Abstract
MPSS (massively parallel signature sequencing) is a sequencing-based technology that uses a unique method to quantify gene expression level, generating millions of short sequence tags per library. We have created a series of databases for four species (Arabidopsis, rice, grape and Magnaporthe grisea, the rice blast fungus). Our MPSS databases measure the expression level of most genes under defined conditions and provide information about potentially novel transcripts (antisense transcripts, alternative splice isoforms and regulatory intergenic transcripts). A modified version of MPSS has been used to perform deep profiling of small RNAs from Arabidopsis, and we have recently adapted our database to display these data. Interpretation of the small RNA MPSS data is facilitated by the inclusion of extensive repeat data in our genome viewer. All the data and the tools introduced in this article are available at http://mpss.udel.edu.
INTRODUCTION
DNA sequencing technologies have improved dramatically in the last decade and numerous whole-genome sequences are now available. These resources include two plant genomes, Arabidopsis and rice, as well as a number of plant pathogen genomes (1–4). Gene predictions and genome annotations are available for these genomes, built using prediction software with integrated experimental data from expressed sequence tags (ESTs) and full-length cDNA sequences (1). However, the experimental data lack the depth required to saturate the identification of mRNA transcripts, and this justifies the development of more advanced technologies. MPSS (massively parallel signature sequencing) sequences 17–20 nt (a ‘signature’) adjacent to the 3′ most DpnII site from millions of molecules in a sample (5,6). This depth provides a quantitative assessment of transcript abundance, while greatly increasing the likelihood of discovering novel transcripts. Gene expression differences can be determined using comparisons across multiple samples, as with DNA microarrays or other gene expression platforms (7).
The recent discovery and analysis of small RNAs (20–25 nt) is exciting and has demonstrated the biological importance of these non-coding molecules. Small RNAs have been isolated from diverse eukaryotic organisms and are typically classified into two major types: small interfering RNAs (siRNAs) and microRNAs (miRNAs) (8,9). Both types of molecules are processed from double-stranded RNA by RNase III enzymes called DICERs, although, their biogenesis and functions differ (10–12). siRNAs can target and degrade complementary mRNA molecules (13) and can trigger transcriptional silencing via histone modifications and/or DNA methylation (14,15). miRNA molecules originate from distinct genomic loci predicted to form ‘hairpin’ structures (11) and can induce degradation of homologous target mRNAs or can prevent mRNA translation. The short length of these small RNAs is more than sufficient to specifically match nearly any given RNA encoded in a genome. The very deep sampling capabilities of MPSS have demonstrated tremendous diversity among Arabidopsis small RNAs (16).
In this report, we describe our databases that facilitate the use of MPSS data. Our databases contain data derived from polyadenylated RNA or size selected small RNAs; we refer to these MPSS datasets as ‘mRNA’ and ‘small RNA’, respectively. We have developed four mRNA MPSS databases from Arabidopsis, rice, grape and the rice blast fungus (Magnaporthe grisea); currently, only our Arabidopsis database has small RNA MPSS data. These databases are built on a common set of web interfaces and are equipped with various graphical and analytical tools that allow the user to retrieve and analyze the data. This article focuses on the essential tools for the first-time user and describes some of the many features that we have added or improved during the past year.
DATABASE CONTENTS
Our databases require two sets of data, one of which comprises genomic information such as chromosomes, genes and potential signatures, the other of which is MPSS expression data derived from different tissues or treatments of the target organism, including signature sequences and abundance values for each signature (17). We obtain the genomic information from outside sources (see below) and build specific tables for each organism. The MPSS data are generated with collaborators in different projects. The database tables are built with Oracle9i and transferred to MySQL for the public web server. The web interface, mainly written in PHP, extracts the data requested by the user and displays the query results in a graphical and analytical output (Figures 1 and 2).
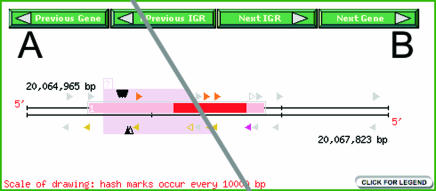
Figure 1.
Images from the ‘Gene Analysis’ output pages showing the annotated UTR regions and exons for the gene, along with the associated genomic signatures. This example has only one exon, and UTRs are indicated with pink shading. All the signatures are linked to the Signature Analysis page. (A) Viewer with small RNA signatures (black triangles pointing toward the DNA) and repetitive sequences (in this example, a retrotransposon-related repeat is shown as a pink block in the background). (B) Viewer with mRNA data only (mRNA signatures appear as colored triangles parallel to the DNA).
