Abstract
Metazoan cell cycle-regulated histone mRNAs are unique cellular mRNAs in that they terminate in a highly conserved stem-loop structure instead of a poly(A) tail. Not only is the stem-loop structure necessary for 3′-end formation but it regulates the stability and translational efficiency of histone mRNAs. The histone stem-loop structure is recognized by the stem-loop-binding protein (SLBP), which is required for the regulation of mRNA processing and turnover. In this study, we show that SLBP is required for the translation of mRNAs containing the histone stem-loop structure. Moreover, we show that the translation of mRNAs ending in the histone stem-loop is stimulated in Saccharomyces cerevisiae cells expressing mammalian SLBP. The translational function of SLBP genetically required eukaryotic initiation factor 4E (eIF4E), eIF4G, and eIF3, and expressed SLBP coisolated with S. cerevisiae initiation factor complexes that bound the 5′ cap in a manner dependent on eIF4G and eIF3. Furthermore, eIF4G coimmunoprecipitated with endogenous SLBP in mammalian cell extracts and recombinant SLBP and eIF4G coisolated. These data indicate that SLBP stimulates the translation of histone mRNAs through a functional interaction with both the mRNA stem-loop and the 5′ cap that is mediated by eIF4G and eIF3.
Most cellular mRNAs terminate in a poly(A) tail to which the poly(A)-binding protein (PABP) binds. PABP is required for translation initiation through its interaction with eukaryotic initiation factor 4G (eIF4G) and eIF4B (7, 30, 32, 33, 44, 55), and this interaction results in an increase in the affinity of PABP for poly(A) RNA and an increase in the affinity of eIF4F for the 5′ cap (m7GpppN, where N represents any nucleotide) (32, 33, 59). Those mRNAs that naturally lack a poly(A) tail present an apparent paradox in that, in the absence of the poly(A) tail, the cap should be virtually nonfunctional, and consequently, the mRNA should be rendered translationally incompetent. Expression of the cell cycle-regulated histone mRNAs is tightly coupled to nuclear DNA synthesis during the S phase of the cell cycle (6), which is achieved through the regulation of their transcription, processing, and mRNA stability (21, 25, 51). Changes in transcription and 3′-end processing account, in part, for the regulation of histone expression mRNA during the G1 phase, whereas the mRNA is specifically destabilized during the G2 phase (24). Cell cycle-regulated histone mRNAs represent the only known class of cellular mRNAs that do not terminate in a poly(A) tail but instead contain a 3′-terminal stem-loop structure that is highly conserved among metazoans (26, 36). A 31-kDa stem-loop-binding protein (SLBP) that is associated with polysomes and specifically binds to the histone stem-loop structure has been identified (23, 35, 43, 58). The requirements for histone mRNA 3′-end processing include the stem-loop and SLBP (13, 14, 57) and a downstream purine-rich region that forms a duplex with a complementary sequence at the 5′ end of U7 snRNA (5, 9, 12, 38, 52). SLBP binds to the histone 3′-terminal stem-loop in the nucleus, remains bound to the mRNA as it is exported to the cytoplasm, and is necessary for the cell cycle regulation of mRNA stability and localization of the mRNA to polysomes (13, 23, 34, 41, 53).
Previous work has demonstrated that the histone stem-loop was necessary and sufficient to support the translation of reporter mRNAs in animal cells when present at the 3′ terminus and, like a poly(A) tail, was functionally dependent on the cap (17). Although SLBP is necessary to mediate the other regulatory aspects associated with the histone stem-loop, its role in translation initiation has not been demonstrated. Moreover, how SLBP might promote efficient translation initiation is unknown.
In this study, we show that SLBP is required for the translation of mRNAs terminating in the histone stem-loop. The stimulatory effect of SLBP can be recapitulated in Saccharomyces cerevisiae cells expressing mammalian SLBP. Genetic analysis indicated that the translational function of SLBP requires eIF4E, eIF4G, and eIF3. SLBP copurified with these initiation factors when they were isolated through their binding either to the 5′ cap or to poly(A)-Sepharose. Coisolation studies of SLBP with cap- or poly(A)-associated initiation factor complexes indicated that the association of SLBP with eIF4F required eIF4G and eIF3. Moreover, eIF4G and SLBP coimmunoprecipitate from mammalian cell extract and copurify as recombinant proteins. These data indicate that SLBP is functionally similar to PABP in that it stimulates the translation of histone mRNAs through an interaction with the 5′ terminus of the mRNA that is mediated by cap-associated initiation factors.
MATERIALS AND METHODS
mRNA constructs, electroporation, and luciferase assays.
The pT7-luc construct, in which the firefly luciferase coding region is under the control of the T7 promoter, has been described previously (16). The histone and related sequences were introduced from synthetic oligonucleotides into the BamHI and KpnI sites of the pT7-luc construct. Specifically, a 32-bp fragment containing the consensus histone stem-loop sequence was introduced 27 bases downstream of the stop codon (similar to the spacing present in histone mRNAs) of the luc reporter gene in a T7-based vector. A restriction site (AflII) incorporated into the construct immediately downstream of the stem-loop (luc-SLWT) allowed the in vitro production of capped luc mRNA terminating in the histone stem-loop. In vitro transcription was carried out as described previously (17). The total length of the 3′ untranslated region was 49 bases. The same luc mRNA construct with the consensus histone stem-loop sequence reversed (luc-SLR) served as the negative control. Yeast electroporation and luciferase assays were performed essentially as described previously (15, 17). For rapamycin treatment, cells were treated for 3 h prior to spheroplasting, during recovery, and during post-RNA delivery. Amino acid starvation was imposed by growing cells in SD medium containing 10 mM 3-aminotriazole for 2 h prior to spheroplasting, during recovery, and during post-RNA delivery.
Functional mRNA half-life analysis.
The rate of luciferase protein production was used as a measure of translational efficiency, and the length of time over which luciferase protein continued to accumulate was used to calculate message stability. Following the delivery of each mRNA construct by electroporation, aliquots of cells were removed at time intervals and luciferase assays were performed. The kinetics of luc mRNA translation were determined by following the appearance of protein as measured by enzyme activity plotted as a function of time. Once loaded onto polysomes, translation proceeds at a rate (i.e., the slope of each curve) that is dictated by its translational efficiency and for a period of time that is determined by the stability of the mRNA. The eventual degradation of the mRNA results in a decreased rate of protein accumulation. Following degradation of the mRNA, further accumulation of luciferase protein ceases, represented by the plateau of each curve. Those forms of an mRNA that are more stable will be translationally active longer, represented in a kinetic analysis by a longer period of time over which the protein will continue to accumulate. The functional half-life is defined as the amount of time needed to complete a 50% decay in the capacity of an mRNA to synthesize protein.
m7GTP-Sepharose 4B and poly(A)-agarose purification.
eIF4F from yeast and mammalian cells was purified with m7GTP-Sepharose as previously described (47). For yeast, the crude extract was made from spheroplasts (from 250 ml of cell culture) in 2.5 ml of buffer M (20 mM Tris [pH 7.5], 0.2 mM EDTA, 100 mM KCl, 5 mM MgCl2, 5% glycerol, 5 mM β-mercaptoethanol, and 1 mM dithiothreitol [DTT] with protease and phosphatase inhibitors). Mammalian 293 cells in buffer M were sonicated for 1 min, the cell debris was pelleted, and the supernatant was used for binding to m7GTP-Sepharose 4B. The m7GTP-Sepharose resin was washed three times with buffer M. Two hundred microliters of resin was used per 250 ml of yeast culture, and 100 μl of resin was used for two 100-mm-diameter plates of 293 cells. Binding was carried out at 4°C for 30 min with gentle shaking. The resin was collected, the supernatant was saved as the flowthrough, and the resin was washed three to five times with buffer M and eluted with 100 μM m7GTP.
