Abstract
Histone deacetylase 1 (HDAC1) is a major regulator of chromatin structure and gene expression. Tight control of HDAC1 expression is essential for normal cell cycle progression of mammalian cells. HDAC1 mRNA levels are regulated by growth factors and by changes in intracellular deacetylase activity levels. Stimulation of the mitogen-activated protein kinase cascade by anisomycin or growth factors, together with inhibition of deacetylases by trichostatin A (TSA), leads to stable histone H3 phosphoacetylation and strongly induced HDAC1 expression. In contrast, activation of the nucleosomal response by anisomycin alone results only in transient phosphoacetylation of histone H3 without affecting HDAC1 mRNA levels. The transcriptional induction of the HDAC1 gene by anisomycin and TSA is efficiently blocked by H89, an inhibitor of the nucleosomal response. Detailed studies of the kinetics of histone acetylation and phosphorylation show that the two modifications are synergistic and essential for induced HDAC1 transcription. Activation of the HDAC1 gene by anisomycin together with TSA or by growth factors is accompanied by phosphoacetylation of HDAC1 promoter-associated histone H3. Our results present evidence for a precise regulatory mechanism which allows induction of the HDAC1 gene in response to proliferation signals and modulation of HDAC1 expression dependent on intracellular deacetylase levels.
The amino-terminal tails of core histones are targets for multiple modifications such as acetylation, phosphorylation, and methylation. Generation of modification-specific antibodies and the identification of some of the modifying enzymes allow us to begin to understand the impact of these modifications on several cellular processes, including DNA replication and transcription. Reversible histone acetylation emerged during recent years as an important mechanism for the chromatin-dependent regulation of gene expression. Acetylation of ɛ-amino groups of lysine residues results in reduced interaction between positively charged histone tails and negatively charged DNA. Local or wide-range histone deacetylation leads to chromatin condensation, while acetylation is believed to increase the accessibility of particular genomic regions for high-molecular-weight protein complexes, thereby setting the stage for transcription.
In addition to acetylation, histone phosphorylation has been recently shown to play an important role for chromatin-associated processes. Distinct sets of kinases have been implicated in these events (references 4 and 41 and references cited therein). On one hand, H3 phosphorylation correlates with entry into mitosis, suggesting a link between chromatin condensation and histone modification by kinases. On the other hand, histone H3 phosphorylation at serine 10 was found to be an important step of the so-called nucleosomal response (26; reviewed in reference 41). This term describes the phosphorylation of histone H3, which leads to the concomitant activation of the immediate-early genes c-fos, c-jun, and c-myc (3, 26). The nucleosomal response can be induced through stimulation of the mitogen-activated protein (MAP) kinase cascade by growth factors, pharmacological agents, or stress. Induction of the MAP kinase pathway leads to the activation of effector kinases (MSK1/Rsk-2) which can phosphorylate histone H3 (33, 40).
Only a small fraction of histone H3 is transiently phosphorylated at the G0/G1 transition in growth factor-stimulated cells (1). This subset of phosphorylated histone H3 proteins is highly susceptible to hyperacetylation induced by histone deacetylase (HDAC) inhibitors. One possible explanation for this finding is the strong preference in in vitro experiments of several acetylating enzymes for histone H3 phosphorylated at serine 10 (5, 24). Indeed, a number of recent observations strongly suggest the presence of cross talk between the different histone modifying mechanisms (reviewed in references 19, 35, and 43). A link between histone acetylation and phosphorylation is provided by studies reporting the association of simultaneously acetylated and phosphorylated (in this report referred to as “phosphoacetylated”) histone H3 with nucleosomes of activated immediate-early genes (5, 6, 23). A concerted action of acetyltransferases and kinases was also demonstrated in yeast (25) and in mammalian cells (28).
An alternative model predicts the independent targeting of histone H3 by kinases and acetyltransferases during the activation of immediate-early genes in mammalian cells (42). Recently published data by Saccani et al. (32) demonstrate a link between histone H3 phosphorylation on specific target promoters in response to inflammatory stimuli and the enhanced recruitment of the transcription factor NF-κB to these sites.
Acetylation of core histones and other proteins is under the control of histone acetyltransferases (HATs) and HDACs. Mammalian HDACs are classified in three groups according to their homology with yeast enzymes (16, 20). The class I enzyme histone deacetylase 1 (HDAC1) was the first identified mammalian deacetylase and is today probably the best-studied HDAC (39). HDAC1 is recruited by a variety of transcriptional regulators to specific genomic regions, thereby mediating the repression of the corresponding target genes (7, 29). We have previously shown that mouse HDAC1 is a late inducible gene which becomes induced when growth factor-stimulated cells traverse the G1/S boundary (2). HDAC1 levels were found to be elevated in highly proliferative tissues, embryonic stem cells, and several transformed cell lines (2, 22). In contrast, HDAC1 expression was downregulated during differentiation in different cell systems such as C2C12 myoblasts, mouse erythroblasts, and F9 teratocarcinoma cells (27; S. Bartl, unpublished observations).
Recent findings indicate the requirement of regulated HDAC expression to ensure unrestricted cell cycle progression. Overexpression of HDAC1 in mouse fibroblasts or disruption of the HDAC1 gene in mouse embryonic stem cells led to impaired cell proliferation (2, 22). The expression of human HDAC1 and certain class II enzymes has been shown to increase in response to deacetylase inhibitor treatment (15, 44). Furthermore, a reduction in the levels of the endogenous class I enzymes HDAC1, HDAC2, and HDAC3 was observed in HDAC1-overexpressing cells (18; B. Schuettengruber, unpublished observations.) In contrast, HDAC1 deficiency resulted in the upregulation of HDAC2 and HDAC3 in mouse embryonic stem cells (22). Interestingly, elevated HDAC2 and HDAC3 levels could not complement the loss of HDAC1 in embryonic stem cells, providing evidence for a specific function of HDAC1 in mammalian cells. Until now, little information concerning the mechanisms controlling class I HDAC expression has been available.
In this report, we show that the mouse HDAC1 gene is activated by a mechanism involving synergistic acetylation and phosphorylation of histone H3. This mechanism allows not only the induction of HDAC1 expression in response to growth factor stimulation but also the fine-tuning of HDAC1 levels dependent on the intracellular acetylase-deacetylase balance.
MATERIALS AND METHODS
Cell culture.
Swiss 3T3 mouse fibroblasts were cultured in Dulbecco's modified Eagle medium (DMEM) containing 10% (vol/vol) fetal calf serum (FCS). Cultures were rendered quiescent by incubation in DMEM containing 0.2% (vol/vol) FCS for 72 h and restimulated to enter the cell cycle by the addition of fresh medium containing 20% (vol/vol) FCS. B6.1 T cells were cultured in DMEM containing 5% (vol/vol) heat-inactivated FCS, 50 mM β-mercaptoethanol, 10 mM l-glutamine, 10 mM HEPES, and 100 U of recombinant human interleukin-2 (IL-2) (kind gift of M. Nabholz, ISREC, Lausanne, Switzerland)/ml. B6.1 cells were growth arrested for 34 h in medium without IL-2 and restimulated to enter the cell cycle by adding fresh medium containing IL-2 (34). The following inhibitors were used in this study: trichostatin A (TSA) (50 ng/ml [0.16 μM]; Wako Pure Chemical Industries), anisomycin (50 ng/ml [0.18 μM]; Sigma), cycloheximide (10 μg/ml [35 μM] or 50 ng/ml [0.18 μM]; Sigma), puromycin (100 ng/ml [0.18 μM]; Sigma), and H89 (10 μM; Alexis Biochemicals).
RNA isolation and Northern blotting.
Isolation of total RNA was done with TRIZOL reagent (GibcoBRL) as specified by the manufacturer. Northern blot hybridization was performed by the sandwich method (37).
Western blot analysis.
Histone preparation and Western blot analyses were performed as previously described (38). All antibodies used in this study were purchased from Upstate Biotechnology, Lake Placid, N.Y.
Nuclear run-on transcription assay.
Transcription reactions were performed for 20 min in the presence of [α-32P]CTP (800 Ci/mmol) as described previously (34). Hybridization and washing was done as reported in Sutterluety et al. (36). HDAC1 cDNA fragments were subcloned in both orientations in M13-derived vectors and used as single-strand probes. The 600-bp probe of β2-microglobulin (10) and sonicated genomic Swiss 3T3 DNA were included as controls.
Chromatin immunoprecipitation.
