Abstract
MfunGD (http://mips.gsf.de/genre/proj/mfungd/) provides a resource for annotated mouse proteins and their occurrence in protein networks. Manual annotation concentrates on proteins which are found to interact physically with other proteins. Accordingly, manually curated information from a protein–protein interaction database (MPPI) and a database of mammalian protein complexes is interconnected with MfunGD. Protein function annotation is performed using the Functional Catalogue (FunCat) annotation scheme which is widely used for the analysis of protein networks. The dataset is also supplemented with information about the literature that was used in the annotation process as well as links to the SIMAP Fasta database, the Pedant protein analysis system and cross-references to external resources. Proteins that so far were not manually inspected are annotated automatically by a graphical probabilistic model and/or superparamagnetic clustering. The database is continuously expanding to include the rapidly growing amount of functional information about gene products from mouse. MfunGD is implemented in GenRE, a J2EE-based component-oriented multi-tier architecture following the separation of concern principle.
INTRODUCTION
The Mouse functional Genome Database (MfunGD) aims to provide a high-quality information resource for the research community incorporating manual annotation of gene products, in particular with respect to the cellular function in the context of their interaction. Mus musculus is one of the most thoroughly studied mammalian model organisms. For thousands of mouse proteins, functional properties have been predicted or experimentally investigated and part of this information is stored in databases like UniProt and MGI (1,2). Due to its exceptional importance as a model organism, the genome sequence of mouse was the second mammalian genome that has been sequenced (3). Mouse is genetically tractable and large collections of mouse mutants exist which yield invaluable insights into the function of mammalian genes (4). Unfortunately, the detection of the genotype of mouse mutants that are obtained by treatment with chemical compounds such as ENU is extremely time-consuming and labour intensive. In order to understand the function of mammalian genes in context and to identify the causes of complex diseases having a genetic background in mammals, bridging the gap between genotype and phenotype will be one of the most important and challenging tasks for the future.
To achieve this goal, the knowledge about the function of isolated proteins needs to be extended to their functional context in the cellular environment. Such an endeavour requires the integration of different sources of information like protein–protein interactions, genetic interactions as well as co-expression data. The integration of these data results in distinct but interconnected networks of proteins responsible for defined functional tasks in cells, so-called functional modules (5). However, so far no reliable data set of functional modules for a mammalian organism exists. As an important step towards this goal, we combine computational methods with manual annotation to the mouse proteome with strong emphasis on the cellular context.
SYSTEM ARCHITECTURE
A comprehensive genome resource must not only be capable to store and display information on gene products but also needs to support manual and semi-automatic annotation. To fulfil these requirements, we implemented MfunGD within the MIPS Genome Research Environment (GenRE). This allows seamless integration of database management systems as well as various components required for a flexible annotation pipeline. GenRE is a J2EE-based component-oriented multi-tier architecture hiding the complexity of the procedures from the user.
For example, the manual annotation process requires not only the access to various data sources, but also its support needs the integration of different algorithms such as clustering of protein family members in a structured way. These databases and applications are typically distributed across physically separated computing resources. We developed an integration tier capable to level the differences between the underlying resources by conversion into so-called data access objects (DAOs). The main advantage of the DAO design pattern within MfunGD is the uniform access of any resource on a JAVA object level. For databases, we used DAOs based on HIBERNATE a high-performance object/relational persistence and query service, whereas for applications the DAOs were explicitly designed. On top of the integration tier, we implemented a so-called business tier based on Enterprise Java Beans (EJBs). EJBs are the core components for any kind of application (business) logic related to complex information processing within the annotation pipeline and advanced queries. For further unification of information, the EJB components accept and deliver results in XML format. The XML format is not only used in the completely separated web-tier for rendering HTML output with XSL style sheets (see Figure 1), but also for the communication with rich-clients for manual annotation hence reducing the time-consuming multiple invocation of EJB methods by the transmission of only one comprehensive XML document.