For poly(A)-agarose purification, cell extract was prepared as described above, except that buffer PA [10 mM HEPES (pH 7.6), 100 mM potassium acetate, 1 mM CaCl2, 1 mM magnesium acetate, 5% glycerol, 1 mM DTT, and protease inhibitors] was used. Purification was carried out as previously described (32). After batch binding for 30 min at 4°C, the resin was collected and washed three times with buffer PA and protein was recovered in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis sample buffer.
Purification of His-tagged SLBP by metal affinity chromatography.
Co2+ resin (Clontech) was used as described in the manufacturer's protocol. Cells were disrupted in binding buffer (50 mM sodium phosphate [pH 7.0], 300 mM NaCl) with sonication. Extract was bound to resin at 4°C for 30 min and washed three times with binding buffer (with 5 mM imidazole), and bound protein was eluted with 150 mM imidazole.
Western analysis.
Anti-eIF4E and anti-PABP antisera were raised against recombinant protein. A human anti-eIF4A antibody was used to detect the yeast homolog. Anti-SLBP antibody was raised against the C-terminal 13-amino-acid region as described previously (58). Following resolution by SDS-polyacrylamide gel electrophoresis, protein was transferred to a nitrocellulose membrane which was blocked in 5% dry milk-phosphate-buffered saline containing 0.1% Tween 20 (PBST) for 2 h. The membrane was incubated with the primary antibody (used at dilutions of 1:1,000 to 1:2,000) for 1 to 2 h at room temperature, washed three times with PBST, incubated with secondary antibody (immunoglobulin G-horseradish peroxidase) for 1 h at room temperature, and washed three times with PBST, and the signal was revealed by an enhanced chemiluminescence reaction. Membranes were reprobed following stripping by incubating the membrane in buffer D (2% SDS, 62.5 mM Tris [pH 6.8], 100 mM β-mercaptoethanol) for 15 min at 60°C with shaking.
Immunoprecipitation.
Cell extract was prepared as described above with buffer IP (20 mM Tris [pH 7.5], 150 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 0.1% Triton X-100, 0.1% SDS, and protease and phosphatase inhibitors) and was precleared with protein A/G-Sepharose 4B resin. Antibody (normally 1 to 5 μl of antibody for 2 ml of extract) was added for 4 to 12 h at 4°C. Two hundred microliters (50% slurry) of protein A/G resin was added; the resin was incubated for 2 h at 4°C, collected, and washed three to five times with buffer IP; and the bound protein was used for Western analysis.
RESULTS
Translational stimulation by the SLBP/stem-loop complex can be recapitulated in yeast.
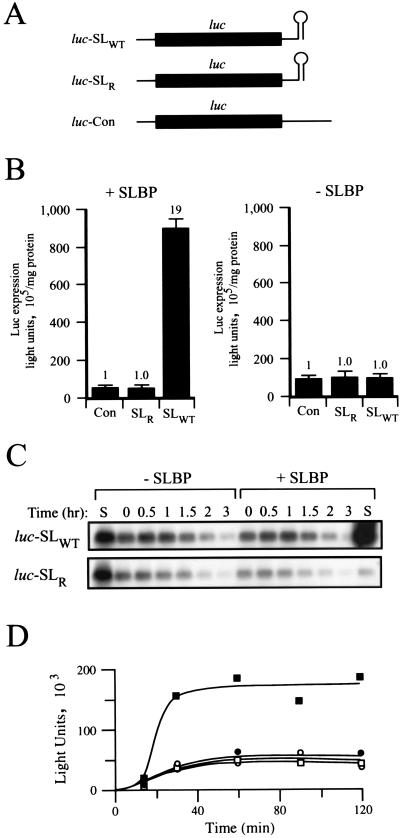
The 3′-terminal stem-loop of metazoan cell cycle-regulated histone mRNAs to which SLBP binds is the necessary and sufficient cis-acting element that directs the efficient translation of this mRNA class (17, 43, 58). S. cerevisiae lacks a homolog of mammalian SLBP, reflecting the finding that yeast histone mRNAs are polyadenylated. To determine whether SLBP is necessary and sufficient to direct efficient translation, human SLBP was expressed in yeast (strain INVSc1). Because yeast cannot correctly process a nonpolyadenlyated mRNA terminating in the histone 3′ stem-loop, mRNA terminating in the 3′-terminal stem-loop (i.e., luc-SLWT [Fig. 1A ]) was synthesized in vitro for introduction by electroporation into yeast. A 32-bp fragment containing the histone 3′ stem-loop was introduced downstream of the luciferase reporter gene (luc) in a T7-based vector (17) from which capped luc mRNA terminating in the histone stem-loop could be synthesized in vitro. The stem-loop was positioned 27 bases downstream of the luc stop codon, similar to the spacing present in histone mRNAs. As controls, luciferase mRNA terminating in the histone stem-loop of reverse orientation (luc-SLR) and a luciferase mRNA containing a 47-nucleotide 3′ untranslated region of random sequence (luc-Con) were included. Following delivery of mRNA (in six replicates) to yeast in the presence or absence of SLBP expression, luciferase activity was measured after 3 h of incubation (time sufficient to allow complete turnover of the introduced mRNA) to determine the extent of translation from each construct. In yeast expressing SLBP, translation from luc-SLWT was 19-fold greater than that from luc-SLR or luc-Con constructs (Fig. 1B, left panel). In the absence of SLBP, expression from mRNA terminating in the histone 3′ stem-loop was essentially identical to that from the control mRNAs (Fig. 1B, right panel).
FIG. 1.
SLBP is necessary and sufficient to direct translation from an mRNA terminating in the histone stem-loop structure. (A) Luciferase (luc) mRNA constructs used in this study: luc-SLWT, which terminates in the wild-type stem-loop; luc-SLR, in which the 3′ stem-loop is inverted; and luc-Con, which contains a 47-nucleotide random sequence that is the same length as that of stem-loop-containing constructs. (B) Expression from the capped luc mRNAs delivered to SLBP-expressing yeast (+SLBP) and control cells (−SLBP). Expression relative to that from the control mRNA luc-Con (set at a value of 1) for each panel is indicated above each histogram. Each mRNA was delivered to yeast as six replicates, and the average and standard deviation are shown. (C) Physical decay of luc-SLWT and luc-SLR mRNAs following their delivery to SLBP-expressing yeast (+SLBP) and control cells (−SLBP) by Northern analysis of total RNA extracted from an equal number of cells at time points after delivery. Lane S, in vitro-synthesized luc mRNA. (D) Functional stability of lucSLWT (• and ▪) mRNAs following their delivery to control yeast (○ and •) or yeast expressing SLBP (□ and ▪). Translation from the mRNAs was followed by measurement of luciferase expression at the time points following mRNA delivery. Once the mRNAs had been degraded, no further translation could occur and the accumulation of luciferase ceased, resulting in the observed plateaus.
Because the histone stem-loop structure regulates the stability as well as the translatability of an mRNA in mammalian cells (17, 34, 41), the physical decay of the introduced luciferase mRNAs in SLBP-expressing and control yeast was examined. Northern analysis of capped luc-SLWT and luc-SLR mRNAs introduced into nonsynchronized cells revealed similar stabilities in yeast in the absence or presence of SLBP (Fig. 1C), indicating that the increased expression from luc-SLWT in SLBP-expressing yeast resulted from an increase in its translation. Because the stability of cell cycle-regulated histone mRNAs in mammalian cells is controlled by the cell cycle, no alteration in stability for an mRNA terminating in the histone stem-loop structure would be expected in nonsynchronized cells. Determination of the functional mRNA half-life confirmed the conclusion drawn from the Northern analysis, in which each mRNA was translationally active for the same length of time (Fig. 1D). These results do not preclude the possibility that the histone stem-loop structure may regulate mRNA stability in yeast expressing SLBP, but rather, they demonstrate that the observed increase in expression from luc mRNA terminating in the histone stem-loop structure in nonsynchronized cells results from increased translatability.
