Abstract
IRESite is an exhaustive, manually annotated non-redundant relational database focused on the IRES elements (Internal Ribosome Entry Site) and containing information not available in the primary public databases. IRES elements were originally found in eukaryotic viruses hijacking initiation of translation of their host. Later on, they were also discovered in 5′-untranslated regions of some eukaryotic mRNA molecules. Currently, IRESite presents up to 92 biologically relevant aspects of every experiment, e.g. the nature of an IRES element, its functionality/defectivity, origin, size, sequence, structure, its relative position with respect to surrounding protein coding regions, positive/negative controls used in the experiment, the reporter genes used to monitor IRES activity, the measured reporter protein yields/activities, and references to original publications as well as cross-references to other databases, and also comments from submitters and our curators. Furthermore, the site presents the known similarities to rRNA sequences as well as RNA–protein interactions. Special care is given to the annotation of promoter-like regions. The annotated data in IRESite are bound to mostly complete, full-length mRNA, and whenever possible, accompanied by original plasmid vector sequences. New data can be submitted through the publicly available web-based interface at http://www.iresite.org and are curated by a team of lab-experienced biologists.
INTRODUCTION
The post-transcriptional control of gene expression is attracting more and more attention at the present time, being seen as a part of the whole process of protein synthesis where both fast and fine tuning of the expression of particular mRNA and control of the overall level of protein synthesis are possible. The initiation of translation is a rate-limiting step of the ribosomal phase of protein synthesis, and thus it is not surprising that both the overall and the targeted control of translation initiation have been found to play an important role in many processes ranging from the embryonic development and control of malignancy, to cellular response to stress and different external or internal stimuli. Generally, translation of all eukaryotic mRNAs is initiated at their 5′-untranslated region (5′-UTR) by binding the initiation complex, comprising of a small ribosomal subunit, other protein factors and the initiator Met-tRNAi. All the eukaryotic cellular mRNAs contain a cap—a methylated guanosine moiety attached to their 5′ terminus, which ensures mRNA stability, and which is recognized by the ribosomal translation initiation complex. Once attached to the cap, the translation initiation complex scans 5′-UTR to the first initiation codon, the complete ribosome is assembled and starts the nascent polypeptide synthesis. This mode of initiation is called the cap-dependent translation initiation.
Some viruses, including several important pathogens of human and livestock, do not bear the methylguanosine cap moiety attached to the 5′ terminus of their RNAs and have evolved a different strategy which allows them to initiate the synthesis of viral proteins by the cap-independent pathway. The alternative strategies of protein synthesis even allow some viruses—e.g. poliovirus, human rhinovirus, foot-and-mouth disease virus—to shut off the host-cell protein synthesis and hence usurp the cellular translational machinery for the efficient synthesis of their own proteins. A common feature of cap-independently translated viral RNAs is the long and highly structured 5′-UTR which mediates the translation initiation complex binding and catalyses the formation of a functional ribosome. The RNA region mediating cap-independent internal binding of the ribosome to viral RNA within the 5′-UTR—and thus internal initiation of protein synthesis—is called Internal Ribosome Entry Site (IRES). From the functional point of view, the viral IRESs vary in primary and higher order structures and in their requirements for canonical translation initiation factors and other cellular or viral proteins which often bind to IRES and facilitate translation initiation complex attachment. Some viral IRESs are able to bind 40S small ribosomal subunit alone (hepatitis C virus) or even the complete 80S ribosome (cricket paralysis virus, Taura syndrome virus) (1,2). IRES RNA elements from 56 distinct viruses have been studied so far. As yet ∼200 scientific publications have been dedicated to the study of the hepatitis C virus (HCV) IRES, thus making the HCV IRES element the most prominent. However, other viral IRES elements, especially from poliovirus (PV), encephalomyelocarditis virus (EMCV), classical swine-fever virus (CSFV), foot-and-mouth disease virus (FMDV), human immunodeficiency virus (HIV), bovine viral diarrhoea virus (BVDV) and cricket paralysis virus (CrPV) are among the frequently studied models. IRESs can be found in viruses belonging to taxonomically distant families. Therefore, we may expect that a similar translation initiation strategy gets utilized also by normal eukaryotic cells.