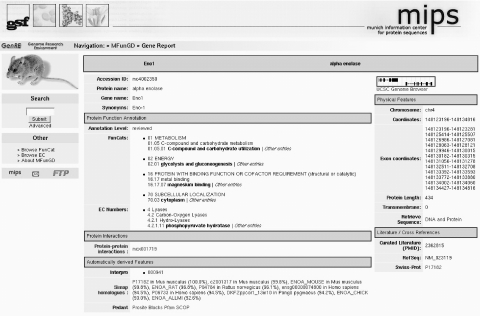
Figure 1.
Screenshot of the MfunGD entry for the enzyme alpha enolase.
A further advantage of the component-oriented approach is the extension of the system with minimal effort. For example, MfunGD has been extended with a configurable advanced query interface component used also by different resources within MIPS. This interface provides the possibility to query the database using logical combinations of terms in a similar way to the Entrez service. Customizable full-text searches across the database are possible without any knowledge of the underlying data structure. Querying indexed information is done by simple expressions allowing wildcards and the combination with logical operators. An example query for searching all mitochondrial proteins (functional category 70.16) with >1000 amino acids is simply performed by the following expression: ‘70.16*[FCC] >1000[PIL]’ instead of a complicated native database query involving several table joins.
DATA CONTENT
An inherent problem in the analysis of mammalian genomes is the lack of a complete and stable set of all exant full-length transcripts. New transcripts and splice variants are published regularly requiring continuous updating of datasets such as RefSeq. Compared to the September 2004 RefSeq mouse release, the dataset of March 2005 contained 1331 new entries, 542 with changes in the transcript sequence whereas 196 that were removed. The MfunGD will also allow updates of genome assembly, transcript data and gene models.
The basis of the MfunGD dataset is a complement of gene products that was obtained by Softberry Inc., which used the FGENESH++C software as gene predictor. This procedure resulted in 42 049 gene products for mouse. Those include 13 259 gene products which were identical or highly similar to RefSeq cDNAs, 18 330 gene models with significant similarity to a non-redundant (NR) database and 10 460 gene models without significant similarity (>90% identity) to the NR database.
Transcripts of this dataset are currently mapped to the curated RefSeq dataset from the mouse strain C57BL/6J by a mapping procedure based on the Blat software (6). Known transcripts from external resources which are not yet present in our dataset are added. The Softberry gene models were based on the Build 30 assembly of the mouse genome (mm3, Feb. 2003). These models were mapped to the May 2004 mouse genome assembly. The UCSC Genome Browser (7) allows visualization of the MfunGD transcripts, gene models and RefSeq data.
ANNOTATION
MfunGD is a resource for manually and automatically annotated proteins and genes from mouse. General protein and gene features like InterPro domains, 3D structure and physical properties are precalculated by the Pedant system (8). InterPro domains and predicted transmembrane domains are shown on the MfunGD web page, other features can be accessed via hyperlinks. Results from Fasta sequence similarity searches against >3 000 000 protein sequences can be retrieved from the SIMAP database (9). Attributes like gene names, protein names and synonyms are retrieved from public resources like UniProt (1), MGD (2) or RefSeq (10). In addition, MfunGD contains information about literature that was used for manual annotation as well as protein ID, FunCat annotation, comments, update information and cross-references to RefSeq, UniProt and MGD.
Manual annotation
A central part of the annotation process is the assignment of functional categories to protein entries. At MIPS, the Functional Catalogue (FunCat) is used for function annotation. This annotation scheme has been applied to the manual annotation of several model organisms (11). FunCat is a hierarchically structured, organism-independent, flexible, controlled and scalable (structured) classification system enabling the functional description of proteins from any organism (11). The capabilities of the FunCat are not only limited to the functional annotation of genomes, but also provide a powerful tool in order to analyse genome- and proteome-wide data generated by large-scale transcriptome/proteome experiments (12–14) as well as the computational analysis of functional networks (15,16). The versatile application makes FunCat a powerful and intensively used tool for integration of protein function data from different sources and thus fulfils the needs of bioinformatics approaches in systems biology.
The assignment of functional categories in the manual annotation process depends primarily on the experimental evidence given in literature. Here, the hierarchical structure of FunCat allows adjusting the specificity of the level of the functional category to the information content of the experiments. In addition to information from literature, data from other sources like InterPro (17) and FunCatDB (11) as well as external resources like SwissProt (1) GenBank (18) and MGI (2) are used in order to obtain a comprehensive overview of the cellular function of respective proteins. Evaluation of the information and the resulting assignment of functional categories lie in the responsibility of trained annotators. So far, ∼4000 mouse proteins have been manually curated. Experimentally investigated proteins are on average associated with 4.6 FunCat categories. Information that exceeds the specificity of FunCat categories is stored as E.C. numbers or is presented in comment fields.