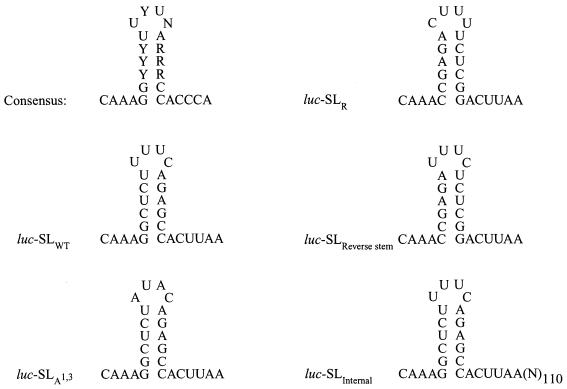
The primary and secondary structure of the metazoan histone stem-loop is highly conserved (Fig. 2). These elements include a 6-bp stem consisting of two GC base pairs at the base, a set of three pyrimidine-purine base pairs forming the central portion of the stem and a UA base pair at the top, and a 4-base loop in which the first and third positions are uridines and the fourth position varies. Mutations to the conserved positions of the loop or stem disrupt its 3′-end processing (14, 42) and translational (17) functions and abolish SLBP binding to the histone stem-loop in animal cells (61). To investigate whether the same mutations would be similarly deleterious to the function of the histone stem-loop in yeast expressing SLBP, the expression from luciferase mRNAs terminating in mutant stem-loops (Fig. 2) was examined. Altering the two conserved uridines in the loop to adenosines (luc-SLA1,3) abolished stem-loop function in SLBP-expressing yeast as did inverting either the entire stem-loop (luc-SLR) or just the stem (luc-SLReverse stem) (Table 1). Internalization of the stem-loop (i.e., luc-SLInternal) also resulted in a substantial, although not complete, loss of function in good agreement with the results obtained in mammalian cells (17). These data demonstrate that, as with mammalian cells, the conserved features of the histone stem-loop that are required for binding SLBP (61) are required for SLBP-mediated translational regulation in yeast.
FIG. 2.
Mutation of conserved positions within the stem-loop disrupts its translational function in yeast. (A) The consensus 3′ stem-loop of histone mRNAs derived from metazoans. Y, pyrimidine; R, purine; N, any nucleotide. The wild-type stem-loop (luc-SLWT) is also shown. In luc-SLA1,3, the two conserved uridines in the loop were changed to adenosines; in luc-SLR, the stem-loop was inverted; and in luc-SLReverse stem, only the stem was inverted. luc-Con served as the negative control. For the luc-SLInternal mRNA, the histone stem-loop was internalized by restricting the luc-SLWT DNA construct at a PvuII 116 bases downstream of the stem-loop.
TABLE 1.
Expression from the luc mRNAs delivered to yeast in the absence or presence of SLBP
| Construct | No SLBP (light units [105]/mg of protein) | SLBP (light units [105]/mg of protein) | Fold increase in expression by SLBPa |
|---|---|---|---|
| luc-SLWT | 27.9 ± 1.7 | 197 ± 6.2 | 7.1 |
| luc-SLA1,3 | 33.1 ± 4.8 | 36.5 ± 1.0 | 1.1 |
| luc-SLR | 25.2 ± 1.4 | 17.7 ± 1.1 | 0.7 |
| luc-SLReverse stem | 17.9 ± 2.9 | 19.2 ± 2.1 | 1.1 |
| luc-SLInternal | 37.5 ± 3.1 | 60.1 ± 10.8 | 1.6 |
| luc-Con | 18.0 ± 0.6 | 14.6 ± 0.6 | 0.8 |
The fold increase conferred by the wild-type and mutant structures was determined as the ratio of luciferase expression in the presence of SLBP to that in the absence of SLBP.
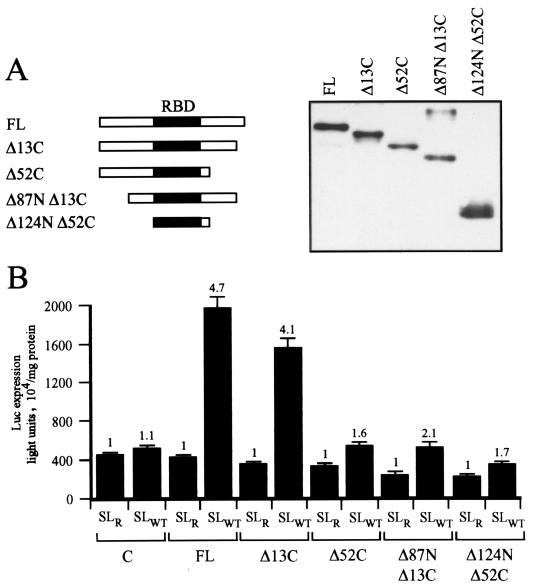
To examine whether mutations to SLBP would affect its translational function, N-terminal- and/or C-terminal-truncated SLBP proteins (58) (Fig. 3A) were expressed in yeast. The RNA-binding domain was retained in all of the deletions, and all mutant proteins remained competent for binding RNA and RNA processing (14, 58). Western analysis using an anti-His-tagged antibody confirmed that all His-tagged proteins were expressed to a similar level (Fig. 3A). The translational regulatory function of the wild-type and mutant SLBP proteins was determined by measuring the expression from capped luc-SLWT and luc-SLR mRNAs delivered to yeast expressing each SLBP construct and calculating the extent to which the histone stem-loop increased luciferase translation. Deletion of the C-terminal 13 amino acids (Δ13C) did not substantially alter SLBP translational function (Fig. 3B). However, deletion of the C-terminal 52 amino acids (Δ52C) or the N-terminal 87 amino acids (Δ87N Δ13C) significantly reduced SLBP translational function (Fig. 3B). These data indicate that the domains on each side of the RNA-binding domain are required for SLBP translational function either for protein interaction or for correct protein folding. Together, these data demonstrate that SLBP-mediated translation can be recapitulated in yeast and that SLBP and the histone stem-loop represent a two-component regulatory complex that is necessary and sufficient for this function.
FIG. 3.
SLBP-mediated translation from the 3′ stem-loop requires intact SLBP. (A) His6-tagged SLBP deletion mutants shown were expressed in yeast, and their expression was determined by Western analysis with an anti-His antibody. FL, full-length SLBP; RBD, RNA-binding domain. (B) Expression from capped luc-SLWT and luc-SLR mRNAs in yeast expressing wild-type and mutant SLBP is shown. Expression from luc-SLWT relative to that from luc-SLR (set at a value of 1) for each pair of mRNAs is indicated above each histogram.
SLBP translational function requires eIF4E, eIF4G, and eIF3.