Chromatin immunoprecipitations were carried out as described previously (3, 5), with a few modifications. Quiescent or stimulated Swiss 3T3 or B6.1 cells were cross-linked for 10 min at 37°C by adding formaldehyde (to a final concentration of 1%) directly to the culture medium in the plate. Cross-linking was stopped by the addition of glycine to a final concentration of 125 mM. Cells were washed twice with ice-cold phosphate-buffered saline and scraped in phosphate-buffered saline containing 10 mM sodium butyrate and 1 mM phenylmethylsulfonyl fluoride (PMSF). Subsequently, the cell pellet was washed in wash buffer I (0.25% Triton X-100, 10 mM EDTA, 0.5 mM EGTA, 10 mM HEPES [pH 7.5], 1 mM PMSF, 10 mM sodium butyrate, 1 μM microcystin, 1 μg/ml each of aprotinin, pepstatin, and leupeptin) and buffer II (0.2 M NaCl, 1 mM EDTA, 0.5 mM EGTA, 10 mM HEPES [pH 7.5], 1 mM PMSF, 10 mM sodium butyrate, 1 μM microcystin, 1 μg/ml each of aprotinin, pepstatin, and leupeptin). Pellets were resuspended in lysis buffer (150 mM NaCl, 25 mM Tris-HCl [pH 7.5], 5 mM EDTA [pH 8.0], 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS], 0.5% sodium deoxycholate, 1 mM PMSF, 10 mM sodium butyrate, 1 μM microcystin, 1 μg/ml each of aprotinin, pepstatin, and leupeptin) and sonicated using a Bandelin UW 70 sonicator. Insoluble material was removed by centrifugation at full speed in a microcentrifuge at 4°C for 15 min. DNA fragments isolated from the lysate are referred to as input. The soluble chromatin was filled up with lysis buffer to a final volume of 1 ml. The chromatin solution was then precleared by addition of 20 μl of protein A/G Sepharose beads (50% slurry, 100 μg of salmon sperm DNA/ml, 500 μg of bovine serum albumin/ml) and rocked at 4°C for 1 h. After centrifugation at 1,000 × g and 4°C for 5 min, the remaining supernatant was incubated with 10 μl of phosphoacetyl histone H3 antibody or myc antibody (as unspecific antibody) at 4°C, with overnight rocking. The following day, chromatin antibody complexes were isolated from the solution by incubation with 30 μl of protein A/G Sepharose beads (50% slurry, 100 μg of salmon sperm DNA/ml, 500 μg of bovine serum albumin/ml), with rocking at 4°C for 2 h. The beads were harvested by centrifugation and washed twice with RIPA buffer (150 mM NaCl, 50 mM Tris-HCl [pH 8.0], 0.1% SDS, 0.5% sodium deoxycholate, 1% NP-40), twice with High Salt buffer (500 mM NaCl, 50 mM Tris [pH 8.0], 0.1% SDS, 1% NP-40), twice with LiCl buffer (25 mM LiCl, 50 mM Tris [H 8.0], 0.5% sodium deoxycholate, 1% NP-40), and two times with TE buffer (10 mM Tris, 1 mM EDTA [pH 8.0]). Chromatin antibody complexes were eluted from the A/G Sepharose beads by addition of 2% SDS, 0.1 M NaHCO3, and 10 mM dithiothreitol to the pellet. Cross-linking was reversed by addition of 0.05 volume of 4 M NaCl and incubation of the eluted samples for 6 h at 65°C. After addition of 0.025 volume of 0.5 M EDTA and 0.05 volume of 1 M Tris-HCl (pH 6.5), proteinase K digestion was performed for 1 h at 45°C. DNA was recovered by a phenol-chloroform-isoamylalcohol extraction followed by a chloroform-isoamylalcohol extraction and precipitated by addition of 0.1 volume of 1 M sodium acetate (pH 5.2), 20 μg of glycogen, and 2.5 volumes of ethanol. Precipitated DNA was dissolved in water, and immunoprecipitated DNA was analyzed for gene sequences of HDAC1, histone H4, and β-globin by PCR.
PCR analysis of immunoprecipitated DNA.
All PCRs were performed on a Biometra D3 thermocycler, using Promega PCR Master Mix. The primer sequences are available upon request. The linear range for each primer pair was determined empirically, using different amounts of Swiss 3T3 and B6.1 genomic DNA. PCRs with increasing amounts of genomic DNA were carried out, along with the PCRs of the immunoprecipitated DNA. PCR products were resolved on 2% agarose-TAE gels and quantified using the ImageQuant program (Molecular Dynamics).
RESULTS
The mouse HDAC1 gene is transcriptionally activated in response to growth factors and HDAC inhibitor treatment.
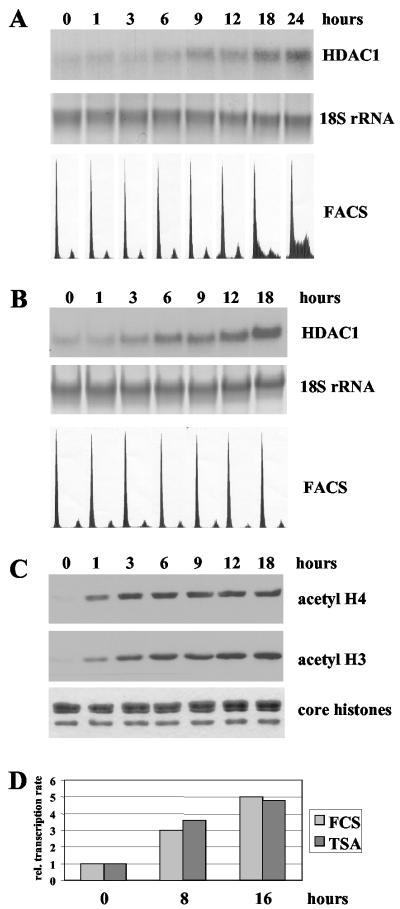
Previously, we reported the identification of HDAC1 as a growth factor-inducible gene in mouse T cells (2). The regulation of the HDAC1 gene in response to growth factors is not restricted to T cells but can be also observed during induction of resting Swiss 3T3 fibroblasts with FCS (Fig. 1A). HDAC1 mRNA expression was low in resting Swiss 3T3 cells and started to rise 6 h after serum induction to reach a fivefold-higher level after 24 h of induction. The increase in HDAC1 expression preceded entry of Swiss 3T3 cells into the S phase (Fig. 1A). These results are in good agreement with previous results obtained with the IL-2-dependent T-cell line B6.1 (2).
FIG. 1.
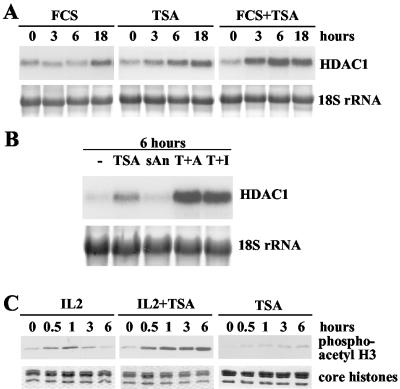
Activation of the mouse HDAC1 gene by growth factors or TSA. (A) Total RNA of quiescent Swiss 3T3 cells, stimulated for different periods (indicated in hours) with 20% FCS, was analyzed on a Northern blot by hybridization with a radiolabeled HDAC1 cDNA. To confirm equal loading and transfer of RNA, 18S rRNA was visualized with methylene blue. Cell cycle arrest and serum stimulation was monitored by FACS analysis. (B) Swiss 3T3 cells were arrested by serum deprivation and treated with 50 ng of TSA/ml for the indicated times. Total RNA was extracted and analyzed for the presence of HDAC1 mRNA as described for panel A. The DNA content of resting and TSA-treated cells was monitored by FACS analysis. (C) Histones were isolated from the cells treated as described for panel B, resolved on a denaturing SDS-16% polyacrylamide gel, transferred to a nitrocellulose membrane, and analyzed on a Western blot with antibodies specific for acetylated histone H3 (acetyl H3) and acetylated histone H4 (acetyl H4). In parallel, a duplicate gel was stained with Coomassie blue to confirm equal loading of histones. (D) In vitro run-on transcription assays. Nuclei of serum-deprived Swiss 3T3 cells or resting cells treated for 8 and 16 h with 20% FCS or 50 ng of TSA/mlwere used to determine the transcription rate of the HDAC1 gene. Equal amounts of radiolabeled newly synthesized RNA were hybridized to single-stranded DNA probes. HDAC1-specific transcription signals for each time point were quantified by densitometric scanning on a Molecular Dynamics Storm 840 Scanner and are depicted as relative transcription rates. The value for HDAC1 transcription in resting cells was arbitrarily set to 1.