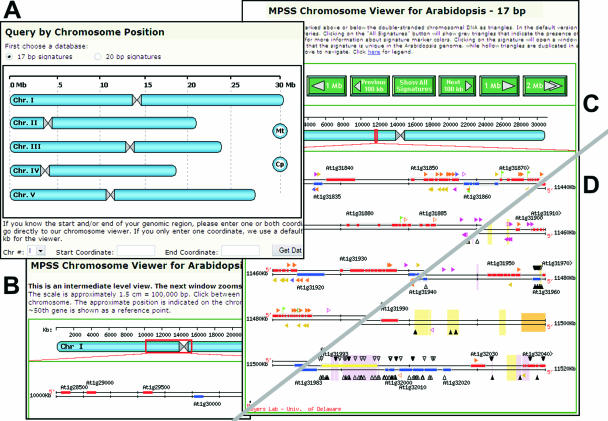
Figure 2.
Accessing a specific chromosomal region using the Arabidopsis MPSS website. (A) The user first clicks on the image of the chromosomes located on the main entry page, or the start or end coordinates can be entered to proceed directly to the primary chromosome viewer (CV). (B) The intermediate CV is launched, and the user can target the region of interest to be displayed more accurately in the primary CV. (C) The primary CV displays the annotated genes and exons along with significantly expressed signatures indicated above and below those genes. The genes and signatures are linked to the Gene Analysis and Signature Analysis pages, respectively. (D) The primary CV with small RNA signatures (black triangles pointing toward the DNA) and repetitive sequences (colored blocks in the background).
ARABIDOPSIS mRNA MPSS DATABASE
The sequence of Arabidopsis is the most complete plant genomic sequence, and The Institute for Genomic Research (TIGR) has for several years provided a comprehensive annotation (1). Our primary database is built on this annotation (TIGR v5.0) with 17 MPSS libraries, representing treated and untreated tissues and flower mutants. The database currently contains 43 810 569 signatures from poly(A) RNA (297 313 distinct signature sequences).
Nearly all sections and tools for the Arabidopsis site can be accessed through an entry page (http://mpss.udel.edu/at). The ‘Simple Query’ (Basic Queries) is useful for the users who are working on a specific gene, specific BAC clone or specific gene family; gene IDs can be used to retrieve all the associated MPSS data (Figure 1B). Briefly, the numbered boxes indicate TIGR-annotated exons and the triangles with colors (not gray) represent the location and strand of the expressed MPSS signatures. Different colors of the triangles indicate different ‘classes’ which are determined based on the position of each signature relative to gene structure (17). The expression level of each signature in each library is displayed in table format at the bottom of the Gene Analysis page.
We have added numerous new features to this page since we first introduced our web interface in 2004 (17). TIGR's v5.0 annotation indicates untranslated regions (UTRs), which we have displayed with pink or light-blue shading (Figure 1B), and splice variants for which we now provide a separate page that displays each variant. The navigation bars at the top of the image linked to adjacent genes and intergenic regions (Figure 1), BLASTP results and a sequence extraction function are also added features that make this website a more comprehensive genome analysis tool. The ability to view intergenic regions was added because many MPSS signatures represent previously unannotated or non-coding transcripts mapped within intergenic regions, particularly the small RNAs described below.
The ‘Query by Chromosome Position’ tool (linked from the main page) is useful for the users who are interested in certain location of a chromosome. Alternatively, users can click on the image map and view specific regions of interest on one of the chromosomes (Figure 2A). This viewer scales to allow the user to pinpoint precisely the target region for display in the primary chromosome viewer (Figure 2B and C). As a combined visual representation of the genomic annotation information and the MPSS expression data, the primary chromosome viewer displays the annotated genes with identifiers on both strands of DNA, along with expressed signatures. The user can view all the annotated genes as well as the potential novel transcripts expressed in the selected region.
RICE mRNA MPSS DATABASE
The rice database includes the most comprehensive set of libraries among our databases, and it can be accessed at http://mpss.udel.edu/rice. With our collaborators, we have generated more than 20 MPSS libraries derived from diverse tissues and abiotically stressed (cold, drought and salt) tissues. These libraries include different growth conditions (light and dark), different developmental stages and several biological replicates. Some of these libraries can be used as control libraries for the abiotic stressed libraries. Numerous additional rice mRNA libraries are underway (see Future Directions).
Similar to the Arabidopsis mRNA MPSS database, the rice MPSS website is built on genomic annotation data from TIGR (currently version 3.1) (4). The web interface includes nearly all of the tools available in Arabidopsis database, with some minor modifications specific to rice. Owing to historical changes in gene identifiers, we added a link to ‘TIGR version converter’ to the site. Since this database uses ‘Os’ gene identifiers from TIGR's annotation, users with different or older TIGR gene IDs may translate their gene IDs, as these ‘Os’ identifiers are likely to be more permanent.