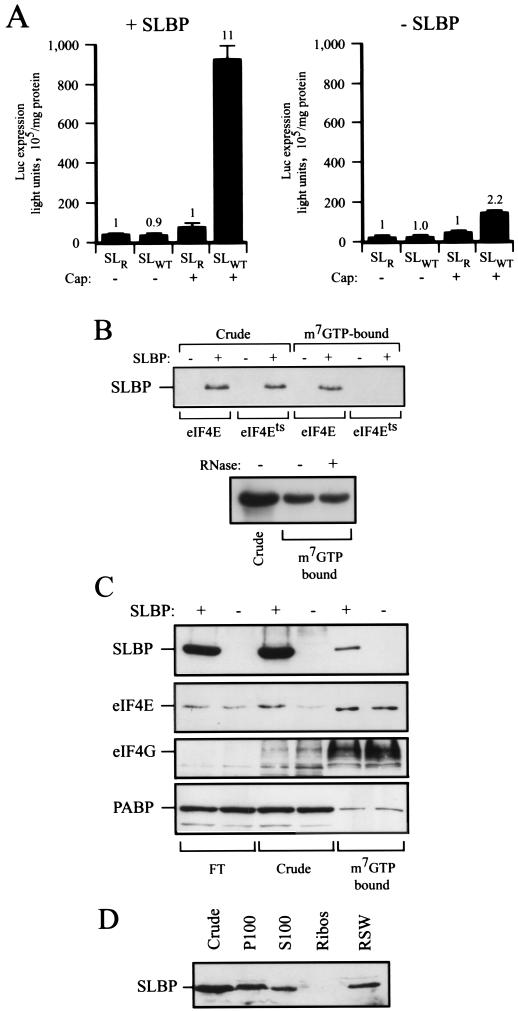
It has been shown previously that the histone stem-loop requires the 5′ cap for its function during translation (17), suggesting an interaction between the SLBP/stem-loop complex and the 5′ cap or the eIF4F that binds to the 5′ cap. Because an examination of the translation of capped or uncapped mRNAs in yeast had demonstrated the functional interaction between the 5′ cap and poly(A) tail (16, 40, 46), the expression from capped or uncapped luc-SLWT and luc-SLR mRNAs was examined in yeast in the presence or absence of SLBP. In the presence of SLBP, the expression from uncapped luc-SLWT mRNA was similar to that of luc-SLR mRNA but was 11-fold greater when the mRNAs were capped (Fig. 4A, left panel), a degree of stimulation that was not observed in the absence of SLBP (Fig. 4A, right panel). The data can also be examined to determine the extent to which the function of the cap is stimulated by the presence of the histone stem-loop. In SLBP-expressing yeast, the addition of a cap to luc-SLR mRNA increased expression 2.0-fold but increased expression from luc-SLWT 21-fold (Fig. 4A, left panel). These data demonstrate that the 5′ cap and histone stem-loop functionally interact in yeast expressing SLBP and are in good agreement with previous observations in mammalian cells (17).
FIG. 4.
SLBP requires a functional interaction with the 5′ cap and copurifies with eIF4F. (A) Expression from capped and uncapped luc-SLWT and -luc-SLR mRNAs in yeast in the presence (+SLBP) or absence (−SLBP) of SLBP is shown. Expression from luc-SLWT relative to that from luc-SLR (set at a value of 1) for each pair of mRNAs is indicated above each histogram. (B) (Top panel) Extract from wild-type and eIF4Ets mutant (strain 4-2) yeast with or without SLBP expression at the nonpermissive temperature (37°C) was bound to m7GTP-Sepharose. The presence of SLBP in crude extract or in the m7GTP-bound fraction was detected by Western analysis. (Bottom panel) Soluble protein from wild-type CW04 yeastexpressing SLBP with or without RNase A treatment was bound to m7GTP-Sepharose, and the presence of SLBP in the m7GTP-bound fraction was detected by Western analysis. (C) Soluble protein from control yeast or yeast expressing SLBP was bound to m7GTP-Sepharose, and the presence of SLBP, eIF4E, eIF4G, and PABP in the crude extract (Crude), flowed through m7GTP-Sepharose (FT), and in the m7GTP-Sepharose-bound fraction (m7GTP bound) was determined by Western analysis. (D) Extract from CW04 expressing SLBP was fractionated into the ribosomal fraction (P100), the supernatant (P100), the ribosomal fraction following the high-salt wash of P100 with 0.5 M NaCl (Ribos), and the ribosomal salt wash fraction representing ribosomal-bound protein that eluted from P100 with 0.5 M NaCl (RSW). The presence of SLBP in the crude extract and each fraction was detected by Western analysis.
To investigate whether SLBP binds to the cap-binding complex, we examined whether SLBP expressed in yeast could be retained on m7GTP-Sepharose. To determine whether SLBP binding to m7GTP-Sepharose was eIF4E dependent, SLBP was expressed in an eIF4Ets mutant strain in which the temperature sensitive (Ts) mutation of eIF4E abolishes its binding at the nonpermissive temperature of 37°C (1, 3). Expression of SLBP was readily detected in crude extract from the wild-type and eIF4Ets mutant strains at 37°C (Fig. 4B). SLBP from wild-type cells was retained on m7GTP-Sepharose, but that from eIF4Ets mutant cells failed to bind at 37°C, suggesting that SLBP binding to m7GTP-Sepharose is eIF4E dependent. Retention of SLBP on m7GTP-Sepharose was RNA independent, as its binding was unaffected by pretreatment of the extract with RNase A (Fig. 4B).
eIF4E is the small subunit of eIF4F in which eIF4E and eIF4A are physically associated with the scaffold protein eIF4G (18, 27, 37). PABP also interacts with eIF4G (55) and copurifies with eIF4F. To determine whether the presence of SLBP in the eIF4F complex affects the composition of this complex, eIF4F was isolated from control and SLBP-expressing yeast on m7GTP-Sepharose in the absence of reporter mRNA. Components of the complex isolated in the absence or presence of SLBP were visualized by Western analysis (Fig. 4C). SLBP copurified with eIF4F on m7GTP-Sepharose and did not alter the amount of eIF4G associated with eIF4E. SLBP copurified with eIF4F even if the extract was pretreated with RNase (data not shown), suggesting that its interaction with eIF4F is RNA independent. SLBP was present in a high ribosomal-salt-wash fraction of yeast ribosomes in the absence of the stem-loop containing reporter mRNA (Fig. 4D), the data supporting the conclusion that SLBP copurifies with eIF4F.
As the association of SLBP with the 5′ cap was eIF4E dependent, we used wild-type yeast and strains harboring mutations in key translation initiation factors to investigate the functional interaction between SLBP and the eIF4F complex. Null mutants were employed for those factors that are not essential, e.g., eIF4B or CAF20, whereas Ts mutants or depletion approaches were employed for those factors that have been shown to have an essential role, including eIF4E, eIF4G, eIF4A, eIF2, eIF3, and PABP. For the analysis, capped luc-SLWT and luc-SLR mRNAs were delivered to each mutant in the absence or presence of SLBP. Since the depletion or loss of an initiation factor would be expected to reduce translation nonspecifically, only the loss of the histone stem-loop-mediated increase in luciferase expression would constitute grounds for concluding that the factor concerned is required for the SLBP-mediated stimulation of translation.
Because SLBP binding to m7GTP was lost in the eIF4Ets mutant strain at the nonpermissive temperature (Fig. 4B), we examined whether the translational function of SLBP required eIF4E. Expression from luc-SLWT mRNA in the wild-type eIF4E strain expressing SLBP was 9.2-fold higher than that from luc-SLR mRNA (Table 2). In agreement with the data presented in Fig. 1, no enhancement was observed in the absence of SLBP expression. Similar results were observed at the nonpermissive temperature (Table 2). Expression from luc-SLWT mRNA in the eIF4Ets mutant strain expressing SLBP was 5.4-fold higher than that from luc-SLR mRNA at the permissive temperature (Table 2). However, at 37°C, translation from the luc-SLWT mRNA was preferentially affected, resulting in a substantial reduction in the SLBP-mediated regulation (Table 2). As cell growth at 37°C did not influence the expression of SLBP, these data suggest that SLBP not only coisolates with eIF4E (Fig. 4) but functionally requires this factor.
TABLE 2.
