouse genes, of which 1188 are alternatively processed. A total of 30 584 introns and 33 816 exons were analyzed, containing a total of 3231 RNA products. These products taken together contain a total of 379 223 putative G-quadruplexes, of which 54 252 are near RNA processing sites [within 120 nt of a splice site or a poly(A) signal]. Note that while GRSDB currently contains data only on human and mouse genes, our computational tools and the database are designed to include other organisms as well.
GRSDB is continuously being updated with new data entries. The database is structured to facilitate a wide variety of queries and to map G-quadruplex sequences relative to the RNA processing sites in both data and graphic formats. The user friendly interface allows comparisons of all the alternative RNA products of individual genes on the same screen. GRSDB is a good resource for researchers interested in investigating the functional relevance of G-quadruplexes, especially in the context of alternative RNA processing.
We are using the database to conduct detailed bioinformatics studies on the distribution patterns of QGRS near RNA processing sites. In particular, we are investigating whether there is a correlation between the distribution pattern of QGRS and alternative processing. Our group is also studying the loop composition of these sequences.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Supplementary Material
Acknowledgments
We thank Fatima Iqbal and Rachel Howitt for assistance with data uploading. We are grateful to Marcelo Halpern for technical assistance with the database server. The authors wish to thank Jeffrey Wilusz of Colorado State University for critically reviewing the database website and providing helpful suggestions. This project was funded in part by grants from the Provost Office and TLTR (The Teaching, Learning and Technology Roundtable) of Ramapo College of New Jersey. Funding to pay the Open Access publication charges for this article was provided by the Divisions of Student Affairs and Academic Affairs of Ramapo College of New Jersey.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gellert M., Lipsett M.N., Davies D.R. Helix formation by guanylic acid. Proc. Natl Acad. Sci. USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zharudnaya M.I., Kolomiets I.M., Potyahaylo A.L., Hovorun D.M. Downstream elements of mammalian pre-mRNA polyadenylation signals: primary, secondary and higher-order structures. Nucleic Acids Res. 2003;31:1375–1386. doi: 10.1093/nar/gkg241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kankia B.I., Barrany G., Musier-Forsyth K. Unfolding of DNA quadruplexes induced by HIV-1 nucleocapsid protein. Nucleic Acids Res. 2005;33:4395–4403. doi: 10.1093/nar/gki741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schaffitzel C., Berer I., Postberg J., Hanes J., Lipps H.J., Plückthun A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl Acad. Sci. USA. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halder K., Chowdhury S. Kinetic resolution of bimolecular hybridization versus intramolecular folding in nucleic acids by surface plasmon resonance: application to G-quadruplex/duplex competition in human c-myc promoter. Nucleic Acids Res. 2005;33:4466–4474. doi: 10.1093/nar/gki750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paeschke K., Simonsson T., Postberg J., Rhodes D., Lipps H.J. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nature Struct. Mol. Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 7.Zaug A.J., Podell E.R., Cech T.R. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc. Natl Acad. Sci. USA. 2005;102:10864–10869. doi: 10.1073/pnas.0504744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simonsson T. G-quadruplex DNA structures—variations on a theme. Biol. Chem. 2001;382:621–628. doi: 10.1515/BC.2001.073. [DOI] [PubMed] [Google Scholar]
- 9.Simonsson T., Pecinka P., Kubista M. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998;26:1167–1172. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phan A.T., Modi Y.S., Patel D.J. Propeller-type parallel-stranded G-quadruplexes in the human c-myc promoter. J. Am. Chem. Soc. 2004;126:8710–8716. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis J.T. G-quartets 40 years later: from 5′-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. Engl. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 12.Liu H., Matsugami A., Katahira M., Uesugi S. A dimeric RNA quadruplex architecture comprised of two G:G(:A):G:G(:A) hexads, G:G:G:G tetrads and UUUU loops. J. Mol. Biol. 2002;322:955–970. doi: 10.1016/s0022-2836(02)00876-8. [DOI] [PubMed] [Google Scholar]