Indeed, 73 eukaryotic mRNAs containing IRES have been reported in the genes of yeast, the fruit fly and mammals during the past few years, and the number is growing rapidly. Proteins encoded by these genes cover a very broad spectrum of functions and take part in many key processes like stress response, embryogenesis and development, angiogenesis, the response to hypoxia and ischemia, cell cycle, oncogenesis, tumorigenesis, apoptosis, the transcription and the translation initiation control and amino acid transport. It was shown that the situation in the cellular IRES field is unlikely to be as simple as had been thought just a few years ago.
Difficulties with the reliable detection and characterization of new IRES stem from both the complexity of genomes and cellular regulatory networks of higher eukaryotes, and the imperfect methodical approaches which are currently used for IRES determination. The most common methods for determination of the IRES activity are based on in vivo production of the 5′-capped bicistronic mRNA containing the desired IRES sequence inserted in between the two reporter genes (3). The calculated expression levels ratio between the second and the first cistron shows the frequency of translation initiation driven by the inserted IRES sequence at the second reporter cistron. The first reporter gene is usually translated by the cap-dependent pathway. The false measurements of the positive IRES activity and other confusions can occur in cases when shorter mRNA species derived from the test bicistronic mRNA appear in the experiment. The IRES activity analyses are usually performed under the conditions in vivo. Such an experimental setup is more prone to produce shorter and consequently ‘more active’ transcripts due to the possible presence of promoters in the DNA master sequence coding for test bicistronic mRNA or, at the mRNA level, due to the presence of sites hypersensitive to breakage or RNases. Cellular IRES sequences are usually much less efficient compared to the larger, highly structured, specialized viral IRESs. The putative IRES sequences come up from DNA regions located in a close proximity to the 5′ coding part of the genes, and thus, due to complexity of cellular transcription regulatory networks one can expect that such sequences will be more inclined to contain weak or cryptic promoters. These promoters might be active only under a particular condition and/or tissue context or might even not be active at all under normal circumstances, but will be active enough when excised from their natural vicinity and inserted into the bicistronic construct in the new molecular context. The set up of suitable experimental conditions and proper controls thus becomes a challenge.
Promoters have already been found in several DNA sequences which had been previously reported to contain IRES, and there are probably more currently recognized cellular IRES sequences that require re-evaluation (4–8). However, clear evidence exists that some mRNAs remain bound to the polysomes and are translated even under circumstances when the cellular cap-dependent translation initiation is reduced, e.g. during stress response [for review, see (9,10)], or even severely impaired, e.g. in poliovirus-infected cells (11). Also many of the currently known cellular IRES segments are well and reliably characterized and their structures, interacting proteins and biological functions are reported [for recent reviews, see (9,10,12–16)]. IRES segments would also be responsible for the efficient translation of downstream cistrons of natural bi- or polycistronic mRNAs whose occurrence has been recently reported in both unicellular and multicellular eukaryotes (17).
We have developed the IRESite database with the aim of comparing all known eukaryotic cellular and viral IRES sequences from the point of view of their primary and secondary structures, their activities under various experimental conditions and their requirements for canonical translation initiation factors and IRES trans-acting factors (ITAFs), as well as with the aim of defining as yet unknown common features of the cellular and viral IRESs, and to answer other important questions concerning the IRES function. We hope that the initial dataset currently available will demonstrate the power of the database and will induce scientists working in the translation initiation field to contribute their results and to compare them with others taking advantage of the provided IRESite tools. IRESite is designed as a non-redundant public database containing a collection of IRES-related data which are not available in any of the publicly available primary databases. We hope that IRESite will be accepted not only as a store of valuable data but will turn into a very helpful analytical tool for molecular biologists.
DATABASE DEVELOPMENT AND DESIGN
The IRESite database has been designed as a very robust storage engine intended for housing the maximum possible scientifically relevant characteristics of every individual experiment ever published in scientific literature in the field of IRES elements. We have collected the methodological characteristics of all previously published experimental setups, characteristics of the molecules involved in the experiments and also identified the data types of measured values. We have also carefully determined several other parameters which all do or could affect the results of the experiments, their interpretation and/or validity. This has led to the accumulation of 92 parameters describing a broad range of experimental attributes (the most important ones are described in detail below), which can be completed in the IRESite record. The database thus accommodates genetic and biochemical data as well as the attributes describing molecules, reagents and methods used in a course of every single experiment. The current database schema is balanced to be succinct while most descriptive. Most of the 92 mentioned attributes are facultative but should be used whenever possible. If some information is not explicitly stated in the original publication, it still can be deduced from the published text or derived from other resources. The data are stored in mysql-4.1 relational database using InnoDB tables and our programs use SQL transactions to ensure maximal data safety. To keep the number of records minimal, the data are organized efficiently and no overly similar rows are accumulated within the very same table. This requires the database to have a certain structure and the direct result of this approach is the separation of the 92 recorded parameters into 25 tables.