GRAPE mRNA MPSS DATABASE
Unlike Arabidopsis and rice, the genomic sequence of grape is not complete. Therefore, this website (http://mpss.udel.edu/grape) is built around an extensive set of Cabernet Sauvignon ESTs created at UC Davis and other institutions (18). The structure of our grape MPSS database is different from that of Arabidopsis and rice because of a lack of data about physical position, introns and intergenic regions. The web interface was modified accordingly, with an added BLAST query function to allow the user query with novel nucleotide sequences and compare these with grape EST collections. The result displays related EST contigs and associated MPSS signatures. This tool enables unique types of queries using the grape MPSS data, such as the identification and transcript abundance of genes expressed in grape berries or the identification of potential alternative splicing and/or antisense transcripts. The dataset also contains substantial numbers of high abundance MPSS signatures not yet corresponding to any identified EST; these data will be increasingly useful for the annotation of previously unidentified genes as grape genomic sequence becomes available.
MAGNAPORTHE mRNA MPSS DATABASE
The Magnaporthe MPSS site (http://mpss.udel.edu/mg) was developed using the whole-genome shotgun sequence assembly from the Broad Institute (version 2.3, October, 2003) (2). An important difference from our other MPSS sites is that this genome is segmented, with many gaps and unordered contigs. Therefore, the Magnaporthe MPSS web interface uses the same contig and super-contig numbers, and it is equipped with the tools described above to analyze genome-matching MPSS signatures derived from mycelium and appressorium libraries.
ARABIDOPSIS SMALL RNA MPSS DATABASE
The most fundamental advance in our website is the addition of small RNA MPSS data. The method that led to the development of this dataset is described elsewhere (16), but the display and interpretation of these data required extensive additions to the mRNA MPSS website. And while the dataset and analysis tools use similar approaches to other plant small RNA databases, our more extensive data has required substantially new analytical tools (19). In addition, the small RNA signatures do not start with a DpnII site (GATC), so they may match anywhere in the genome.
As an introduction to our small RNA database, we have added a set of links to specific types or sources of siRNAs or miRNAs, leading the user to our modified viewer (Figures 1A and 2D). In this viewer, small RNAs appear as black triangles, pointing toward the DNA strands and representing each genomic match of an endogenous small RNA sequenced by MPSS. Because repetitive sequences are known to be sources of siRNAs, we used low-stringency analyses with RepeatMasker (http://repeatmasker.org) (20), einverted and etandem (21) to identify and display these genomic regions. The low stringency identifies many transposon- or retrotransposon-related sequences not annotated by TIGR, and these predictions are often supported by the experimentally derived small RNA data. We indicate different repeat classes in pastel-shaded background colors (e.g. light pink, yellow, blue, etc.) (Figures 1A and 2D). For individual genes or intergenic regions, we provide links to the FindMiRNA site that can provide supporting data for novel miRNAs (22).
FUTURE DIRECTIONS
Although we anticipate additional small RNA MPSS data for Arabidopsis, rice and Magnaporthe, and we are therefore adding repeat data to our rice and Magnaporthe websites, most of the new growth in our database is likely to be rice mRNA MPSS data. In collaboration with the laboratory of Dr Guo-liang Wang (Ohio State University), we currently have ∼40 rice mRNA libraries in preparation. These libraries include analyses of indica rice and F1 hybrids with Nipponbare, as well as resistant and susceptible responses to Magnaporthe or Xanthomonas oryzae pv oryzae treatments. We are currently working on novel tools to dissect allele-specific expression level polymorphisms in the japonica and indica rice subspecies, because the MPSS signatures will vary based on SNPs or indels. The inclusion of Magnaporthe-infected rice will necessitate direct connections between our rice and Magnaporthe databases. These rice libraries will contain a small proportion of Magnaporthe-derived signatures, facilitating the identification and measurement of host and pathogen transcription during infection.
We also anticipate the release of a library analysis tool (‘LIBAN’) to facilitate and automate the characterization of entire libraries, queries based on library comparisons, and the production of lists of signatures or genes with defined characteristics or expression patterns. This tool uses a Java-based web interface to customize and generate database queries without requiring bioinformatics skills. It represents a significant advance over our existing ‘advanced tools’ page which will be superseded by LIBAN.
Acknowledgments
We are grateful to David Lee and Huizhuan Wu for their contributions to earlier versions of several of our MPSS databases. Our collaborators who contributed plant materials, RNA or MPSS libraries include Drs Venu Reddyvari-Channa and Guo-liang Wang of the Ohio State University (rice and Magnaporthe materials), Drs Cheng Lu and Pam Green of the University of Delaware (small RNA materials) and Dr Alberto Iandolino of the University of California-Davis (grape materials). This work was supported primarily by awards from the NSF Plant Genome Program (#0321437) and USDA (2005-35604-15326). Funding to pay the Open Access publication charges for this article was provided by the NSF Plant Genome Research award.
Conflict of interest statement. None declared.