To investigate whether reversible histone acetylation plays a role in the regulation of HDAC1 gene expression, we examined the effect of the HDAC inhibitor TSA on HDAC1 mRNA levels in resting fibroblasts. Northern blot analysis of serum-starved Swiss 3T3 fibroblasts treated with TSA (50 ng/ml) for various periods of time revealed a five- to sixfold increase in HDAC1 expression after 18 h of drug treatment, showing a kinetics similar to that observed after serum induction (Fig. 1B). The activation of the HDAC1 gene was not due to TSA-induced reentry into the cell cycle, since the deacetylase inhibitor did not affect the cell cycle arrest, as shown by fluorescence-activated cell sorter (FACS) analysis (Fig. 1B, lower panel). Simultaneous analysis of core histone acetylation with acetyl-specific antibodies showed that TSA treatment increased acetylation of histones H3 and H4 (Fig. 1C). Similarly, TSA induced both HDAC1 mRNA expression and histone acetylation in resting B6.1 cells (reference 38 and data not shown). These results suggest that HDACs might be involved in the repression of HDAC1 transcription in resting fibroblasts. Therefore, we next determined whether the observed activation of the HDAC1 gene by TSA or serum occurred at the transcriptional level. To this end, we performed run-on transcription assays of nuclei isolated from serum-arrested, serum-induced, or TSA-treated Swiss 3T3 cells (Fig. 1D). We observed a fivefold induction in the HDAC1 transcription rate after 16 h of TSA treatment and a similar induction 16 h after serum stimulation. These data are consistent with results from luciferase reporter analyses showing a comparable induction of the HDAC1 promoter by these stimuli in mouse fibroblasts (B. Schuettengruber and C. Seiser, unpublished data). Our results indicate that increased HDAC1 expression in response to growth factors or histone hyperacetylation is due at least in part to transcriptional activation of the HDAC1 gene.
Inducers of the MAP kinase pathway and the HDAC inhibitor TSA synergize in the activation of the HDAC1 gene.
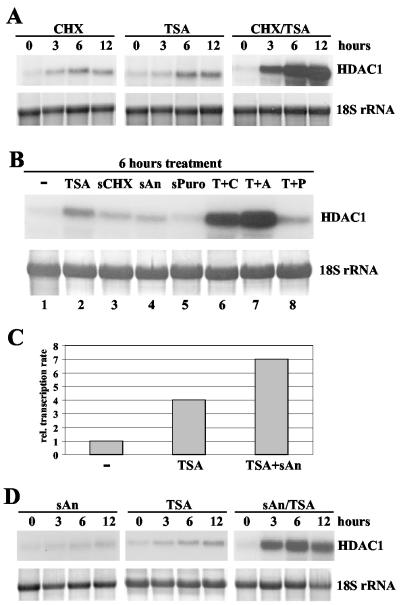
To investigate whether TSA induces HDAC1 mRNA levels via a newly synthesized protein or directly affects HDAC1 mRNA levels, we assessed the effect of TSA in the absence or presence of the translation inhibitor cycloheximide. These experiments revealed that cycloheximide (10 μg/ml) alone exhibited a weak effect on HDAC1 mRNA expression. In contrast, combinatorial treatment of resting cells with TSA and cycloheximide led to superinduction of HDAC1 expression, suggesting a cooperative activation of the HDAC1 gene by the two agents (Fig. 2A).
FIG. 2.
Cooperative activation of the HDAC1 gene by inducers of the MAP kinase pathway and TSA. (A) Cycloheximide cooperates with TSA in the induction of HDAC1 mRNA expression. Swiss 3T3 cells were arrested by serum deprivation and treated with 10 μg of cycloheximide (CHX)/ml or 50 ng of TSA/ml or both agents together for the times (in hours) indicated. Total RNA was extracted and analyzed on Northern blots for HDAC1 expression. 18S rRNA was stained with methylene blue. (B) Subinhibitory concentrations of anisomycin are sufficient to synergize with TSA in the activation of the HDAC1 gene. Resting Swiss 3T3 cells were left untreated (−) or treated for 6 h with 50 ng of TSA/ml, 50 ng of cycloheximide (sCHX)/ml, 50 ng of anisomycin (sAn)/ml, 100 ng of puromycin (sPuro)/ml, or TSA in combination with a translation inhibitor (TSA plus cycloheximide [T+C], TSA plus anisomycin [T+A], or TSA plus puromycin [T+P]). Total RNA was prepared and probed with the radiolabeled HDAC1 cDNA. The 18S rRNA was visualized by staining of the membrane with methylene blue. (C) Transcriptional activation of HDAC1 by cooperation between TSA and the MAP kinase pathway inducer anisomycin. The nuclei of Swiss 3T3 cells, after being subjected to serum arrest treatment for 48 h, were treated for 6 h with 50 ng of TSA/ml or TSA plus 50 ng of anisomycin (TSA+sAn)/ml and used to determine the transcription rate of the HDAC1 gene. Values for each time point were obtained by densitometric scanning of HDAC1-specific transcription signals. Severalfold induction values were calculated as the ratio of transcriptional activity in resting cells to that in stimulated cells. (D) Resting Swiss 3T3 cells were stimulated with subinhibitory concentrations of anisomycin (sAn; 50 ng/ml) or TSA (50 ng/ml) or both drugs simultaneously (sAn/TSA) for the indicated periods. Total RNA was prepared and probed as previously described. To confirm equal loading and transfer of RNA, 18S rRNA was stained with methylene blue.
Importantly, translation inhibitors such as cycloheximide and anisomycin have been shown to activate the MAP kinase pathway at low concentrations which fail to affect the eukaryotic translation machinery (referred to as subinhibitory concentrations [11]). Consequently, we next examined whether the growth factor-inducible HDAC1 gene could be activated by subinhibitory concentrations of translation inhibitors via the MAP kinase pathway. Treatment of quiescent Swiss 3T3 cells with subinhibitory concentrations of anisomycin and cycloheximide (50 ng/ml) showed only a very weak effect on HDAC1 mRNA levels (1.5-fold), while another translation inhibitor, puromycin, had no influence on HDAC1 expression (Fig. 2B, lanes 3 to 5). In contrast, combination of TSA with cycloheximide or anisomycin at subinhibitory concentrations led to a 10-fold superinduction of HDAC1 mRNA expression (Fig. 2B, lanes 6 and 7). No synergistic effect was observed for TSA and puromycin (Fig. 2B, lane 8). Since anisomycin and cycloheximide, but not puromycin, are able to activate the MAP kinase pathway (11), this result suggests that activation of the MAP kinase cascade in combination with inhibition of HDACs leads to superinduction of the HDAC1 gene. Stimulation of the MAP kinase pathway alone is not sufficient to efficiently activate HDAC1 gene expression.
To confirm that superinduction of the HDAC1 mRNA during combinatorial drug treatment is due to transcriptional activation, we performed run-on transcription assays of nuclei isolated from serum-deprived Swiss 3T3 cells or from resting cells treated for 6 h with TSA or for 6 h with TSA and anisomycin (Fig. 2C). Levels of HDAC1 sense transcripts increased fourfold after 6 h of TSA treatment. In combination, TSA and anisomycin lead to a sevenfold increase in transcription of the HDAC1 gene. These data clearly indicate a transcriptional activation of the HDAC1 gene during the combinatorial TSA-anisomycin treatment of resting Swiss 3T3 cells.
To define the exact kinetics of HDAC1 mRNA superinduction, quiescent Swiss 3T3 cells were stimulated for different time periods with TSA or anisomycin alone or with both agents simultaneously (Fig. 2D). Northern blot analysis again revealed a weak induction of HDAC1 mRNA levels during anisomycin stimulation (1.5-fold) and a moderate increase after TSA treatment (compare Fig. 1B). During combinatorial drug treatment, HDAC1 mRNA levels started to rise prior 3 h after induction (8-fold) and reached 11-fold-higher levels at 6 h. Thus, stimulation of resting cells with a combination of TSA and anisomycin leads to an accelerated induction of HDAC1 mRNA expression, which reaches levels higher than that observed after treatment with one stimulus alone. Taken together, these findings suggest a cooperativity of phosphorylating and acetylating activities in the activation of the mouse HDAC1 gene.