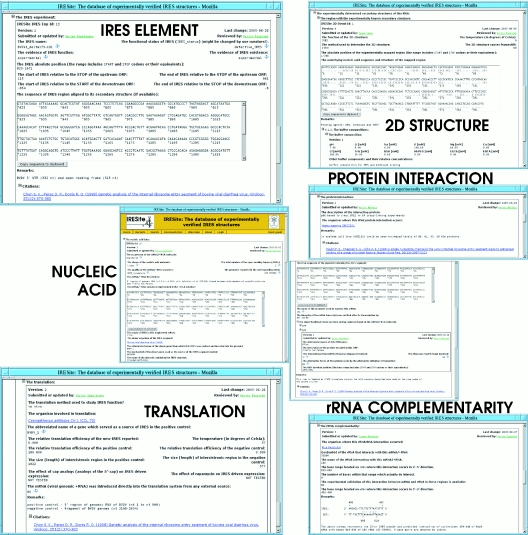
The IRESite database can accommodate any scientific report describing either a new IRES segment, its primary or secondary structure, interaction with proteins or rRNA or even reports improving the functional characterization (Figure 1). The database can also cope with records which were shown in more recent publications not to be IRESs at all (for various reasons). This rapid development in IRES research has led to the introduction of the ‘IRES status’ attribute (explained in the text below) which allows us to rate the IRES elements using a relatively fine-grained scale. Thus, it is possible to re-annotate already existing data in IRESite when new findings appear and it is possible to append new results to previously reported entries while keeping the new data separated from the former and retaining proper citations. For example, it is very simple for a user to add new results describing a new RNA–protein interaction or yet another secondary structure determined on some formerly reported RNA molecule or even add new expression data obtained by translation of the very same bicistronic reporter mRNA molecule in another host organism or another in vitro system.
Figure 1.
Every individual IRESite record consists of two facultative sections (NUCLEIC ACID and IRES ELEMENT) and up to four optional sections (rRNA COMPLEMENTARITY, TRANSLATION, PROTEIN INTERACTION and 2D STRUCTURE). Whenever possible, the data are hyperlinked to external databases including GenBank, NCBI Taxonomy and protein-oriented databases. Every section provides Remarks field for the additional data and link to original publication(s). For simplicity, the data shown here are representative pieces of several distinct records (non-existing chimera). All sections of every IRESite record except the NUCLEIC ACID section allow embedded unlimited amount of child sections. Thus, multiple regions of the mRNA/+RNA can be annotated on single molecule as an IRES containing region, RNA–protein interacting region (and multiple interacting proteins too), rRNA complementary region and secondary structures. In the case of ‘engineered’ records, IRES activity within these regions is described in section TRANSLATION by the experimentally measured values.
The IRESite is a curated database with an extra value added by curators in respect of the data contained in the original publications. Such new data include for example full-length sequence information, evaluation of the methodological approach used in the original publications, recalculation of experimentally measured values into relative scale, etc. All IRESite records consist of several parts which have their own version numbers. This allows the precise tracking of changes by future third party software using the data (Figure 1).
CONTENT OF THE DATABASE
The IRESite database distinguishes two basic types of records—‘natural’ and ‘engineered’. IRES records referred to herein as ‘natural’ contain data describing the features of the naturally occurring RNA molecule inherently possessing the particular IRES sequence. Remarkable attributes of ‘natural’ records are as follows:
Description of the mRNA/+RNA molecule.
Occurrence of promoters in donor DNA.
List of open-reading frames, their coordinates, description, the presence of frameshift or translational read-through sites and the existence of multiple protein variants as the result of alternative initiation of translation.