FunCat annotation using hRMN/gSPC
The high number of gene products in mammals requires supporting manual annotation by automated prediction of protein functions. The relation between sequence similarity and functional conservation has been well established for protein domains and complete proteins. Since transfer of functional annotation given high sequence conservation is reliable, MfunGD data sources for human, mouse and rat as well as other mammalian proteins annotated in SwissProt were used. For the human genome, manual annotation was obtained from Biomax Informatics AG. Using conservative thresholds, FunCat information has been transferred to 13 193 mouse proteins. If any of these protein entries is subsequently subjected to manual inspection, information of the automated process is supplemented with literature information and modified, if necessary. Available in-house manual annotation was complemented by an automated mapping of available manual GO-annotations to FunCat categories. Based on sequence similarity data and InterPro domains, further sequence-associated information was compiled. Automated annotation support is provided by two different systems, gSPC and hRMN. gSPC stands for ‘global SuperParamagnetic Clustering’ (19) in which sequences are clustered in a Monte Carlo process according to a sequence similarity score. Functional annotation is then transferred within a cluster from known to unclassified proteins by a consensus process among the known proteins in the same cluster. An internal parameter of the process determines the granularity, specificity and coverage of clusters. SPC has been further developed into globalSPC by systematic variation of the parameter settings (19). The hRMN method (heterogeneous Relational Markov Network) generates confidence values for the assignment of functional classification. hRMN is based on a network graph able to employ any parameter that can be assigned to a pair of sequences such as sequence similarity, InterPro domains or quantitative data such as correlation of transcript regulation. Independent graphs formed by independent data sources are connected to form a Markov network, taking advantage of the synergistic effects between them (20). In the graph, nodes represent proteins and the edges are weighted according to the strength of the relation between adjacent nodes. Nodes may have FunCat labels as attributes whose propagation from known to unknown proteins is assessed utilizing Belief Propagation (19)/Generalized BP (20). Note that Belief Propagation does not require the sources to be uncorrelated or have similar distribution. Moreover, BP allows us to simultaneously calculate the marginal beliefs for all (not only one) attributes, and solves conflicts incurred by the inherent property of FunCat to allow more than one functional label for each protein, which confronts classifiers with a non-standard, soft classification task.
INVESTIGATION OF PROTEINS IN THEIR CELLULAR CONTEXT
While the primary goal of any sequencing effort is the identification of the genetic elements of an organism, the ultimate perspective in the functional analysis is a better understanding of the molecular function to uncover the molecular cause of human diseases. With the first mammalian genomes at hand, it becomes obvious that a gene-centric view is fundamentally insufficient to understand complex cellular processes such as signal transduction, gene regulation or cell differentiation. An understanding of life processes requires the integration of genome as well as transcriptome, metabolome and other -omics sciences. Any quantitative model of cellular networks must combine different types of information.
The integration of our Mammalian Protein–Protein Interaction Database (21) and the Mammalian Protein Complex database (22) with public protein–protein interaction data, gene expression data and text mining results will form the basis for the compilation of functional modules from mouse. This data set will be manually curated in order to serve as a reference data set for functional modules for a mammalian model organism and moreover provide a useful resource on the way to close the gap between genotype and phenotype.
