Abstract
The Mouse Genome Database (MGD) integrates genetic and genomic data for the mouse in order to facilitate the use of the mouse as a model system for understanding human biology and disease processes. A core component of the MGD effort is the acquisition and integration of genomic, genetic, functional and phenotypic information about mouse genes and gene products. MGD works within the broader bioinformatics community to define referential and semantic standards to facilitate data exchange between resources including the incorporation of information from the biomedical literature. MGD is also a platform for computational assessment of integrated biological data with the goal of identifying candidate genes associated with complex phenotypes. MGD is web accessible at http://www.informatics.jax.org. Recent improvements in MGD described here include the incorporation of an interactive genome browser, the enhancement of phenotype resources and the further development of functional annotation resources.
INTRODUCTION
The Mouse Genome Database (MGD) provides a comprehensive and integrated view of genetic, genomic and biological information for the laboratory mouse (1,2). The primary mission of MGD is to facilitate the use of the laboratory mouse as a model for studying human biology and disease. MGD contains information on mouse genes, genetic markers and genomic features and includes comprehensive information for these features including sequence sets, mapping details, Gene Ontology (GO) annotations, allele descriptions and mutant phenotype characteristics (Table 1). MGD integrates sequence with biology through the curated association of genome, transcript and protein sequence sets with mouse genes, and work done in collaboration with other large genome informatics resources. A primary goal of MGD is to be the hub for mouse phenotype information and to provide robust access to phenotypic data for human users and for computational applications.
Table 1.
Snapshot of data content in MGD: September 12, 2005
| MGD data statistics | September 12, 2005 |
|---|---|
| Number of genes with sequence data | 27 617 |
| Number of genes (incl. unmapped mutants) | 30 881 |
| Number of markers (including genes) | 57 368 |
| Number of markers mapped | 53 212 |
| Number of genes with protein sequence information | 19 580 |
| Number of genes with GO annotations | 16 572 |
| Number of mouse/human orthologies | 15 849 |
| Number of mouse/rat orthologies | 15 532 |
| Number of genes with one or more phenotypic alleles | 6191 |
| Number of cataloged phenotypic alleles | 14 338 |
| Number of references | 94 891 |
| Number of mouse nucleotide sequences integrated into the MGI system (includes expressed sequence tags) | >7 600 000 |
MGD is updated daily and there are weekly data exchanges with other major genomics resources, such as NCBI and UniProt. A recent snapshot of MGD content is shown in Table 1. MGD continues to evolve, expanding its data coverage, improving data access, and providing new data query, analysis and display tools. Current efforts focus on the incorporation of genome visualization tools and the explicit representation of mouse models in the context of their relationship to human disease presentations.
MGD is a core component of the Mouse Genome Informatics (MGI) database resource (http://www.informatics.jax.org) hosted at The Jackson Laboratory (http://www.jax.org). Other projects and resources that are part of the MGI system include the Gene Expression Database (GXD) (3) (http://www.informatics.jax.org/mgihome/GXD/aboutGXD.shtml) and the Mouse Tumor Biology Database (MTB) (4) (http://tumor.informatics.jax.org). All MGI component groups participate actively in the development and application of the Gene Ontology (GO) (5) (http://www.geneontology.org).
IMPROVEMENTS DURING 2004
Implementation of Mouse Genome Browser
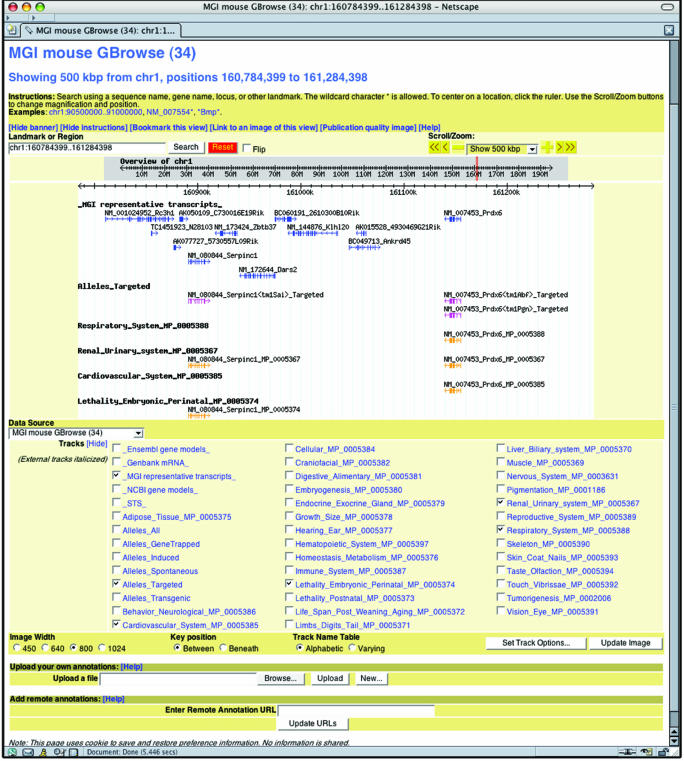
A mouse implementation of the Generic Genome Browser (6) has been installed at MGD. A number of unique MGD tracks are available on the Mouse GBrowse (http://gbrowse.informatics.jax.org/cgi-bin/gbrowse/mouse_build_34), including MGI representative transcripts, SNPs, alleles, QTLs and phenotype annotations (Figure 1). Also available are the Ensembl and NCBI genome annotation, GenBank mouse mRNAs and STS markers. GBrowse is also used to generate thumbnail images of the intron–exon structure of genes on the MGD gene detail pages. GBrowse allows users to upload their own genome annotations to be viewed in the context of existing annotation tracks. GBrowse is accessible from the MGI home pages as well as from Gene Detail pages.