IRES status describes whether the recorded element is currently considered as functional IRES. This field can clearly separate functional IRESs from their aberrant forms.
mRNA status similarly describes the quality of the mRNA/+RNA sequence as it is available in the database. It clearly highlights records containing experimentally verified full-length sequence information.
The rRNA complementarity section helps to separate IRES elements containing regions complementary to some of the rRNAs.
The RNA–protein interaction data section is used to collect maximum information about the IRES trans-acting factors (ITAFs).
The RNA secondary structure section is used to describe experimentally verified secondary structures. We use bracket notation (18) to demonstrate paired and unpaired nucleic acid regions.
IRES records referred to as ‘engineered’ describe IRES containing RNA molecules, which were artificially constructed by researchers and used in some experiments. Besides containing essentially the same descriptive set of data which are used for the annotation of ‘natural’ RNA, the ‘engineered’ records contain additional entries. These entries describe the experiment and/or set of experiments where particular artificially constructed (meaning engineered) RNA was used. This kind of organization of the database allows careful monitoring of experimental conditions, detailed evaluation of positive and negative controls used during the experiment and mainly allows comparison of the experimentally obtained data with other already published experiments. In addition to the features of ‘natural’ records shown above, the ‘engineered’ records have the following attributes:
Name and sequence of the plasmid vector including the inserted IRES element.
Promoter name used to drive transcription from the plasmid.
Analyses taken to prove that mRNA was transcribed in its expected size and integrity.
The effect of various inhibitors of cap-dependent translation on yields/activity of the reporter proteins.
Actual yields/activity of the reporter proteins related to positive and negative control whenever applicable.
Size of tested intercistronic region and position of the IRES within it.
Size of intercistronic region of the positive and the negative controls.
Type of translation system (either in vitro or in vivo) including further specification (cell line, organism, temperature, etc.) of the experimental conditions.
The ‘natural’ records are expected to be updated by curators when new findings are reported. On the other hand, the ‘engineered’ records are not expected to change in this case as they refer to an individual experiment carried out in the past. The only exception could be changes in the interpretation of the recorded experimental results reflected by the IRES status and Remarks columns. It is very important to note that not all ‘engineered’ records describe the functional IRES element as the IRES function is typically demonstrated hand-in-hand with studies of IRES variants displaying somehow impaired activity and supported by negative and positive controls. This will lead to over-representation of the ‘engineered’ records in the database. Their increasing number will make them the most valuable due to their potential to be used in direct comparison of function, e.g. of the particular IRES segment under different experimental conditions or inversely of different IRES segments in the same cell line, vector, translation lysate, etc.
DATABASE ACCESS AND INTERFACE
The IRESite database is accessible freely through the Internet at http://www.iresite.org. Its user-friendly interface is made of dynamically generated HTML/JavaScript based web-pages. The database contents can be easily browsed using any web browser or searched for by an extremely powerful search interface which allows the search by a combination of multiple parameters. Whenever applicable, the IRESite entries are cross-linked to several external databases including GenBank (19), PubMed (20), NCBI Taxonomy (20), PDB (21) and SCOP (22).
At the moment, the search tool offers 50 distinct query fields which cover the most relevant fields available in the database. For example, a user can query the IRESite by any substring appearing in the name of an organism, the name of a gene, the name of an IRES, the description, the function or the remarks columns and in any combination of these. The user can also filter the data by the size of an mRNA/+RNA molecule, by the size of an IRES, by relative position of the IRES within the underlying mRNA/+RNA, by the promoter used for transcription from plasmid vector and even by the yields of the translation reactions. Further, it is possible to include/exclude from the result such experiments where cap analogs were (or were not) supplemented into the translational system (or introduced into the host cells). Importantly, one can find experiments where mRNA was introduced ready-made into the translational system. The user can also filter experiments where secondary structure was characterized, where RNA–protein interaction was demonstrated or where base-pairing between IRES containing RNA and rRNA was reported.