SLBP translation function requires eIF4E but not caf20 or eIF4B
| Temp and factor | No SLBP (light units, 106)
|
SLBP (light units, 106)
|
||||
|---|---|---|---|---|---|---|
| SLR | SLWT | SLWT/SLR | SLR | SLWT | SLWT/SLR | |
| 24°C | ||||||
| eIF4E | 2.04 ± 0.27 | 2.38 ± 0.27 | 1.2 | 2.71 ± 0.60 | 25.0 ± 0.96 | 9.2 |
| eIF4Ets | 1.49 ± 0.12 | 1.91 ± 0.20 | 1.3 | 2.26 ± 0.43 | 12.2 ± 0.85 | 5.4 |
| 37°C | ||||||
| eIF4E | 1.97 ± 0.24 | 2.26 ± 0.33 | 1.1 | 1.61 ± 0.23 | 13.8 ± 0.64 | 8.6 |
| eIF4Ets | 1.15 ± 0.24 | 1.01 ± 0.15 | 0.9 | 1.18 ± 0.15 | 2.09 ± 0.34 | 1.8 |
| 24°C | ||||||
| caf20 | 0.26 ± 0.04 | 0.26 ± 0.05 | 1.0 | 0.24 ± 0.04 | 1.96 ± 0.31 | 8.2 |
| eIF4B | 0.17 ± 0.02 | 0.16 ± 0.01 | 0.9 | 0.16 ± 0.02 | 2.08 ± 0.29 | 1.3 |
The requirement for nonessential factors involved in the translation initiation in SLBP-mediated translational enhancement was also examined. CAF20 inhibits eIF4G binding to eIF4E (2, 10), whereas eIF4B promotes the RNA helicase function of eIF4A and eIF4F (8). Expression from luc-SLWT and luc-SLR mRNAs was examined in caf20- or eIF4B-null strains expressing SLBP (Table 2). In each case, the degree of SLBP-mediated translational enhancement was similar to that determined for the wild-type strain CW04 (see below), suggesting that neither factor is specifically required for SLBP function.
We next investigated whether SLBP functionally requires eIF4G. eIF4G is expressed from two genes in yeast, and the proteins (eIF4G1 and eIF4G2) are only 53% homologous (19). Mutant strains expressing only eIF4G1 (tif4632) or eIF4G2 (tif4631) are viable, but the double null is inviable (19). Because the depletion of both isoforms of eIF4G can be achieved by treatment of the yeast with rapamycin (4), we examined the function of SLBP in wild-type or mutant strains expressing only eIF4G1 or eIF4G2 before or after rapamycin treatment. Treatment of yeast with rapamycin results in the rapid degradation of eIF4G without affecting the levels of other eIF4F subunits, i.e., eIF4E and eIF4A (4). Although rapamycin treatment has been reported to affect the transcription of some genes in yeast (45), rapamycin selectively reduces the level of eIF4G through the destabilization of the existing eIF4G protein (4). We confirmed that no degradation of eIF4E (see below) or eIF4A (data not shown) was observed following rapamycin treatment. Moreover, rapamycin treatment did not reduce the level of PABP (see below) or eIF3 (data not shown). Successful depletion of eIF4G following rapamycin treatment was confirmed (data not shown). Expression from luc-SLWT mRNA in the wild-type CW04 strain expressing SLBP was 9.2-fold higher than that from luc-SLR mRNA (Table 3). This SLBP-mediated enhancement was reduced to control levels in cells treated with rapamycin (Table 3) despite the fact that no alteration in the level of SLBP expression was observed (data not shown). The translational enhancement by SLBP was observed in both the eIF4G1-deleted and eIF4G2-deleted strains (Table 3), suggesting that either isoform of eIF4G was capable alone of supporting SLBP function. The lower level of translational enhancement observed in each mutant was reproducible, possibly resulting from reduced levels of total eIF4G. As with the wild-type cells, the translational function of SLBP was virtually lost in the eIF4G1 or eIF4G2 mutant strains following rapamycin treatment (Table 3), confirming that SLBP function is dependent on eIF4G.
TABLE 3.
SLBP translation function requires eIF4G
| Condition and factor or strain | No SLBP (light units 106)
|
SLBP (light units 106)
|
||||
|---|---|---|---|---|---|---|
| SLR | SLWT | SLWT/SLR | SLR | SLWT | SLWT/SLR | |
| No rapamycin | ||||||
| CW04 | 1.32 ± 0.08 | 1.39 ± 0.26 | 1.1 | 1.59 ± 0.29 | 14.7 ± 0.63 | 9.2 |
| tif4631 | 1.24 ± 0.20 | 0.96 ± 0.11 | 0.8 | 0.96 ± 0.13 | 7.53 ± 0.33 | 7.8 |
| tif4632 | 1.03 ± 0.16 | 1.02 ± 0.19 | 1.0 | 1.32 ± 0.12 | 5.54 ± 0.19 | 4.2 |
| With rapamycin | ||||||
| CW04 | 2.02 ± 0.40 | 2.21 ± 0.48 | 1.1 | 1.56 ± 0.19 | 3.47 ± 0.14 | 2.2 |
| tif4631 | 0.70 ± 0.08 | 0.81 ± 0.22 | 1.2 | 0.37 ± 0.11 | 0.62 ± 0.37 | 1.7 |
| tif4632 | 0.27 ± 0.18 | 0.39 ± 0.17 | 1.4 | 0.43 ± 0.17 | 0.67 ± 0.29 | 1.6 |
| 24°C | ||||||
| eIF4Ats | 1.19 ± 0.42 | 1.23 ± 0.19 | 1.0 | 0.97 ± 0.13 | 3.28 ± 0.66 | 3.4 |
| eIF4AWT | 1.89 ± 0.35 | 1.70 ± 0.45 | 0.9 | 1.79 ± 0.10 | 5.89 ± 0.92 | 3.3 |
| 37°C | ||||||
| eIF4Ats | 0.58 ± 0.08 | 1.12 ± 0.09 | 1.9 | 0.83 ± 0.10 | 3.30 ± 0.28 | 4.0 |
| eIF4AWT | 1.48 ± 0.26 | 2.19 ± 0.59 | 1.5 | 1.37 ± 0.37 | 7.50 ± 0.84 | 5.4 |
To determine whether eIF4A is required for SLBP-mediated translational enhancement, expression from luc-SLWT and luc-SLR mRNAs was measured in an eIF4Ats mutant strain which is isogenic with CW04. In strains expressing wild-type or mutant eIF4A from a multicopy vector, a lower level of SLBP-mediated enhancement of translation from luc-SLWT mRNA relative to that from luc-SLR mRNA was observed even at the permissive temperature (Table 3). However, in contrast to the findings with eIF4E (Table 2), no reduction in the SLBP-mediated enhancement of translation was observed in the eIF4Ats mutant strain at the nonpermissive temperature (Table 3), suggesting that eIF4A may not be specifically required for the SLBP-mediated regulation of translation. The reproducible decrease in SLBP-mediated enhancement observed in strains expressing wild-type and mutant eIF4A may be a consequence of their expression from a multicopy vector.
eIF4G functions to recruit eIF3, which in turn is responsible for promoting binding of the 40S ribosomal subunit to an mRNA (31). To determine whether SLBP requires eIF3, a mutant strain containing a Ts mutation in the Prt1 (eIF3b) subunit (22, 39) was employed. Expression from luc-SLWT mRNA in the wild-type eIF3 strain expressing SLBP was 9.1-fold higher than that from luc-SLR mRNA, which was altered little when the cells were shifted to 37°C (Table 4). SLBP-mediated translational enhancement was reproducibly lower in the prt1-1 mutant strain at the permissive temperature and was abolished when the cells were shifted to 37°C (Table 4) despite the fact that no alteration in the level of SLBP expression was observed, the data suggesting that SLBP function is dependent on eIF3.
TABLE 4.