Figure 1.
Mouse GBrowse: a screen shot of the MGI Mouse GBrowse interface. Using Mouse GBrowse, users can display MGD-specific annotations about genes, allele types (e.g. spontaneous, targeted, transgenic, etc.) and phenotype to gene associations. Each glyph displayed in the browser is hypertext linked to the appropriate detail page in MGD. Particularly note the phenotype tracks.
Enhanced representation of relationship between mouse models and human diseases
For some time, MGD curators have associated mouse mutant alleles with OMIM, a text-based compendium of human genes and diseases maintained by the Johns Hopkins University (7). This was accomplished by inserting simple hypertext links into mouse mutant allele descriptions where mice carrying these alleles had been reported as disease models. These links were not searchable and could be viewed only at the level of the allele description.
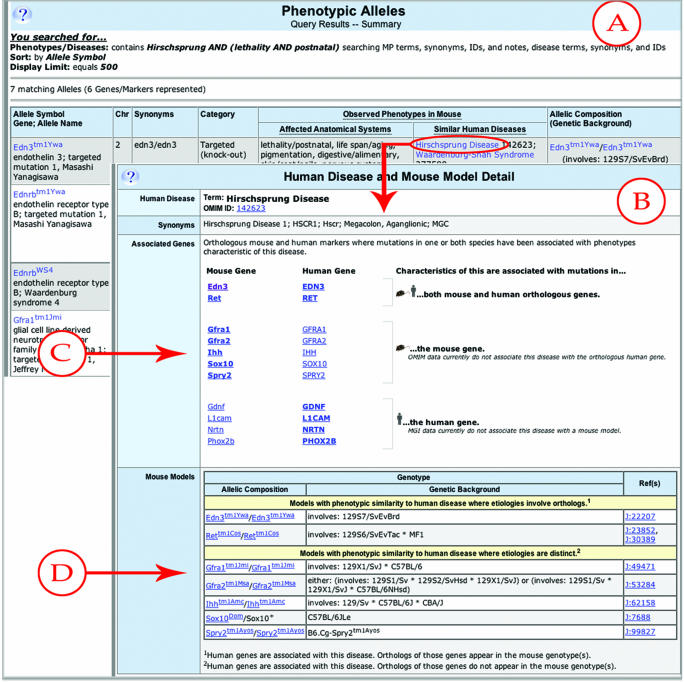
We have made these more explicit to show the relationship between the actual mouse genotype and the human disease it models. We have added OMIM disease terms to MGD as a vocabulary to allow users to access these data from a human-centric as well as a mouse-centric view (Figure 2). The vocabulary of OMIM disease terms is stored and maintained in the same way as the other controlled vocabularies (GO, Anatomy, etc.). Currently, 5920 OMIM terms are contained in MGD and loads are refreshed nightly.
Figure 2.
Mouse models and human diseases. Panel A: Search summary page of phenotypic alleles indicating observed phenotypes in mouse. The human diseases given in the column ‘Similar human diseases’ link to the page describing the relationship of this human disease to mouse (Panel B). Panel B: Detail page of human disease and mouse models for Hirschsprung disease. The associated gene section (arrow C) shows where orthologous mouse and human genes have been associated with this disease phenotype. The mouse models section (arrow D) specifies the genotypes of mouse models for this disease that have etiologies involving orthologs or etiologies which are distinct.