New data can be submitted through the IRESite web interface after creating a personal account. The submission form contains extensive help text for every field directly accessible from the browser window, thus the submission procedure is very simple. Conveniently, for a submitter sequence information including the names of the genes encoded within ORFs, their synonyms, source host organism and several other parameters are fetched from the GenBank when the GI:# number is supplied. Similarly, citation data are fetched from NCBI PubMed when a PMID number is provided. Individual account holders can store their own, as yet unfinished submission data on the server and therefore postpone the submission. During any subsequent session, the user may continue any of the formerly unfinished submissions and eventually submit the record to the curators. They verify completeness of the record, the logical content and especially ensure that the IRES is properly labelled as either being ‘true IRES’ (read functional) or being a ‘defective’ mutant variant of the former, or will mark the record as ‘not IRES’ (e.g. when a cryptic promoter is found in the mRNA). Similarly, curators can postpone their work and thus only approved records are released. Any submitted data can be delayed and made available to the public after a certain date in the future upon request. The curators contact submitters by email.
DISCUSSION AND FUTURE DEVELOPMENT
We present here fully a functional database solution which is dedicated to IRES segments of viral and cellular origin. By the end of October 2005, the IRESite has been filled with the initial set of data consisting of 30 ‘natural’ and 40 ‘engineered’ entries. Those already available ‘natural’ mRNA records constitute more than one-fifth of all known viral and cellular IRESs and thus represent a considerable portion of all the data published so far. The IRESite database further extends the idea of Bonnal and co-authors who clearly showed the necessity of the database aimed at viral and cellular IRESs (23). However, the databases do not overlap to any considerable degree. Furthermore, the IRESite is designed to accommodate RNA secondary structure information and information about the experiments carried out with the aim of both studying and utilizing the IRES elements. As we have already mentioned herein, most of the information presented in the IRESite database is manually extracted from published literature and with the help of public databases. Many independent pieces of information had to be creatively put together in order to obtain the final records which have subsequently been evaluated by a team of experienced biologists. As an example of this kind of laborious work, we use herein one of the 92 available attributes—the mRNA sequence. Our aim is to collect the full-length sequences of all mRNAs manifesting IRES activity. Many of these sequences do not exist in any database and they have to be either manually extracted from the literature or mostly taken from public sequence databanks and manipulated to match the description in literature. A similar task to collect experimentally verified data was also faced by UTRdb curators. UTRdb is a valuable database of 5′ and 3′ untranslated sequences of eukaryotic mRNAs which contains as few as 52 UTRexp records with experimentally investigated 5′ and 3′-UTRs of ∼300.000 entries automatically derived from primary sequence databanks (24). Some of the UTRexp sequences like the IRESite entries had to be manually extracted from literature.
In the near future, we plan to provide IRESite with a blast interface and a graphical representation of the currently available secondary structure data. We are also preparing the implementation of a new tool which will allow us to search the database by secondary structure motifs. We will continue to fill the database with new data and expect that most, if not all, currently known cellular IRES sequences will appear in the database by the beginning of 2006. We hope that the IRESite will help to clarify the situation in the eukaryotic cellular IRES field and will help to formulate new operational criteria more suitable for the distinction of eukaryotic cellular IRES sequences. Finally, because of the content and the regular updates of primary data, we believe that the IRESite database will become a respected source of information.
IRESite is an open database and thus we would like to invite the scientific community not only to use and analyse its content but also to kindly contribute their results and provide feedback. Our team is ready to help any researcher with the data submission procedure.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We would like to thank Klara Cisarova for helpful discussions and Steve Button for his language assistance. This work was supported by the Czech Grant Agency (Grant No. 204/03/1487), by the Grant Agency of the Charles University (Grant No. 251/2004/B-BIO/PrF) and by the Ministry of Education (Grant No. MSM 0021620813). Funding to pay the Open Access publication charges for this article was provided by Czech Grant Agency (Grant No. 204/03/1487).
Conflict of interest statement. None declared.