Anisomycin-induced phosphoacetylation of histone H3 is stabilized by inhibition of HDAC activity.
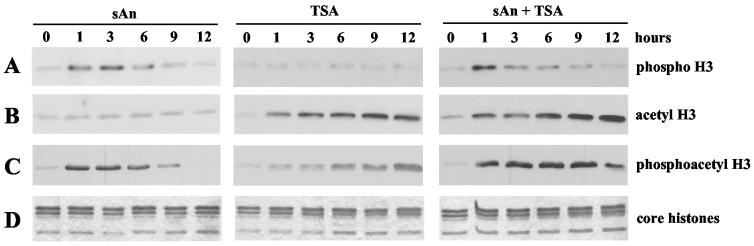
Our previous results have demonstrated that inhibition of HDACs and stimulation of the MAP kinase pathway result in a synergistic induction of HDAC1 transcription. Next, we investigated the modification state of bulk chromatin under conditions in which the HDAC1 gene is activated. To study the effect of anisomycin and TSA on histone H3 modification in vivo, histones were extracted from resting Swiss 3T3 cells stimulated with either anisomycin or TSA or with both agents together for various periods of time. Core histones were examined by Western blot analysis using a panel of modification-specific antibodies. As expected (see reference 41 for a review and references therein), serine 10 of histone H3 was rapidly and transiently phosphorylated in response to the presence of anisomycin (Fig. 3A). In contrast, no significant change in the acetylation of histone H3 was observed after anisomycin treatment (Fig. 3B). TSA treatment of resting Swiss 3T3 cells did not affect phosphorylation of histone H3, as detected by the phospho-H3 antibody (Fig. 3A), but induced the acetylation of bulk histone H3 (Fig. 3B). During combinatorial drug treatment, phosphorylation of histone H3 seemed also to be rapidly and transiently induced (Fig. 3A).
FIG. 3.
TSA-mediated acetylation stabilizes serine 10 phosphorylation at histone H3 in anisomycin-stimulated cells. Serum-starved Swiss 3T3 cells were stimulated with 50 ng of anisomycin (sAn)/ml or 50 ng of TSA/ml or with anisomycin and TSA simultaneously (sAn + TSA) for the indicated periods. Acid-soluble nuclear proteins were isolated in the presence of phosphatase and deacetylase inhibitors and analyzed on a Western blot, using phospho-H3 antibodies (A) or acetyl-H3 antibodies (B) or phosphoacetyl-H3 antibodies (C). (D) Duplicate gels were stained with Coomassie blue to ensure equal loading.
Previously, it was suggested that phospho-H3 antibodies could be impaired in recognition of phosphorylated serine 10 when adjacent lysine groups are acetylated (6). To overcome the potential problem of antibody occlusion, Western blots were analyzed with an antibody recognizing only dimodified histone H3, which is phosphorylated on serine 10 and acetylated on lysine 14. Stimulation of cells with anisomycin led to a rapid and transient increase of dimodified histones H3 (Fig. 3C). After simultaneous treatment of quiescent cells with TSA and anisomycin, we observed a stabilizing effect of TSA on histone H3 phosphoacetylation (compare sAn with sAn+TSA in Fig. 3C). This finding was in striking contrast with the results obtained with the phospho-H3 antibody, with which only a transient phosphorylation of histones H3 was seen (Fig. 3A). This result would suggest that the subfraction of phosphoacetylated histone H3 was not recognized by the phospho-H3 antibody due to reduced affinity for the dimodified histone H3 isoform.
Similarly, an impaired recognition of phosphoacetylated histone H3 was recently reported for the acetyl-H3 antibody (42). Furthermore, EGF-induced dimodification of histone H3 was detected with the phosphoacetyl-H3 antibody but not with the acetyl-H3 antibody (5). In agreement with these previous findings, the subpopulation of histone H3 that was transiently phosphoacetylated in response to anisomycin was not detected by the acetyl-H3 antibody (Fig. 3B).
Surprisingly, prolonged treatment of cells with TSA also induced phosphoacetylated histone H3 (Fig. 3C). This observation is in accordance with data showing that changes in the acetyltransferase-deacetylase balance by deletion of the HDAC1 gene in embryonic stem cells resulted in increased phosphoacetylation of histone H3 (22). In conclusion, this set of data shows that a small population of phosphoacetylated histone H3 induced by activators of the MAP kinase pathway (1) can be stabilized by inhibition by TSA of deacetylases.
Acetylation has to meet phosphorylation to act synergistically on HDAC1 gene induction.
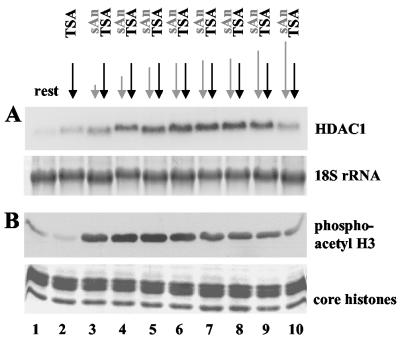
Next we investigated whether the acetylating and phosphorylating stimuli have to occur in a certain order. Swiss 3T3 cells were treated with TSA for 6 h, and anisomycin was added before, simultaneously with, or during the TSA treatment. RNA and histones were isolated and analyzed by Northern and Western blot analysis, respectively (Fig. 4A and B, respectively). Northern blot analysis revealed a moderate increase in HDAC1 mRNA levels after 6 h of TSA treatment (2.5-fold; Fig. 4A, lane 2). Simultaneous treatment of arrested cells with anisomycin and TSA for 6 h led to superinduction of HDAC1 mRNA levels (11.5-fold increase; Fig. 4A, lane 6). When cells were stimulated with anisomycin 1 h before TSA treatment, the synergistic effect decreased (8.5-fold; Fig. 4A, lane 9); when cells were stimulated with anisomycin 3 h before TSA treatment, the synergistic effect nearly disappeared (4-fold; Fig. 4A, lane 10). Cells treated with anisomycin for 1, 3, or 5 h in the presence of TSA displayed elevated HDAC1 mRNA levels, which increased with the length of anisomycin treatment (Fig. 4A, lanes 3 to 5).
FIG. 4.
Simultaneous induction of histone H3 acetylation and phosphorylation is required for efficient HDAC1 gene activation. (A) Northern blot analysis of resting Swiss 3T3 cells treated with 50 ng of TSA/ml for 6 h. Anisomycin (50 ng/ml) was added either during the last 1, 3, 5, or 6 h (lanes 3 to 6) after TSA treatment or 15 min, 30 min, 1 h, or 3 h before TSA treatment (lanes 7 to 10). As controls, cells were left untreated (lane 1) or treated only with TSA (lane 2). The Northern blot was hybridized with the radiolabeled HDAC1 cDNA. To confirm equal loading of RNA, the nylon membrane was stained with methylene blue after transfer. (B) In parallel, Western blot analysis of core histones, which were isolated from cells treated as described in panel A, was performed. Levels of dimodified histone H3 were determined with phosphoacetyl-H3 antibodies. To ensure equal loading, duplicate gels were processed and stained with Coomassie blue.
Using Western blot analysis with the phosphoacetyl-H3 antibody, the highest levels of dimodified histone H3 could be detected in cells treated with anisomycin in the presence of TSA for 1, 3, 5, or 6 h (Fig. 4B, lanes 3 to 6). When anisomycin was added before TSA, the levels of phosphoacetylated histone H3 were found to be reduced to a degree which was dependent on the length of the anisomycin pretreatment (Fig. 4B, lanes 7 to 10).
These results demonstrate a correlation between phosphoacetylation of histone H3 and HDAC1 gene induction. As shown in Fig. 3A, treatment of cells with anisomycin led to the rapid and transient phosphoacetylation of histone H3. Simultaneous inhibition of HDAC activity leads to stabilization of histone H3 phosphoacetylation and subsequent induction of HDAC1 mRNA expression. In the case of a longer time gap between the two stimuli, TSA cannot stabilize the dimodification and consequently fails to superinduce HDAC1 transcription. We concluded that acetylating and phosphorylating signals have to occur within a relatively small window of time to stabilize phosphoacetylation of histone H3 and to activate the HDAC1 gene.
The nucleosomal response is required for induction of the HDAC1 gene.