SLBP translation function requires eIF3
| Temp and factor | No SLBP (light units, 106)
|
SLBP (light units, 106)
|
||||
|---|---|---|---|---|---|---|
| SLR | SLWT | SLWT/SLR | SLR | SLWT | SLWT/SLR | |
| 24°C | ||||||
| eIF3b | 0.94 ± 0.11 | 1.17 ± 0.09 | 1.2 | 1.01 ± 0.18 | 9.18 ± 0.29 | 9.1 |
| eIF3bts | 0.73 ± 0.08 | 0.87 ± 0.08 | 1.2 | 0.70 ± 0.20 | 3.76 ± 0.36 | 5.4 |
| 37°C | ||||||
| eIF3b | 0.65 ± 0.06 | 0.76 ± 0.05 | 1.2 | 0.54 ± 0.04 | 4.71 ± 0.28 | 8.7 |
| eIF3bts | 0.39 ± 0.05 | 0.34 ± 0.06 | 0.9 | 0.28 ± 0.03 | 0.38 ± 0.04 | 1.4 |
Translation initiation is also controlled at the level of initiator tRNA binding to the 40S ribosomal subunit, mediated by the physiological regulation of eIF2 activity (27, 28, 29). For example, increased phosphorylation of the α subunit of eIF2 (eIF2α) at serine 51 by GCN2 in response to amino acid starvation reduces eIF2 activity in S. cerevisiae (28, 29). Mutation of Ser-51 to alanine (eIF2α-S51A) prevents phosphorylation and results in constitutive activity, whereas mutation of the serine to aspartic acid (eIF2α-S51D) mimics phosphorylation and results in constitutively reduced activity (11). To examine whether changes in eIF2α function affect SLBP function, expression from luc-SLWT and luc-SLR mRNAs was measured in the absence or presence of SLBP in wild-type, GCN2 mutant yeast and strains expressing eIF2α containing either the S51A or S51D mutations (Table 5). SLBP function in these strains was examined in cells not experiencing amino acid starvation (i.e., for histidine) and following starvation for histidine, which induces GCN2 activity. SLBP was expressed to a similar level in all strains (data not shown). In nonstarved cells, translation from luc-SLWT mRNA in wild-type eIF2α/GCN2 cells expressing SLBP was 13-fold higher than that from luc-SLR mRNA (Table 5). No reduction in the degree of SLBP-mediated enhancement of translation was observed in eIF2α-S51A/GCN2 or eIF2α/gcn2 cells (Table 5). Furthermore, in eIF2α-S51D/GCN2 cells, the degree of SLBP-mediated enhancement was not reduced (Table 5). In starved cells, the SLBP-mediated enhancement of translation was abolished in eIF2α/GCN2 cells as well as in eIF2α-S51A/GCN2 and eIF2α-S51D/GCN2 cells. A similar loss in SLBP function was observed in stationary-phase cells (data not shown). Partial retention of SLBP activity was observed in eIF2α/gcn2 cells, possibly indicating that eIF2 may be partially functionally required for SLBP or that the loss of GCN2 function may lead to changes in other components of the translational machinery that are involved in the activity of SLBP. These data suggest that amino acid starvation results in changes in the translation machinery that result in the loss of SLBP-mediated regulation and are partially controlled by GCN2 but may not involve changes in eIF2 phosphorylation.
TABLE 5.
eIF2 is not specifically required for SLBP translation function
| Condition and cell type | No SLBP (light units, 106)
|
SLBP (light units, 106)
|
||||
|---|---|---|---|---|---|---|
| SLR | SLWT | SLWT/SLR | SLR | SLWT | SLWT/SLR | |
| Nonstarved | ||||||
| eIF2α/GCN2 | 0.17 ± 0.04 | 0.20 ± 0.04 | 1.2 | 0.13 ± 0.03 | 1.67 ± 0.58 | 13 |
| eIF2α/gcn2 | 0.29 ± 0.05 | 0.31 ± 0.07 | 1.1 | 0.10 ± 0.06 | 1.79 ± 0.72 | 18 |
| eIF2α-S51A/GCN2 | 0.12 ± 0.03 | 0.15 ± 0.04 | 1.3 | 0.07 ± 0.04 | 1.53 ± 0.56 | 22 |
| eIF2α-S51D/GCN2 | 0.19 ± 0.03 | 0.25 ± 0.02 | 1.3 | 0.07 ± 0.02 | 1.19 ± 0.61 | 17 |
| Starved | ||||||
| eIF2α/GCN2 | 0.48 ± 0.09 | 0.68 ± 0.07 | 1.4 | 0.80 ± 0.17 | 0.90 ± 0.60 | 1.1 |
| eIF2α/gcn2 | 0.26 ± 0.05 | 0.22 ± 0.02 | 0.8 | 0.72 ± 0.17 | 2.54 ± 0.60 | 3.5 |
| eIF2α-S51A/GCN2 | 1.45 ± 0.30 | 1.48 ± 0.10 | 1.0 | 0.97 ± 0.12 | 1.14 ± 0.28 | 1.2 |
| eIF2α-S51D/GCN2 | 1.37 ± 0.25 | 0.88 ± 0.38 | 0.6 | 2.34 ± 0.15 | 3.02 ± 0.99 | 1.3 |
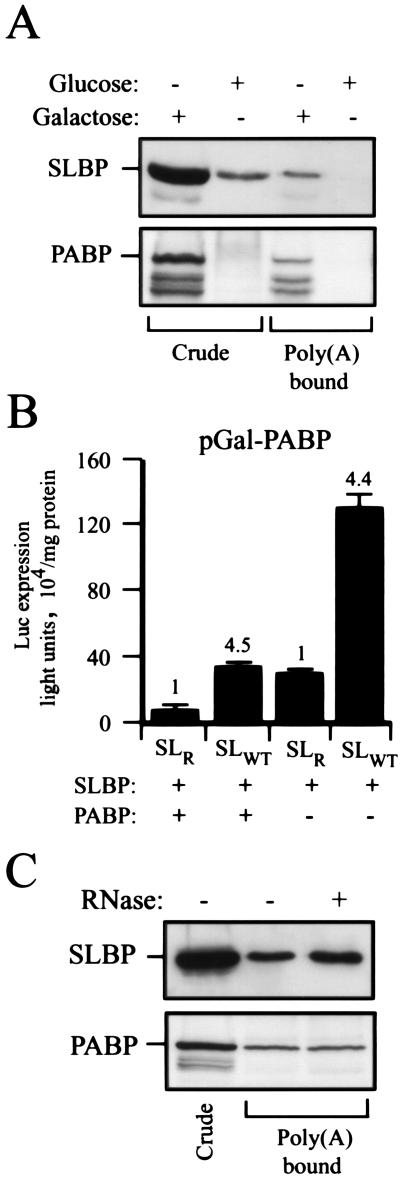
PABP interacts with eIF4G and functions to promote 48S complex formation (48, 54). Although cell cycle-regulated histone mRNAs do not contain a poly(A) tail, SLBP may still interact with and require PABP. To determine whether SLBP functionally requires PABP, expression from luc-SLWT and luc-SLR mRNAs was measured in a mutant strain expressing PABP under the control of a galactose-inducible (Gal1) promoter (49). Yeast were grown in galactose-containing medium and switched to glucose for 20 h to deplete PABP in the cells, which was confirmed by Western blotting of both the crude extracts (Fig. 5A, first two lanes) and of poly(A)-agarose bound fractions (Fig. 5A, last two lanes). In yeast grown in the presence of galactose, SLBP was recovered on poly(A)-agarose in association with PABP in the absence of added mRNA. Expression from luc-SLWT mRNA in yeast expressing SLBP and grown in galactose-containing medium was 4.5-fold higher than that from luc-SLR mRNA (Fig. 5B) and was altered little following the depletion of PABP, suggesting that SLBP does not require PABP. Despite the fact that a substantial decrease in the level of PABP had no effect on SLBP-mediated regulation, we cannot rule out the possibility that PABP may be required if the small amount of PABP remaining after the depletion is sufficient to fulfill its role in this mechanism.