REFERENCES
- 1.Cevallos R.C., Sarnow P. Factor-independent assembly of elongation-competent ribosomes by an internal ribosome entry site located in an RNA virus that infects penaeid shrimp. J. Virol. 2005;79:677–683. doi: 10.1128/JVI.79.2.677-683.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pisarev A.V., Shirokikh N.E., Hellen C.U. Translation initiation by factor-independent binding of eukaryotic ribosomes to internal ribosomal entry sites. C. R. Biol. 2005;328:589–605. doi: 10.1016/j.crvi.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 3.Pelletier J., Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 4.Han B., Zhang J.T. Regulation of gene expression by internal ribosome entry sites or cryptic promoters: the eIF4G story. Mol. Cell. Biol. 2002;22:7372–7384. doi: 10.1128/MCB.22.21.7372-7384.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verge V., Vonlanthen M., Masson J.M., Trachsel H., Altmann M. Localization of a promoter in the putative internal ribosome entry site of the Saccharomyces cerevisiae TIF4631 gene. RNA. 2004;10:277–286. doi: 10.1261/rna.5910104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mauro V.P., Edelman G.M., Zhou W. Reevaluation of the conclusion that IRES-activity reported within the 5′ leader of the TIF4631 gene is due to promoter activity. RNA. 2004;10:895–897. doi: 10.1261/rna.7160404. discussion 898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Z., Dong Z., Han B., Yang Y., Liu Y., Zhang J.T. Regulation of expression by promoters versus internal ribosome entry site in the 5′-untranslated sequence of the human cyclin-dependent kinase inhibitor p27kip1. Nucleic Acids Res. 2005;33:3763–3771. doi: 10.1093/nar/gki680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hecht K., Bailey J.E., Minas W. Polycistronic gene expression in yeast versus cryptic promoter elements. FEMS Yeast Res. 2002;2:215–224. doi: 10.1111/j.1567-1364.2002.tb00086.x. [DOI] [PubMed] [Google Scholar]
- 9.Holcik M., Sonenberg N. Translational control in stress and apoptosis. Nature Rev. Mol. Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 10.Komar A.A., Hatzoglou M. Internal ribosome entry sites in cellular mRNAs: mystery of their existence. J. Biol. Chem. 2005;280:23425–23428. doi: 10.1074/jbc.R400041200. [DOI] [PubMed] [Google Scholar]
- 11.Johannes G., Carter M.S., Eisen M.B., Brown P.O., Sarnow P. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc. Natl Acad. Sci. USA. 1999;96:13118–13123. doi: 10.1073/pnas.96.23.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stoneley M., Willis A.E. Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;23:3200–3207. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]
- 13.Pickering B.M., Willis A.E. The implications of structured 5′ untranslated regions on translation and disease. Semin. Cell Dev. Biol. 2005;16:39–47. doi: 10.1016/j.semcdb.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 14.Marash L., Kimchi A. DAP5 and IRES-mediated translation during programmed cell death. Cell Death Differ. 2005;12:554–562. doi: 10.1038/sj.cdd.4401609. [DOI] [PubMed] [Google Scholar]
- 15.Spriggs K.A., Bushell M., Mitchell S.A., Willis A.E. Internal ribosome entry segment-mediated translation during apoptosis: the role of IRES-trans-acting factors. Cell Death Differ. 2005;12:585–591. doi: 10.1038/sj.cdd.4401642. [DOI] [PubMed] [Google Scholar]
- 16.Holcik M. Targeting translation for treatment of cancer—a novel role for IRES? Curr. Cancer Drug Targets. 2004;4:299–311. doi: 10.2174/1568009043333005. [DOI] [PubMed] [Google Scholar]
- 17.Blumenthal T. Operons in eukaryotes. Brief Funct. Genomic Proteomic. 2004;3:199–211. doi: 10.1093/bfgp/3.3.199. [DOI] [PubMed] [Google Scholar]
- 18.Hofacker I.L., Fontana W., Stadler P.F., Bonhoeffer S., Tacker M., Schuster P. Fast folding and comparison of RNA secondary structures. Monatsheft für Chemie. 1994;125:167–188. [Google Scholar]
- 19.Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L. GenBank. Nucleic Acids Res. 2005;33:D34–D38. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wheeler D.L., Barrett T., Benson D.A., Bryant S.H., Canese K., Church D.M., DiCuccio M., Edgar R., Federhen S., Helmberg W., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2005;33:D39–D45. doi: 10.1093/nar/gki062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andreeva A., Howorth D., Brenner S.E., Hubbard T.J., Chothia C., Murzin A.G. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonnal S., Boutonnet C., Prado-Lourenco L., Vagner S. IRESdb: the Internal Ribosome Entry Site database. Nucleic Acids Res. 2003;31:427–428. doi: 10.1093/nar/gkg003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mignone F., Grillo G., Licciulli F., Iacono M., Liuni S., Kersey P.J., Duarte J., Saccone C., Pesole G. UTRdb and UTRsite: a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 2005;33:D141–D146. doi: 10.1093/nar/gki021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.