To substantiate the correlation between phosphoacetylation of histone H3 and HDAC1 gene induction, we analyzed the effect of the protein kinase inhibitor H89. The compound is a member of the H-series of protein kinase inhibitors (12) and was found to be an inhibitor of the nucleosomal response, whereas phosphorylation of certain transcription factors remains largely unaffected by H89 (40).
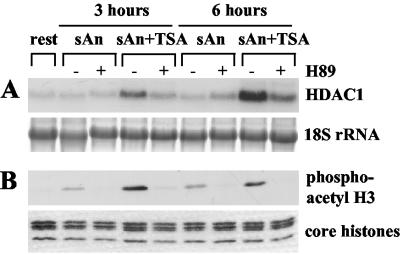
Serum-starved Swiss 3T3 cells were stimulated for 3 or 6 h with anisomycin alone or with anisomycin in combination with TSA with or without pretreatment with 10 μM H89 for 15 min. This concentration was shown to be optimal for inhibiting the nucleosomal response in vivo without affecting transcription factor phosphorylation (40). Northern blot analysis showed that pretreatment with H89 efficiently inhibited the activation of the HDAC1 gene by anisomycin and TSA (Fig. 5A). At the same time, H89 interfered with anisomycin- or TSA-anisomycin-stimulated phosphoacetylation of histone H3 (Fig. 5B). Thus, the correlation between the nucleosomal response and HDAC1 gene expression suggests a mechanistic link between these two processes.
FIG. 5.
H89 inhibits the induction of the HDAC1 gene during combinatorial TSA and anisomycin treatment. (A) Northern blot analysis of resting Swiss 3T3 cells left untreated or pretreated with 10 μM H89 for 15 min. Cells were then left untreated (−) or stimulated (+) for the indicated time periods with 50 ng of anisomycin (sAn)/ml alone or with anisomycin in combination with TSA (sAn+TSA). The Northern blot was hybridized with the radiolabeled HDAC1 cDNA (HDAC1). Transferred 18S rRNA was stained with methylene blue to confirm loading and transfer of similar amounts of RNA. (B) Phosphoacetylation of histone H3 is inhibited by H89. Western blot analysis of core histones isolated from cells treated as described for panel A was performed with a phosphoacetyl-H3 antibody. To confirm equal loading, a duplicate gel was stained with Coomassie blue.
Physiological stimuli in combination with the presence of TSA are capable of superinducing HDAC1 mRNA levels.
If anisomycin mediates its effect on HDAC1 gene expression via the MAP kinase pathway, physiological agents also known to activate the MAP kinase cascade should exert a similar effect in combination with TSA. Consequently, we replaced the pharmacological agent anisomycin with FCS to test its ability to act synergistically with TSA on HDAC1 expression. Simultaneous treatment of arrested Swiss 3T3 cells with TSA and FCS led to a faster and more pronounced induction of HDAC1 mRNA levels compared to that seen with FCS or TSA alone (Fig. 6A). Nevertheless, it is important to mention that superinduction of the HDAC1 gene during simultaneous serum and TSA treatment was less efficient than during combinatorial treatment with TSA and anisomycin.
FIG. 6.
Superinduction of HDAC1 mRNA levels by TSA in combination with growth factors. (A) Resting S 3T3 cells were treated for the indicated time periods with 20% FCS or TSA (50 ng/ml) or TSA and serum simultaneously (FCS+TSA). Total RNA was prepared, and HDAC1 mRNA was detected by Northern blot hybridization with a radiolabeled HDAC1 cDNA probe. The 18S rRNA was visualized by staining of the membranes with methylene blue. (B) IL-2-deprived B6.1 cells were left untreated (−) or stimulated for 6 h with 50 ng of TSA/ml alone or 50 ng of anisomycin (sAn)/ml alone or TSA in combination with anisomycin (T+A) or IL-2 (T+I). Northern blot analysis was done as described for panel A. (C) Histones from cells treated for the indicated periods with IL-2, TSA, or TSA in combination with IL-2 (IL-2+TSA) were isolated in the presence of phosphatase inhibitors and deacetylase inhibitors and analyzed on a Western blot, using phosphoacetyl-H3 antibodies.
HDAC1 mRNA expression was previously shown to be induced by growth stimulation with IL-2 of the T-cell line B6.1 (2). B6.1 cells can be arrested by IL-2 deprivation in the presence of all other growth factors and restimulated by recombinant IL-2. Therefore, we tested next whether TSA also cooperates with the growth factor IL-2 in the activation of the HDAC1 gene in B6.1 cells. HDAC1 mRNA levels were unchanged after a short treatment with anisomycin or IL-2 (Fig. 6B) (2). TSA treatment for 6 h of resting B6.1 cells led to a two- to threefold increase in HDAC1 mRNA levels (Fig. 6B). Combinatorial treatment with TSA and IL-2 resulted in the superinduction of HDAC1 mRNA levels. Similarly, TSA in combination with anisomycin led to strongly elevated HDAC1 mRNA levels (Fig. 6B).
In parallel, we analyzed the phosphoacetylation pattern of histone H3 in B6.1 cells treated for up to 6 h with IL-2 alone or TSA alone or a combination of IL-2 and TSA. As shown in Fig. 6C, IL-2 treatment led to rapid and transient phosphoacetylation of histone H3, while TSA alone had only a weak effect. Simultaneous addition of TSA and IL-2 stabilized the dimodification of histone H3 in B6.1 cells. Thus, in two different cell systems, TSA cooperates with physiological inducers of the MAP kinase pathway in the stabilization of histone H3 phosphoacetylation and in the induction of the HDAC1 gene.
Phosphoacetyl histone H3 is associated with the HDAC1 promoter upon transcriptional activation with a physiological stimulus.
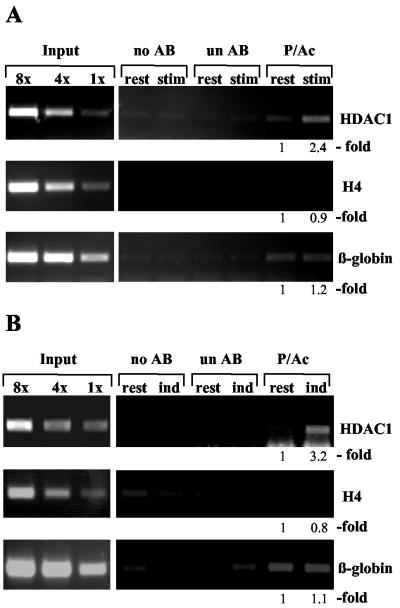
To investigate whether HDAC1 gene activation involves phosphoacetylation of nucleosomes associated with the HDAC1 promoter region, we performed chromatin immunoprecipitation assays using phosphoacetyl-H3 antibodies. Serum-deprived Swiss 3T3 cells were left untreated or were stimulated for 3 h with anisomycin in combination with TSA. After cross-linking of cells with formaldehyde, sonicated chromatin was immunoprecipitated with phosphoacetyl-H3 antibodies. In parallel, chromatin immunoprecipitation assays were performed without antibodies and with nonspecific antibodies to control the specificity of the immunoprecipitation. The amount of HDAC1 promoter DNA present in the immunoprecipitated chromatin fractions was analyzed by quantitative PCR, using primer pairs derived from the HDAC1 promoter. As controls, PCRs with primers specific for the histone H4 gene and the β-globin gene were performed in parallel. Histone H3 associated with these genes has been previously shown to be unaffected by anisomycin-induced phosphoacetylation (42). Further PCRs were carried out with standards comprising fixed amounts of genomic DNA as templates to ensure that amplification reactions remain in the linear range. Immunoprecipitation with the phosphoacetyl-H3 antibody of equivalent amounts of chromatin from arrested and stimulated cells (Fig. 7A) revealed a reproducible 2.4-fold increase in the association of dimodified histone H3 with the HDAC1 promoter after stimulation of cells with TSA and anisomycin. In contrast, the amounts of precipitated β-globin DNA and histone H4 DNA did not change significantly after stimulation of cells, suggesting that the increased association of dimodified histone H3 with the HDAC1 gene is significant. These data prove for the first time that phosphoacetylated histone H3 can be associated not only with immediate-early gene induction but also with the activation of a late inducible gene. Furthermore, we asked whether the HDAC1 gene is also associated with dimodified histone H3 under physiological conditions. Chromatin immunoprecipitation assays were performed with chromatin prepared from resting or IL-2-stimulated B6.1 cells as described above. While only small amounts of HDAC1 promoter DNA were immunoprecipitated from resting cells, IL-2 stimulation resulted in a 3.2-fold increased association of the HDAC1 promoter with phosphoacetylated H3 (Fig. 7B). No enrichment of dimodified histone H3 was seen at the control genes (histone H4 and β-globin). Together, these studies show that phosphoacetylated histone H3 is associated in vivo with the activated mouse HDAC1 gene.