- 13.Bashkirov V.I., Scherthan H., Solinger J.A., Buerstedde J.M., Heyer W.D. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J. Cell. Biol. 1997;136:761–773. doi: 10.1083/jcb.136.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darnell J.C., Jensen K.B., Jin P., Brown V., Warren S.T., Darnell R.B. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 15.Bonnal S., Schaeffer C., Creancier L., Clamens S., Moine H., Prats A.-C., Vagner S. A single internal ribosome entry site containing a G quartet RNA structure drives fibroblast growth factor 2 gene expression at four alternative translation initiation codons. J. Biol. Chem. 2003;278:39330–39336. doi: 10.1074/jbc.M305580200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oliver A., Bogdarina I., Schroeder E., Taylor I.A., Kneale G.G. Preferential binding of fd gene5 protein to tetraplex nucleic acid structures. J. Mol. Biol. 2000;301:575–584. doi: 10.1006/jmbi.2000.3991. [DOI] [PubMed] [Google Scholar]
- 17.Bagga P.S., Ford L.P., Chen F., Wilusz J. The G-rich auxiliary downstream element has distinct sequence and position requirements and mediates efficient 3′ end pre-mRNA processing through a trans-acting factor. Nucleic Acids Res. 1995;23:1625–1631. doi: 10.1093/nar/23.9.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bagga P.S., Arhin G.K., Wilusz J. DSEF-1 is a member of the hnRNP H family of RNA binding proteins and stimulates pre-mRNA cleavage and polyadenylation. Nucleic Acids Res. 1998;26:5343–5350. doi: 10.1093/nar/26.23.5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arhin G.K., Boots M., Bagga P.S., Milcarek C., Wilusz J. Downstream sequence elements with different affinities for the hnRNP H/H′ protein influence the processing efficiency of mammalian polyadenylation signals. Nucleic Acids Res. 2002;30:1842–1850. doi: 10.1093/nar/30.8.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Veraldi K.L., Arhin G.K., Martincic K., Chung-Ganster L.-H., Wilusz J., Milcarek C. hnRNP F influences binding of a 64 kDa subunit of cleavage stimulation factor to mRNA precursors in mouse B cells. Mol. Cell. Biol. 2001;21:1228–1238. doi: 10.1128/MCB.21.4.1228-1238.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian B., Hu J., Lutz C. A large-scale analysis pf mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruce S.R., Dingle R.W.C., Peterson M.L. B-cell and plasma-cell splicing differences: a potential role in regulated immunoglobulin RNA processing. RNA. 2003;9:1264–1273. doi: 10.1261/rna.5820103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faustino N.A., Cooper T.A. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 24.Johnson J.M., Castle J., Garrett-Engele P., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 25.Min H., Chan R.C., Black D.L. The generally expressed hnRNP F is involved in a neural-specific pre-mRNA splicing event. Genes Dev. 1995;9:2659–2671. doi: 10.1101/gad.9.21.2659. [DOI] [PubMed] [Google Scholar]
- 26.Chou M.-Y., Rooke N., Turck C.W., Black D.L. hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol. Cell. Biol. 1999;19:69–77. doi: 10.1128/mcb.19.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caputi M., Zahler A.M. SR proteins and hnRNP H regulate the splicing of the HIV-1 tev-specific exon 6-D. EMBO J. 2002;21:845–855. doi: 10.1093/emboj/21.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han K., Yeo G., An P., Burge C., Grabowski P. A combinatorial code for splicing silencing: UAGG and GGG motifs. PLoS Biol. 2005;3:0843–0860. doi: 10.1371/journal.pbio.0030158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garnea D., Revil T., Fisette J.-F., Chabot B. Heterogeneous nuclear ribonucleoprotein F/H proteins modulate the alternative splicing of the apoptotic mediator Bcl-x. J. Biol. Chem. 2005;24:22641–22650. doi: 10.1074/jbc.M501070200. [DOI] [PubMed] [Google Scholar]
- 30.D'Antonio L., Bagga P.S. Computational methods for predicting intramolecular G-quadruplexes in nucleotide sequences. Proceedings of the IEEE Computational Systems Bioinformatics Conference (CSB'04); August 16–19; Stanford, CA. 2004. pp. 561–562. [Google Scholar]
- 31.Huppert J.L., Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Todd A.K., Johnston M., Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Risitano A., Fox K. Influence of loop size on the stability of intramolecular DNA quadruplexes. Nucleic Acids Res. 2004;32:2598–2606. doi: 10.1093/nar/gkh598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Črnugelj M., Šket P., Plavec J. Small change in a G-rich sequence, a dramatic change in topology: new dimeric G-quadruplex folding motif with unique loop orientations. J. Am. Chem. Soc. 2003;125:7866–7871. doi: 10.1021/ja0348694. [DOI] [PubMed] [Google Scholar]
- 35.Hazel P., Huppert J., Balasubramanian S., Neidle S. Loop-length-dependent folding of G-quadruplexes. J. Am. Chem. Soc. 2004;126:16405–16415. doi: 10.1021/ja045154j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.