New presentation of Gene Ontology annotations
We have responded to biologists' requests to provide functional annotation information in human-readable form by providing Gene Ontology annotations in a templated text representation (Figure 3). A lexicon of grammar rules and templates provides the structure for the automated construction of the GO text from the GO annotation files. Users may view GO annotations via their web browsers either as text or in a tabular presentation. Presently, the text presentation is updated weekly, whereas the data in the tables are updated daily. We will continue to improve the text generation scripts to incorporate paragraphs generated from other structured data in MGD.
Figure 3.
GO annotations as text. In response to user requests, we have created a lexicon and templates that enable computational generation of GO annotations in text form. This is provided to our web interface users in conjunction with the tabular presentation of the GO annotation information. Here, the GO text for gene Ednrb annotations is presented.
OTHER ACTIVITIES OF NOTE IN 2005
Harmonization of mouse and rat nomenclatures. MGD initiated the merging of mouse and rat gene, allele and strain nomenclature guidelines via the Committee for Standardized Nomenclature in Mice and the Rat Genome and Nomenclature Committee. There is now a common standard for nomenclatures in rodent species that provides a simplified system for researchers and should lessen the redundancy of names of strains in mice and rats and encourage the co-naming of gene orthologs.
FANTOM 3 data loads. MGD staff have participated in each of the three FANTOM (Functional Annotation of the Mouse) projects as part of a consortium working to curate a large cDNA clone sequences. The FANTOM 3 project provides a comprehensive view of the transcriptional landscape of the mouse genome (8). More than 100 000 cDNA clone sequences have been released to date by the FANTOM consortium and they have been integrated into MGD.
Electronic publication. We now have posted an electronic publication of The Anatomy of the Laboratory Mouse by Margaret J. Cook. This out-of-print book is a classic anatomy text first published in 1965. This book joins other out-of-print classics of mouse genetics in electronic publication at MGD.
OTHER INFORMATION
Mouse gene nomenclature
The MGD gene annotation group assigns unique symbols and names to mouse genes under the guidelines set by the International Committee on Standardized Genetic Nomenclature for Mice (http://www.informatics.jax.org/mgihome/nomen/index.shtml). Through curation of shared links between MGI and other bioinformatics resources, the official nomenclature for mouse genes is becoming widely disseminated. The MGI nomenclature group works closely with nomenclature specialists for human (http://www.gene.ucl.ac.uk/cgi-bin/nomenclature/searchgenes.pl) and rat (http://rgd.mcw.edu) to provide consistent nomenclature for mammalian species. The mouse and human nomenclature committees collaborate with scientists prominent in a specialist area to develop a nomenclature scheme for those genes that should be grouped together in a family, such as the ARID family (10) or to revise the nomenclature for an already established gene family, such as the ACOT family (11). Scientists can contact the MGD nomenclature coordinator by email (nomen@informatics.jax.org) and can reserve symbols prior to publication using the electronic nomenclature submission form (http://www.informatics.jax.org/mgihome/nomen/nomen_submit_form.shtml).
Electronic data submission
Any type of data that MGD maintains can be submitted as an electronic contribution. Over the last year, the most frequent submissions have been of mutant and phenotypic allele information originating with the large mouse mutagenesis centers. Other common types of submission include mutant and QTL mapping data. Each electronic submission receives a permanent database accession ID. All datasets are associated with either a publication or an electronic submission reference. MGD reference pages provide links to associated datasets. On-line information about data submission procedures is found at http://www.informatics.jax.org/mgihome/submissions/submissions_menu.shtml.
Community outreach and user support
MGD provides extensive user support through on-line documentation and easy email or phone access to User Support Staff. User Support WWW access: http://www.informatics.jax.org/mgihome/support/support.shtml; Email access: mgi-help@informatics.jax.org; Telephone access: +1 207 288 6445; Fax access: +1 207 288 6132.
Other outreach
MGI-LIST (http://www.informatics.jax.org/mgihome/lists/lists.shtml) is a moderated and active email bulletin board supported by the MGI Users Support group.
HIGH LEVEL OVERVIEW OF THE MAIN COMPONENTS AND IMPLEMENTATION
The MGD database and software system was first released on the web in 1994. Here, we summarize the high level functionality of the major components of the system. At the heart of MGD is the database itself. MGD is implemented in the Sybase relational database management system. Its many tables form the core within which the biological information is stored. Additional sequence data such as BLAST-able databases and genome assembly files are stored outside the relational database. There are two primary vehicles by which data are entered into MGD: the editing interface and automated load programs. The editing interface (EI) is an interactive, graphical application. Curators use the EI to enter new data from the literature, to verify the results of automated loads and to correct errors. The automated load programs integrate larger datasets from many sources into the database. Automated loads involve QC checks and processing algorithms that integrate the bulk of the data automatically and identify issues to be resolved by curators or the data provider. Through these two vehicles, the EI and automated loads, we are able to acquire and integrate large amounts of data into high-quality, curated information.