REFERENCES
- 1.Haas B., Wortman J., Ronning C., Hannick L., Smith R., Maiti R., Chan A., Yu C., Farzad M., Wu D., et al. Complete reannotation of the Arabidopsis genome: methods, tools, protocols and the final release. BMC Biol. 2005;3:7. doi: 10.1186/1741-7007-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dean R.A., Talbot N.J., Ebbole D.J., Farman M.L., Mitchell T.K., Orbach M.J., Thon M., Kulkarni R., Xu J.-R., Pan H., et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature. 2005;434:980–986. doi: 10.1038/nature03449. [DOI] [PubMed] [Google Scholar]
- 3.Buell C.R., Joardar V., Lindeberg M., Selengut J., Paulsen I.T., Gwinn M.L., Dodson R.J., Deboy R.T., Durkin A.S., Kolonay J.F., et al. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl Acad. Sci. USA. 2003;100:10181–10186. doi: 10.1073/pnas.1731982100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yuan Q., Ouyang S., Wang A., Zhu W., Maiti R., Lin H., Hamilton J., Haas B., Sultana R., Cheung F., et al. The Institute for Genomic Research Osa1 rice genome annotation database. Plant Physiol. 2005;138:18–26. doi: 10.1104/pp.104.059063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brenner S., Johnson M., Bridgham J., Golda G., Lloyd D.H., Johnson D., Luo S., McCurdy S., Foy M., Ewan M., et al. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol. 2000;18:630–634. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]
- 6.Brenner S., Williams S.R., Vermaas E.H., Storck T., Moon K., McCollum C., Mao J.I., Luo S., Kirchner J.J., Eletr S., et al. In vitro cloning of complex mixtures of DNA on microbeads: physical separation of differentially expressed cDNAs. Proc. Natl Acad. Sci. USA. 2000;97:1665–1670. doi: 10.1073/pnas.97.4.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oudes A.J., Roach J.C., Walashek L.S., Eichner L.J., True L.D., Vessella R.L., Liu A.Y. Application of Affymetrix array and massively parallel signature sequencing for identification of genes involved in prostate cancer progression. BMC Cancer. 2005;5:86. doi: 10.1186/1471-2407-5-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grishok A., Pasquinelli A.E., Conte D., Li N., Parrish S., Ha I., Baillie D.L., Fire A., Ruvkun G., Mello C.C. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C.elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 9.Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 10.Mallory A.C., Dugas D.V., Bartel D.P., Bartel B. MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Current Biol. 2004;14:1035–1046. doi: 10.1016/j.cub.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 11.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 12.Xie Z., Johansen L.K., Gustafson A.M., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:e104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hannon G.J. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 14.Matzke M., Matzke A.J.M., Kooter J.M. RNA: guiding gene silencing. Science. 2001;293:1080–1083. doi: 10.1126/science.1063051. [DOI] [PubMed] [Google Scholar]
- 15.Lippman Z., Gendrel A.-V., Black M., Vaughn M.W., Dedhia N., Richard McCombie W., Lavine K., Mittal V., May B., Kasschau K.D., et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- 16.Lu C., Tej S.S., Luo S., Haudenschild C.D., Meyers B.C., Green P.J. Elucidation of the small RNA component of the transcriptome. Science. 2005;309:1567–1569. doi: 10.1126/science.1114112. [DOI] [PubMed] [Google Scholar]
- 17.Meyers B.C., Lee D.K., Vu T.H., Tej S.S., Edberg S.B., Matvienko M., Tindell L.D. Arabidopsis MPSS. An online resource for quantitative expression analysis. Plant Physiol. 2004;135:801–813. doi: 10.1104/pp.104.039495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goes da Silva F., Iandolino A., Al-Kayal F., Bohlmann M.C., Cushman M.A., Lim H., Ergul A.R.F., Kabuloglu E.K., Osborne C., et al. Characterizing the grape transcriptome: Analysis of expressed sequence tags from multiple Vitis species and development of compendium of gene expression during berry development. Plant Physiol. 2005;139:574–597. doi: 10.1104/pp.105.065748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gustafson A.M., Allen E., Givan S., Smith D., Carrington J.C., Kasschau K.D. ASRP: the Arabidopsis small RNA project database. Nucleic Acids Res. 2005;33:D637–D640. doi: 10.1093/nar/gki127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smit A.F.F., Hubley R., Green P. 1996–2004. RepeatMasker open-3.0.
- 21.Rice P., Longden I., Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 22.Adai A., Johnson C., Mlotshwa S., Archer-Evans S., Manocha V., Vance V., Sundaresan V. Computational prediction of miRNAs in Arabidopsis thaliana. Genome Res. 2005;15:78–91. doi: 10.1101/gr.2908205. [DOI] [PMC free article] [PubMed] [Google Scholar]