FIG. 7.
Increased phosphoacetylation of promoter-associated histone H3 during activation of the HDAC1 gene. (A) Formaldehyde cross-linked chromatin was prepared from arrested (rest) and TSA-plus-anisomycin (50 ng/ml each) (stim)-treated Swiss 3T3 cells and was precipitated without antibody (no AB), with an unspecific antibody (un AB), or with a phosphoacetylated H3 antibody (P/Ac). DNA from the antibody-bound fraction and total input DNA isolated from chromatin used for the immunoprecipitation were analyzed by quantitative PCR. PCR products were quantified using the ImageQuant program, and relative signal intensities are indicated. (B) Formaldehyde cross-linked chromatin from IL-2-deprived cells (rest) and from B6.1 cells subjected to induction with IL-2 for 16 h (ind) was left untreated (no AB) or was immunoprecipitated with a phosphoacetyl-H3 antibody (P/Ac) or an unspecific antibody (un AB). PCR analysis was performed as described for panel A. The results shown are representative of three independent experiments.
DISCUSSION
HDAC1 and proliferation.
For this report, we analyzed the mechanisms involved in the regulation of the class I HDAC HDAC1. We show here that the mouse HDAC1 gene is activated by a novel mechanism which involves synergistic phosphorylation and acetylation of histone H3. HDAC1 was previously found to be upregulated in proliferating mouse cells such as IL-2-induced T cells and serum-stimulated fibroblasts (2). There are at least two potential important functions of HDAC1 in proliferating cells. On one hand, increasing amounts of HDAC1 might be required for the deacetylation of de novo synthesized acetylated histones during replication. Indeed, the increase of HDAC1 upon S phase entry correlates with the deacetylation of particular lysine residues of histone H4 in IL-2-induced B6.1 cells (38). A role of the class I enzymes HDAC1 and HDAC2 during histone deposition is plausible, since both proteins have been found to be associated with the methyltransferase DNMT1 (14, 30, 31). Furthermore, HDAC2 has been shown to be present at late-replication foci (31).
On the other hand, HDAC1 might be required for the repression of cell cycle inhibitors in cycling cells. This idea is supported by the recent finding that disruption of the HDAC1 gene in mouse embryonic stem cells results in increased expression of a subset of CDK inhibitors and reduced proliferation capacity of HDAC1 null cells (22). As a repressor of growth-inhibitory functions, therefore, HDAC1 seems to be a crucial target for HDAC inhibitors in cancer therapy (7, 21).
Transcriptional regulation of HDAC1 expression.
The outcome of previous studies suggests that the regulation of deacetylase levels is important for the unrestricted cell cycle progression of mammalian cells (2, 9, 22). The requirement for tight control of HDAC function is also reflected by the growing number of reports describing the regulation of class II enzymes (recently reviewed in references 16 and 20). Little is known about the mechanisms controlling the expression and activities of class I HDACs. The results presented in this report demonstrate the induction of mouse HDAC1 by different stimuli such as growth factors and the HDAC inhibitor TSA or combinatorial treatment with anisomycin and TSA. Run-on assays (Fig. 1D and 2C) clearly prove the transcriptional regulation of the murine HDAC1 gene. This finding is substantiated further by reporter assays showing the activation of the HDAC1 promoter by growth factors or TSA (B. Schuettengruber, unpublished observations). Our data do not exclude an additional effect of anisomycin and TSA on HDAC1 mRNA stability. However, HDAC1 mRNA is relatively stable, with a half-life of about 6 h in resting Swiss 3T3 cells (S. Bartl and B. Schuettengruber, unpublished data). Therefore, changes in HDAC1 mRNA stability could hardly contribute to the 10-fold induction increase of HDAC1 expression after 6 h of treatment with anisomycin and TSA. In conclusion, our data indicate that HDAC1 mRNA expression is predominantly regulated on the transcriptional level by both growth factors and HDAC inhibitors.
Histone acetylation and HDAC1 expression.
Our finding that inhibition of HDACs by TSA induces HDAC1 mRNA expression in resting mouse cells suggests an involvement of HDACs in the repression of the HDAC1 gene. Similarly, HDAC inhibitors have been shown to affect the expression of class I HDACs in Hep3B cells and human lymphocytes (8, 17). Taken together, these results suggest a potential feedback loop controlling HDAC expression. In accordance with this idea, class I deacetylase expression was changed in response to overexpression or loss of HDAC1 (see the introduction). However, from these previous results it was not clear if these effects are direct or are indirect effects via the induction of transcriptional regulators. Treatment with the HDAC inhibitor TSA of IL-2-deprived B6.1 cells or serum-starved Swiss 3T3 fibroblasts induces a significant increase in both HDAC1 mRNA levels and transcription rate (Fig. 1 and 6). Simultaneous treatment with cycloheximide or anisomycin at inhibitory or subinhibitory concentrations leads to an accelerated and enhanced increase in HDAC1 transcription (Fig. 2). The fact that activation of HDAC1 by TSA and anisomycin also occurs in the absence of protein synthesis demonstrates a direct effect of the HDAC inhibitor on the transcription of the HDAC1 gene. In agreement with these results, coexpression of HATs also leads to the activation of luciferase reporter genes driven by the murine HDAC1 promoter (B. Schuettengruber, unpublished observations). Thus, the N-terminal tails of HDAC1 promoter-associated histones might act as a sensor for the intracellular balance between acetylases and deacetylases.
Effect of TSA and anisomycin on histone modifications.
Growth factor stimulation of resting cells is accompanied by the rapid and transient phosphorylation and phosphoacetylation of histone H3 (4, 41). As previously reported, this response can also be induced by subinhibitory concentrations of anisomycin. Our study not only confirmed these results but led to two novel findings.
First, we showed that simultaneous treatment of resting cells with anisomycin and TSA stabilizes the phosphoacetylated form of histone H3. Until now it has not been clear whether phosphorylation and acetylation are individually targeted to the same histone tail or whether one modification is a prerequisite for the other modification. A model of Cheung et al. (4) proposes that phosphorylation at serine 10 precedes and is required for acetylation at lysine 14. In contrast, metabolic labeling studies from Barratt et al. (1) suggest that the two modifications are independently targeted to nucleosomes. A recent report from the same group (42) provides more support for independent acetylation and phosphorylation of nucleosomes associated with certain immediate-early genes.
Our results cannot distinguish between these two models but strongly suggest that inhibition of deacetylases affects the phosphorylation of serine 10 at histone H3. One could imagine different scenarios for the observed effects. TSA-induced acetylation of core histones may lead to enhanced recruitment of kinases. Alternatively, it is possible that phosphatases can be prevented from recognizing their substrate (e.g., phospho-S10) by adjacent modifications on neighboring residues due to steric hindrance. Finally, kinases or phosphatases regulating the reversible phosphorylation of histone H3 possibly can be modulated in their activities by acetylation.
In addition, we showed that prolonged treatment with the HDAC inhibitor alone also leads to phosphoacetylation of histone H3. However, the response to TSA alone is significantly delayed in comparison to the TSA-mediated activation in the presence of growth factors (IL-2) or anisomycin. These data suggest that, in the absence of growth factor signals, TSA indirectly induces phosphoacetylation of histone H3. In principle, this could be due to activation of MAP kinases by deacetylase inhibitors (13). However, H89 does not interfere with the delayed phosphacetylation induced by TSA alone (B. Schuettengruber, unpublished observations). Alternatively, TSA might cause the slow accumulation of phosphoacetylated histone H3 by affecting chromatin modifications either directly or via the involved enzymes, as discussed above.
Links between histone modifications and HDAC1 regulation.