FIG. 5.
SLBP copurifies with PABP on poly(A)-agarose. (A) Yeast, in which expression of PABP is under the control of the galactose-inducible promoter (pGAL-PABP), was grown in galactose and switched to either galactose- or glucose-containing medium and grown for a further 20 h. Soluble protein from an equal number of cells was bound to poly(A)-agarose. The presence of SLBP or PABP in crude extract (Crude) or in the poly(A)-bound fraction was determined by Western analysis. (B) Luciferase expression from capped luc-SLWT and luc-SLR mRNAs was examined in the galactose- or glucose-grown yeast shown to the left. Expression from luc-SLWT relative to that from luc-SLR (set at a value of 1) for each pair of mRNAs is indicated above each histogram. (C) Extract from yeast expressing SLBP treated with or without RNase A was bound to poly(A)-agarose, and SLBP and PABP were detected by Western analysis.
To further investigate SLBP coisolation with PABP, PABP from yeast expressing SLBP was allowed to bind to poly(A)-agarose. To eliminate the possibility of any RNA tethering between SLBP and PABP, the extract was treated with RNase A, which was inactivated with DTT (data not shown) prior to loading onto poly(A)-agarose. As shown in Fig. 5C, SLBP was present in the poly(A)-bound fraction whether or not the extract was pretreated with RNase A but did not bind poly(A)-agarose in the absence of PABP (Fig. 5A). These data suggest that, although SLBP can coisolate in a complex containing PABP, the latter is not required for the SLBP-mediated enhancement of translation with reporter mRNAs.
Interaction of SLBP with eIF4F requires eIF4G and eIF3 but not eIF4E or eIF4A.
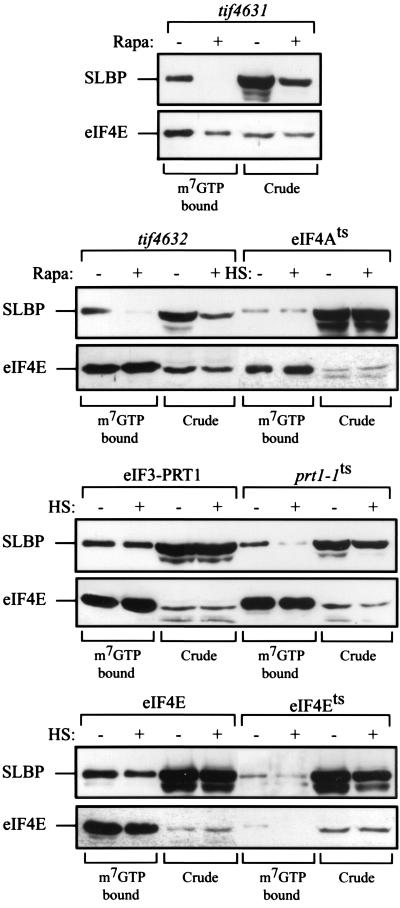
As the genetic analysis in yeast revealed a functional requirement for eIF4E, eIF4G, and eIF3 for the SLBP-mediated regulation of translation and SLBP can be retained on m7GTP-Sepharose (Fig. 4), these data suggest that SLBP may physically interact with one or more of these proteins. Such an interaction may promote a physical interaction between the termini of histone mRNAs, functionally circularizing the mRNA and facilitating its translation. The requirements for the interaction of SLBP with eIF4F were further investigated by examining the binding of SLBP to m7GTP-Sepharose when SLBP was expressed in the same strains described above (eIF4G2 [tif4631], eIF4G1 [tif4632], eIF4Ets, eIF4Ats, or prt1-1). In addition, the requirement for eIF4G was investigated by examining whether SLBP from control or rapamycin-treated tif4631 and tif4632 cells was retained on m7GTP-Sepharose. As observed in Fig. 4, SLBP was retained on m7GTP-Sepharose from extract prepared from tif4631 and tif4632 mutants (Fig. 6). However, retention of SLBP on m7GTP-Sepharose was lost when either mutant was treated with rapamycin (Fig. 6). The effect of rapamycin treatment on reducing the level of eIF4G appeared to be selective, as the treatment had no effect on the level of eIF4E in crude extract (Fig. 6), as has been reported previously (4) as well as on eIF4A, eIF3 (see above), and PABP (see below), suggesting that the reduction in the coisolation of SLBP was not a result of a decrease in eIF4E or these other factors. The observation that SLBP was retained on m7GTP-Sepharose in tif4631 or tif4632 mutants prior to but not following their treatment with rapamycin suggests that the association of SLBP with eIF4F requires eIF4G. Retention of SLBP on m7GTP-Sepharose was observed in the eIF4Ets mutant at the permissive temperature but not at the nonpermissive temperature (Fig. 6), confirming the requirement for eIF4E in mediating the binding of SLBP to m7GTP-Sepharose, observed in Fig. 4. In contrast, the retention of SLBP on m7GTP-Sepharose when expressed in the eIF4Ats mutant was similar at the permissive and nonpermissive temperatures (Fig. 6), suggesting that eIF4A is not required for SLBP to copurify with eIF4F. Retention of SLBP on m7GTP-Sepharose was unaffected in the prt1-1 (eIF3bts) mutant strain at the permissive temperature but not at 37°C (Fig. 6), indicating that active eIF3 is at least partially required for SLBP to copurify with eIF4F.
FIG. 6.
eIF4G and eIF3 are required for SLBP copurification with eIF4E. Soluble protein from each indicated yeast strain expressing SLBP was bound to m7GTP-Sepharose. The presence of SLBP and eIF4E in crude extract (Crude) or in the m7GTP-bound fraction was determined by Western analysis. Rapamycin (Rapa) treatment of tif4631 and tif4632 mutants to deplete eIF4G or treatment at 37°C of the eIF4Ats, prt1-1, and eIF4Ets mutant strains is as indicated. HS, heat stress at the nonpermissive temperature.
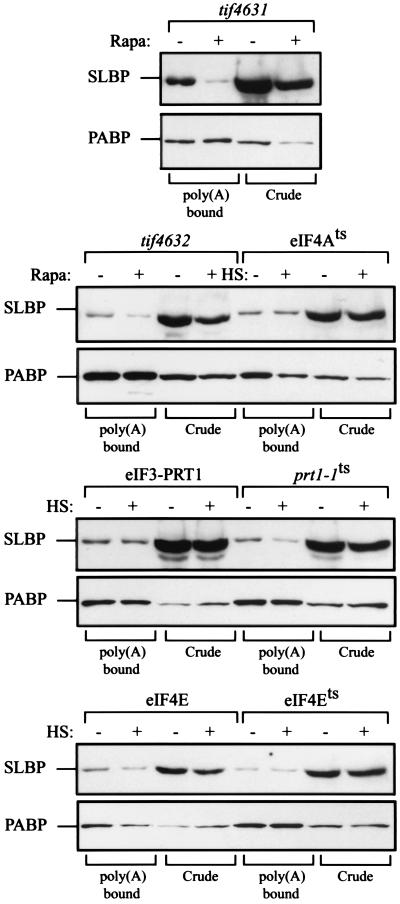
We also examined whether the coisolation of SLBP with PABP observed in Fig. 5C was dependent on eIF4E, eIF4G, or eIF3. To this end, extracts from the same strains expressing SLBP were loaded onto poly(A)-agarose resin and the retention of SLBP was determined by Western analysis. Figure 7 shows that, while SLBP was recovered with PABP in untreated tif4631 or tif4632 mutants cells, its coisolation with PABP was substantially reduced following rapamycin treatment, particularly in the tif4631 mutant. Rapamycin treatment had little to no effect on the level of PABP and had no effect on the amount of PABP recovered on poly(A)-agarose resin (Fig. 7), suggesting that the reduction in the coisolation of SLBP in rapamycin-treated cells was not a result of a decrease in PABP (or eIF4E, eIF4A, or eIF3). Moreover, once normalized to the level of PABP in untreated crude extract, the level of SLBP in extract from rapamycin-treated cells was not substantially different from that from nontreated cells. These data suggest that eIF4G was required for the coisolation of SLBP with PABP. Retention of SLBP on poly(A)-agarose resin was not dependent on eIF4E or eIF4A but did require eIF3 (Fig. 7). These data suggest that SLBP does not interact directly with PABP or eIF4E, but rather, it associates with the eIF4F/PABP complex via its association with eIF4G and, in part, with eIF3. Despite detecting no decrease in eIF4E, PABP, eIF4A, or eIF3 in rapamycin-treated cells, it is formally possible that a reduction in a protein not examined may also be involved in the coisolation of SLBP in this assay.