Acknowledgments
This work was supported by 031U212C BFAM (BMBF) to H.W.M. and TE 380/1-1 (DFG) to I.V.T./H.W.M. Funding to pay the Open Access publication charges for this article was provided by the GSF-National Research Center for Environment and Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bairoch A., Apweiler R., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eppig J.T., Bult C.J., Kadin J.A., Richardson J.E., Blake J.A., Anagnostopoulos A., Baldarelli R.M., Baya M., Beal J.S., Bello S.M., et al. The Mouse Genome Database (MGD): from genes to mice—a community resource for mouse biology. Nucleic Acids Res. 2005;33:D471–D475. doi: 10.1093/nar/gki113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 4.Auwerx J., Avner P., Baldock R., Ballabio A., Balling R., Barbacid M., Berns A., Bradley A., Brown S., Carmeliet P., et al. The European dimension for the mouse genome mutagenesis program. Nature Genet. 2004;36:925–927. doi: 10.1038/ng0904-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartwell L.H., Hopfield J.J., Leibler S., Murray A.W. From molecular to modular cell biology. Nature. 1999;402:C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 6.Kent W.J. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karolchik D., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riley M.L., Schmidt T., Wagner C., Mewes H.W., Frishman D. The PEDANT genome database in 2005. Nucleic Acids Res. 2005;33:D308–D310. doi: 10.1093/nar/gki019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mewes H.W., Amid C., Arnold R., Frishman D., Güldener U., Mannhaupt G., Münsterkotter M., Pagel P., Strack N., Stümpflen V., et al. MIPS: analysis and annotation of proteins from whole genomes. Nucleic Acids Res. 2004;32:D41–D44. doi: 10.1093/nar/gkh092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pruitt K.D., Tatusova T., Maglott D.R. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruepp A., Zollner A., Maier D., Albermann K., Hani J., Mokrejs M., Tetko I., Güldener U., Mannhaupt G., Münsterkötter M., et al. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 2004;32:5539–5545. doi: 10.1093/nar/gkh894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balazsi G., Kay K.A., Barabasi A.L., Oltvai Z.N. Spurious spatial periodicity of co-expression in microarray data due to printing design. Nucleic Acids Res. 2003;31:4425–4433. doi: 10.1093/nar/gkg485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clare A., King R.D. How well do we understand the clusters found in microarray data? In Silico Biol. 2002;2:511–522. [PubMed] [Google Scholar]
- 14.Tavazoie S., Hughes J.D., Campbell M.J., Cho R.J., Church G.M. Systematic determination of genetic network architecture. Nature Genet. 1999;22:281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- 15.Dobrin R., Beg Q.K., Barabasi A.L., Oltvai Z.N. Aggregation of topological motifs in the Escherichia colitranscriptional regulatory network. BMC Bioinformatics. 2004;5:10. doi: 10.1186/1471-2105-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeger-Lotem E., Sattath S., Kashtan N., Itzkovitz S., Milo R., Pinter R.Y., Alon U., Margalit H. Network motifs in integrated cellular networks of transcription-regulation and protein–protein interaction. Proc. Natl Acad. Sci. USA. 2004;101:5934–5939. doi: 10.1073/pnas.0306752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mulder N.J., Apweiler R., Attwood T.K., Bairoch A., Bateman A., Binns D., Bradley P., Bork P., Bucher P., Cerutti L., et al. InterPro, progress and status in 2005. Nucleic Acids Res. 2005;33:D201–D205. doi: 10.1093/nar/gki106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L. GenBank. Nucleic Acids Res. 2005;33:D34–D38. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Surmeli D., Ratman O., Tetko I., Mewes H.W. 2005. FunCat functional assignment by Belief Propagation. ISMB05 Poster. [Google Scholar]
- 20.Yedidia J., Freeman W., Weiss Y. 2004. Constructing free energy approximations and generalized belief propagation algorithms. TR-2004-040. 2004. Mitsubishi Electric Research Laboratories Technical Report. [Google Scholar]
- 21.Pagel P., Kovac S., Oesterheld M., Brauner B., Dunger-Kaltenbach I., Frishman G., Montrone C., Mark P., Stümpflen V., Mewes H.W., et al. The MIPS mammalian protein–protein interaction database. Bioinformatics. 2005;21:832–834. doi: 10.1093/bioinformatics/bti115. [DOI] [PubMed] [Google Scholar]
- 22.Mewes H.W., Frishman D., Mayer K.F.X., Münsterkötter M., Noubibou O., Pagel P., Rattei T., Oesterheld M., Ruepp A., Stümpflen V. MIPS: analysis and annotation of proteins from whole genomes in 2005. Nucleic Acids Res. 2006;33:D169–D172. doi: 10.1093/nar/gkj148. [DOI] [PMC free article] [PubMed] [Google Scholar]