Stimulation of histone hyperacetylation by TSA treatment of resting cells leads to activation of the HDAC1 gene. Surprisingly, the rise of HDAC1 mRNA levels is delayed compared to the increase in core histone acetylation in response to the HDAC inhibitor (Fig. 1). The kinetics of HDAC1 induction in response to the HDAC inhibitor resembles more the time course of histone H3 phosphoacetylation (Fig. 3). This finding indicates that histone H3 phosphoacetylation is in fact the more relevant consequence of TSA treatment for HDAC1 expression. This hypothesis is based on the following observations. Timing of acetylating and phosphorylating signals to coincide is essential for both stable phosphoacetylation and HDAC1 induction (Fig. 4). Inhibition of the nucleosomal response by the inhibitor H89 blocks both histone H3 phosphoacetylation and HDAC1 gene activation. Finally, increased HDAC1 promoter phosphoacetylation is linked to HDAC1 gene activation by anisomycin and TSA or IL-2. Therefore, we suggest that HDAC1 is activated by the cooperation of acetylating and phosphorylating signals, which ultimately results in phosphoacetylation of histone H3 on the HDAC1 promoter. Until now, phosphoacetylation of histone H3 was presumed to be associated only with activation of immediate-early genes (5, 6, 23).
The model: one-step activation versus two-step activation.
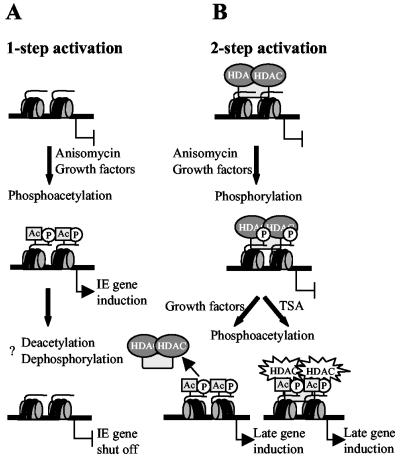
According to our model, a single signal conferred by the MAP kinase cascade is sufficient to activate immediate-early genes by a mechanism which involves phosphoacetylation of histone H3. On these promoters, the local acetylase-deacetylase balance favors acetylation. Therefore, the MAP kinase signal is sufficient to induce phosphoacetylation of histone H3 on these promoters (Fig. 8A). This mechanism, referred to as one-step activation, perfectly fulfills the requirement of a rapid transmission of the signal from the cell surface to the chromatin-embedded immediate-early gene.
FIG. 8.
Model for the activation of immediate-early genes and the HDAC1 gene by histone H3 phosphoacetylation. (A) One-step model for immediate-early genes. The default state of these genes favors acetylation. Activation of the MAP kinase pathway by either growth factors or pharmacological agents like anisomycin leads to the rapid and transient phosphoacetylation of histone H3 and simultaneous activation of immediate-early gene transcription. The mechanisms to turn off gene expression are not yet elucidated but might involve deacetylation and dephosphorylation of histone H3. (B) Two-step model for the HDAC1 gene. Due to the presence of deacetylases, the default state of this gene favors deacetylation. Induction of the nucleosomal response can induce histone H3 phosphorylation but not phosphoacetylation at the promoter. In a second step, the local balance between deacetylases and acetylases is changed either by inhibition of HDACs by TSA or by dissociation of deacetylases in response to growth signals. Both mechanisms result in phosphoacetylation of histone H3 at the HDAC1 gene locus. HATs might already be present but dominated by HDACs or specifically recruited. Active HATs prevent dephosphorylation of serine 10, thereby stabilizing the phosphoacetylation mark on histone H3. This mechanism would ensure transcription for as long as the cells proliferate.
In contrast, the activation of a late gene such as HDAC1 requires two signals (Fig. 8B). Activation of the nucleosomal response is not sufficient to induce HDAC1 (compare Fig. 2 and 3). This is most probably due to a local predominance of HDACs on the HDAC1 promoter. Inhibition of deacetylase activity by TSA can shift the balance towards acetylation, resulting in phosphoacetylation of histone H3 at the HDAC1 promoter and transcriptional induction.
Under physiological conditions, growth factor-mediated activation of the MAP kinase pathway might result in phosphorylation of histone H3 on the HDAC1 promoter. However, due to the predominance of HDACs, histone H3 phosphoacetylation at the HDAC1 promoter is inhibited. In a second step, HATs have to be recruited (and/or HDACs have to be removed), thereby inducing H3 phosphoacetylation and activation of the target promoter (two-step activation). The presence of HATs at the HDAC1 gene might prevent dephosphorylation and in this way might stabilize the phosphoacetylation mark on histone H3 to allow HDAC1 transcription throughout the cell cycle, as previously observed (2). Our model would also account for the differences between the transient expression of immediate-early genes and the stimulated persisting expression of HDAC1 in growth factor-induced cells. Indeed, a number of recent observations strongly suggest a cross talk between the different histone-modifying mechanisms (reviewed in reference 19).
Several open questions remain to be answered in the future.
How general is this model? Until now, only a limited number of immediate-early genes have been shown to be activated by the nucleosomal response. Preliminary data from our group indicate that HDAC1 is not the only gene that can be activated by acetylating and phosphorylating signals. A specific set of genes was found to be superinduced by combinatorial treatment with TSA and anisomycin (C. Hauser and B. Schuettengruber, unpublished observations). Furthermore, the HDACs HDAC2 and HDAC3 have been shown to be regulated by both growth factors and deacetylase levels and therefore would be good candidates for genes responsive to histone H3 phosphoacetylation (8, 9; S. Bartl and G. Lagger, unpublished data).
What are the factors conferring the signal to the target promoters? The acetylation signal is most likely mediated by individual transcription factors recruiting HATs, since a large number of HAT-associated regulators have been identified during recent years. The recruitment of the kinase signal is less clear. Only a subset of histone H3 becomes phosphorylated upon growth factor stimulation, suggesting a high specificity for this reaction. In future experiments, we plan to examine H3 modification patterns throughout the entire HDAC1 gene to ask whether H3 phosphoacetylation is limited to the promoter region. Reporter assays and chromatin immunoprecipitation experiments with transgenes will clarify whether transcription factors mediate histone H3 phosphorylation or whether the modification is locus dependent and is regulated by epigenetic marks.
Acknowledgments
We are grateful to T. Sauer for FACS analyses and to P. Cheung for phosphoacetyl-H3 antibodies. We also thank E. Wintersberger, G. Ammerer, and M. Posch for critical reading of the manuscript.
This work was supported by the Austrian FWF (P13068-GEN, P13638-GEN, and P14909-GEN) and the Herzfelder Stiftung.
C.H. and B.S. contributed equally to this work.