There are several different routes for public data access. The web interface (WI) is the main tool allowing users to query interactively and display our data through a web browser. MouseBLAST allows users to do sequence similarity searches against a variety of rodent-relevant sequence databases that are culled weekly from NCBI, UniProt and other providers. Mouse GBrowse (see above) allows users to visualize mouse datasets against the genome as a series of linear tracks. Supplementing these three interactive tools is a large collection of public database reports generated nightly. These reports provide concise listings of large subsets of data, for example, all mouse/human homologies. They are a major source for other data providers who link to or use MGD data in their products and for computational biologists who use MGD data in their analyses. Direct SQL access to the database is available for sophisticated users who wish to construct custom queries or to perform analyses not possible through the WI. Finally, we have an initial web services API that is providing programmatic access to MGD. This API is designed for specific groups outside of MGI that access our data. A more robust and general API is planned for the near future.
CITING MGD
The following citation format is suggested when referring to datasets specific to the MGD component of MGI: Mouse Genome Database (MGD), Mouse Genome Informatics, The Jackson Laboratory, Bar Harbor, Maine (URL: http://www.informatics.jax.org). (Type in date (month, year) when you retrieved the data cited.). For general citation of the Mouse Genome Informatics (MGI) resource please cite this article.
Acknowledgments
The Mouse Genome Database is supported by NIH/NHGRI grant HG00330. Funding to pay the Open Access publication charges for this article was provided by NHGRI P41-00330.
Conflict of interest statement. None declared.
REFERENCES
- 1.Eppig J.T., Bult C.J., Kadin J.A., Richardson J.E., Blake J.A., the Mouse Genome Database Group The Mouse Genome Database (MGD): from genes to mice—a community resource for mouse biology. Nucleic Acids Res. 2004. 2005;33:D471–D475. doi: 10.1093/nar/gki113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bult C.J., Blake J.A., Richardson J.E., Kadin J.A., Eppig J.T., the Mouse Genome Database Group The Mouse Genome Database (MGD): integrating biology with the genome. Nucleic Acids Res. 2004;32:D476–D481. doi: 10.1093/nar/gkh125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hill D.P., Begley D.A., Finger J.H., Hayamizu T.F., McCright I.J., Smith C.M., Beal J.S., Corbani L.E., Blake J.A., Eppig J.T., et al. The Mouse Gene Expression Database (GXD): updates and enhancements. Nucleic Acids Res. 2004;32:D568–D571. doi: 10.1093/nar/gkh069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Näf D., Krupke D.M., Sundberg J.P., Eppig J.T., Bult C.J. The mouse tumor biology database: a public resource for cancer genetics and pathology of the mouse. Cancer Res. 2002;62:1235–1240. [PubMed] [Google Scholar]
- 5.The Gene Ontology Consortium. The Gene Ontology (GO) Database and Informatics Resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stein L.D., Mungall C., Shu S., Caudy M., Mangone M., Day A., Nicherson E., Stajich J.E., Harris T.W., Arva A., et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Online Mendelian Inheritance in Man, OMIM (TM). McKusick-Nathans Institute for Genetic Medicine; Johns Hopkins University, Baltimore, MD; National Center for Biotechnology Information, National Library of Medicine, Bethesda, MD. 2000. World Wide Web URL: http://www.ncbi.nlm.nih.gov/omim/ [Google Scholar]
- 8.The FANTOM Consortium and the RIKEN Genome Exploration Research Group. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 9.Doolittle D.P., Davisson M.T., Guidi J.N., Green M.C. Catalog of Mutant Genes and Polymorphic Loci. In: Lyon M.F., Raston S., Brown S.D.M., editors. Genetic Variants and Strains of the Laboratory Mouse, 3rd edn. Vol 1. Oxford: Oxford University Press; 1996. pp. 1–16. 17–854. [Google Scholar]
- 10.Wilsker D., Probst L., Wain H.M., Maltais L., Tucker P.W., Moran E. Nomenclature of the ARID family of DNA-binding proteins. Genomics. 2005;86:242–251. doi: 10.1016/j.ygeno.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 11.Hunt M.C., Yamada J., Maltais L.J., Wright M.W., Podesta E.J., Alexson S.E. A revised nomenclature for mammalian acyl-CoA thioesterases/hydrolases. J. Lipid Res. 2005;46:2029–2032. doi: 10.1194/jlr.E500003-JLR200. [DOI] [PubMed] [Google Scholar]