REFERENCES
- 1.Barratt, M. J., C. A. Hazzalin, E. Cano, and L. C. Mahadevan. 1994. Mitogen-stimulated phosphorylation of histone H3 is targeted to a small hyperacetylation-sensitive fraction. Proc. Natl. Acad. Sci. USA 91:4781-4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartl, S., J. Taplick, G. Lagger, H. Khier, K. Kuchler, and C. Seiser. 1997. Identification of mouse histone deacetylase 1 as a growth factor-inducible gene. Mol. Cell. Biol. 17:5033-5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chadee, D. N., M. J. Hendzel, C. P. Tylipski, C. D. Allis, D. P. Bazett Jones, J. A. Wright, and J. R. Davie. 1999. Increased Ser-10 phosphorylation of histone H3 in mitogen-stimulated and oncogene-transformed mouse fibroblasts. J. Biol. Chem. 274:24914-24920. [DOI] [PubMed] [Google Scholar]
- 4.Cheung, P., C. D. Allis, and P. Sassone-Corsi. 2000. Signaling to chromatin through histone modifications. Cell 103:263-271. [DOI] [PubMed] [Google Scholar]
- 5.Cheung, P., K. G. Tanner, W. L. Cheung, P. Sassone Corsi, J. M. Denu, and C. D. Allis. 2000. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell 5:905-915. [DOI] [PubMed] [Google Scholar]
- 6.Clayton, A. L., S. Rose, M. J. Barratt, and L. C. Mahadevan. 2000. Phosphoacetylation of histone H3 on c-fos- and c-jun-associated nucleosomes upon gene activation. EMBO J. 19:3714-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cress, W. D., and E. Seto. 2000. Histone deacetylases, transcriptional control, and cancer. J. Cell. Physiol. 184:1-16. [DOI] [PubMed] [Google Scholar]
- 8.Dangond, F., and S. R. Gullans. 1998. Differential expression of human histone deacetylase mRNAs in response to immune cell apoptosis induction by trichostatin A and butyrate. Biochem. Biophys. Res. Commun. 247:833-837. [DOI] [PubMed] [Google Scholar]
- 9.Dangond, F., D. A. Hafler, J. K. Tong, J. Randall, R. Kojima, N. Utku, and S. R. Gullans. 1998. Differential display cloning of a novel human histone deacetylase (HDAC3) cDNA from PHA-activated immune cells. Biochem. Biophys. Res. Commun. 242:648-652. [DOI] [PubMed] [Google Scholar]
- 10.Daniel, F., D. Morello, O. Le-Bail, P. Chambon, Y. Cayre, and P. Kourilsky. 1983. Structure and expression of the mouse beta 2-microglobulin gene isolated from somatic and non-expressing teratocarcinoma cells. EMBO J. 2:1061-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edwards, D. R., and L. C. Mahadevan. 1992. Protein synthesis inhibitors differentially superinduce c-fos and c-jun by three distinct mechanisms: lack of evidence for labile repressors. EMBO J. 11:2415-2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engh, R. A., A. Girod, V. Kinzel, R. Huber, and D. Bossemeyer. 1996. Crystal structures of catalytic subunit of cAMP-dependent protein kinase in complex with isoquinolinesulfonyl protein kinase inhibitors H7, H8, and H89. Structural implications for selectivity. J. Biol. Chem. 271:26157-26164. [DOI] [PubMed] [Google Scholar]
- 13.Espinos, E., A. Le Van Thai, C. Pomies, and M. J. Weber. 1999. Cooperation between phosphorylation and acetylation processes in transcriptional control. Mol. Cell. Biol. 19:3474-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuks, F., W. A. Burgers, A. Brehm, L. Hughes Davies, and T. Kouzarides. 2000. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat. Genet. 24:88-91. [DOI] [PubMed] [Google Scholar]
- 15.Gray, S. G., and T. J. Ekstrom. 1998. Effects of cell density and trichostatin A on the expression of HDAC1 and p57Kip2 in Hep 3B cells. Biochem. Biophys. Res. Commun. 245:423-427. [DOI] [PubMed] [Google Scholar]
- 16.Gray, S. G., and T. J. Ekström. 2001. The human histone deacetylase family. Exp. Cell Res. 262:75-83. [DOI] [PubMed] [Google Scholar]
- 17.Gray, S. G., I. Svechnikova, W. Hartmann, L. O'Connor, M. Aguilar Santelises, and T. J. Ekstrom. 1998. IGF-II and IL-2 act synergistically to alter HDAC1 expression following treatments with trichostatin A. Cytokine 12:1104-1109. [DOI] [PubMed] [Google Scholar]
- 18.Humphrey, G. W., Y. Wang, V. R. Russanova, T. Hirai, J. Qin, Y. Nakatani, and B. H. Howard. 2001. Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J. Biol. Chem. 276:6817-6824. [DOI] [PubMed] [Google Scholar]
- 19.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 20.Khochbin, S., A. Verdel, C. Lemercier, and D. Seigneurin Berny. 2001. Functional significance of histone deacetylase diversity. Curr. Opin. Genet. Dev. 11:162-166. [DOI] [PubMed] [Google Scholar]
- 21.Kramer, O. H., M. Gottlicher, and T. Heinzel. 2001. Histone deacetylase as a therapeutic target. Trends Endocrinol. Metab. 12:294-300. [DOI] [PubMed] [Google Scholar]
- 22.Lagger, G., D. O'Carroll, M. Rembold, H. Khier, J. Tischler, G. Weitzer, B. Schuettengruber, C. Hauser, R. Brunmeir, T. Jenuwein, and C. Seiser. 2002. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 21:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, J., M. Gorospe, D. Hutter, J. Barnes, S. M. Keyse, and Y. Liu. 2001. Transcriptional induction of MKP-1 in response to stress is associated with histone H3 phosphorylation-acetylation. Mol. Cell. Biol. 21:8213-8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lo, W. S., R. C. Trievel, J. R. Rojas, L. Duggan, J. Y. Hsu, C. D. Allis, R. Marmorstein, and S. L. Berger. 2000. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol. Cell 5:917-926. [DOI] [PubMed] [Google Scholar]
- 25.Lo, W. S., L. Duggan, N. C. Tolga Emre, R. Belotserkovskya, W. S. Lane, R. Shiekhattar, and S. L. Berger. 2001. Snf1—a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science 293:1142-1146. [DOI] [PubMed] [Google Scholar]
- 26.Mahadevan, L. C., A. C. Willis, and M. J. Barratt. 1991. Rapid histone H3 phosphorylation in response to growth factors, phorbol esters, okadaic acid, and protein synthesis inhibitors. Cell 65:775-783. [DOI] [PubMed] [Google Scholar]
- 27.Mal, A., M. Sturniolo, R. L. Schiltz, M. K. Ghosh, and M. L. Harter. 2001. A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD: inhibition of the myogenic program. EMBO J. 20:1739-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merienne, K., S. Pannetier, A. Harel-Bellan, and P. Sassone Corsi. 2001. Mitogen-regulated RSK2-CBP interaction controls their kinase and acetylase activities. Mol. Cell. Biol. 21:7089-7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ng, H. H., and A. Bird. 2000. Histone deacetylases: silencers for hire. Trends Biochem. Sci. 25:121-126. [DOI] [PubMed] [Google Scholar]
- 30.Robertson, K. D., S. Ait Si Ali, T. Yokochi, P. A. Wade, P. L. Jones, and A. P. Wolffe. 2000. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 25:338-342. [DOI] [PubMed] [Google Scholar]
- 31.Rountree, M. R., K. E. Bachman, and S. B. Baylin. 2000. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 25:269-277. [DOI] [PubMed] [Google Scholar]
- 32.Saccani, S., S. Pantano, and G. Natoli. 2001. p38-dependent marking of inflammatory genes for increased NF-κB recruitment. Nat. Immunol. 3:69-75. [DOI] [PubMed] [Google Scholar]
- 33.Sassone Corsi, P., C. A. Mizzen, P. Cheung, C. Crosio, L. Monaco, S. Jacquot, A. Hanauer, and C. D. Allis. 1999. Requirement of Rsk-2 for epidermal growth factor-activated phosphorylation of histone H3. Science 285:886-891. [DOI] [PubMed] [Google Scholar]
- 34.Seiser, C., S. Teixeira, and L. C. Kuhn. 1993. Interleukin-2-dependent transcriptional and post-transcriptional regulation of transferrin receptor mRNA. J. Biol. Chem. 268:13074-13080. [PubMed] [Google Scholar]
- 35.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 36.Sutterluety, H., S. Bartl, A. Doetzlhofer, H. Khier, E. Wintersberger, and C. Seiser. 1998. Growth-regulated antisense transcription of the mouse thymidine kinase gene. Nucleic Acids Res. 26:4989-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sutterluety, H., S. Bartl, J. Karlseder, E. Wintersberger, and C. Seiser. 1996. Carboxy-terminal residues of mouse thymidine kinase are essential for the rapid degradation in quiescent cells. J. Mol. Biol. 259:383-392. [DOI] [PubMed] [Google Scholar]
- 38.Taplick, J., V. Kurtev, G. Lagger, and C. Seiser. 1998. Histone H4 acetylation during interleukin-2 stimulation of mouse T cells. FEBS Lett. 436:349-352. [DOI] [PubMed] [Google Scholar]
- 39.Taunton, J., C. A. Hassig, and S. L. Schreiber. 1996. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272:408-411. [DOI] [PubMed] [Google Scholar]
- 40.Thomson, S., A. L. Clayton, C. A. Hazzalin, S. Rose, M. J. Barratt, and L. C. Mahadevan. 1999. The nucleosomal response associated with immediate-early gene induction is mediated via alternative MAP kinase cascades: MSK1 as a potential histone H3/HMG-14 kinase. EMBO J. 18:4779-4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomson, S., L. C. Mahadevan, and A. L. Clayton. 1999. MAP kinase-mediated signalling to nucleosomes and immediate-early gene induction. Semin. Cell Dev. Biol. 10:205-214. [DOI] [PubMed] [Google Scholar]
- 42.Thomson, S., A. L. Clayton, and L. C. Mahadevan. 2001. Independent dynamic regulation of histone phosphorylation and acetylation during immediate-early gene induction. Mol. Cell 8:1231-1241. [DOI] [PubMed] [Google Scholar]
- 43.Turner, B. M. 2000. Histone acetylation and an epigenetic code. Bioessays 22:836-845. [DOI] [PubMed] [Google Scholar]
- 44.Verdel, A., and S. Khochbin. 1999. Identification of a new family of higher eukaryotic histone deacetylases. Coordinate expression of differentiation-dependent chromatin modifiers. J. Biol. Chem. 274:2440-2445. [DOI] [PubMed] [Google Scholar]