FIG. 7.
eIF4G and eIF3 are required for SLBP copurification with PABP. Soluble protein from each indicated yeast strain expressing SLBP was bound to poly(A)-agarose. The presence of SLBP and PABP in crude extract (Crude) or in the poly(A)-bound fraction was determined by Western analysis. Rapamycin treatment of tif4631 and tif4632 mutants to deplete eIF4G or treatment at 37°C of the eIF4Ats, prt1-1, and eIF4Ets mutant strains is as indicated. HS, heat stress at the nonpermissive temperature.
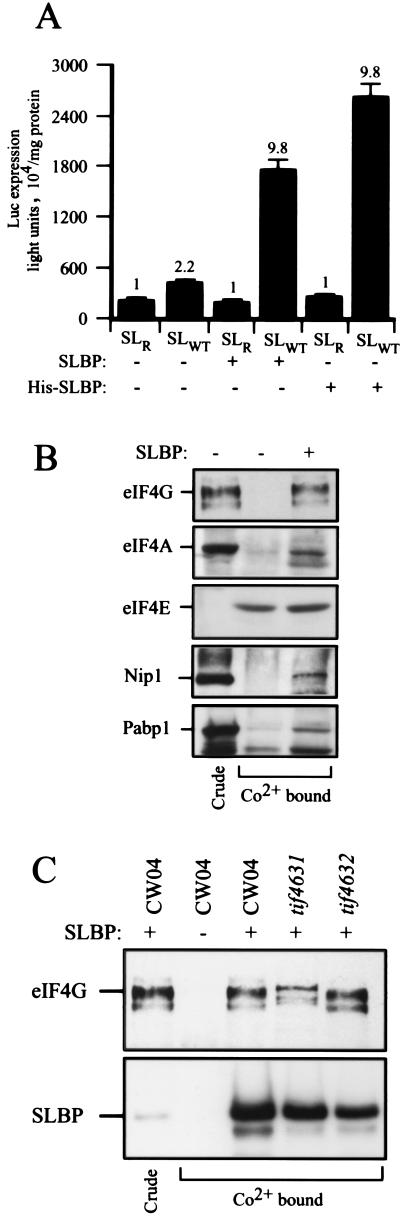
Purification of His6-tagged SLBP was also used to investigate its in vivo interactions. As shown in Fig. 8A, H is6-tagged SLBP was as active as the untagged protein in promoting translation from luc-SLWT mRNA relative to that from luc-SLR mRNA. eIF4G, eIF4A, Nip1, and Pab1p specifically copurified with His6-tagged SLBP in the absence of the histone stem-loop (Fig. 8B, last lane versus middle lane), while eIF4E was retained nonspecifically by the resin in the absence of SLBP (Fig. 8B, middle lane). Furthermore, eIF4G1 and eIF4G2 each copurified with His6-tagged SLBP (Fig. 8C), suggesting that SLBP can assemble with either isoform of eIF4G, correlating with the ability of each eIF4G mutant strain to support SLBP translational function (Table 3). These data support the conclusion that SLBP assembles into a cap-binding complex via eIF4G.
FIG. 8.
eIF4G copurifies with SLBP from yeast. (A) Luciferase expression from capped luc-SLWT and luc-SLR mRNAs was examined in control yeast or yeast expressing wild-type or N-terminal His6-tagged SLBP. Expression from luc-SLWT relative to that from luc-SLR (set at a value of 1) for each pair of mRNAs is indicated above each histogram. (B) His6-tagged SLBP was isolated by Co2+-affinity chromatography (last lane). Yeast containing the empty His6-tagged vector was used as a control (middle lane). The presence of SLBP, eIF4G,eIF4A, eIF4E, the Nip1 subunit of eIF3, and Pap1p in crude extract (lane 1) or in the Co2+-bound fractions was determined by Western analysis with factor-specific antibodies. (C) His6-tagged SLBP was isolated from CW04 and tif4631 and tif4632 mutants by Co2+-affinity chromatography. Yeast containing the empty His6-tagged vector (second lane) was used as a control. The presence of eIF4G and SLBP was determined by Western analysis.
SLBP associates with eIF4G in mammalian cells.
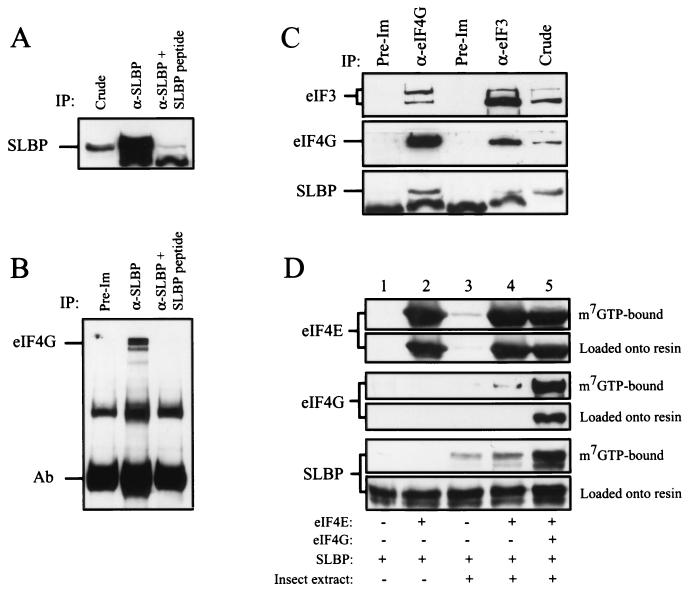
To examine whether SLBP assembles with eIF4G in mammalian cells, immunoprecipitation of endogenous SLBP from 293 cells was performed with an antibody raised against the SLBP C-terminal 13-amino acid peptide (Fig. 9A, middle lane), with recovery blocked by the corresponding competing peptide (Fig. 9A, last lane versus middle lane). eIF4G specifically coimmunoprecipitated with SLBP (Fig. 9B, middle lane), supporting the observations from yeast that SLBP assembles into a complex with eIF4G. eIF3 was not detected in these immunoprecipitates (data not shown), suggesting that the interaction between SLBP and eIF4G may be direct and stable. In a reciprocal experiment, SLBP was observed to coimmunoprecipitate with eIF4G (Fig. 9C). The use of anti-eIF3 serum resulted in the coimmunoprecipitation of SLBP but to a level that was significantly lower than that obtained with anti-eIF4G serum (Fig. 9C). The recovery of SLBP in the immunoprecipitate correlated more closely with that of eIF4G rather than that of eIF3, suggesting that eIF4G may be more important for the assembly of SLBP with eIF4F.
FIG. 9.
SLBP and eIF4G copurify from mammalian cells. (A) SLBP was subject to immunoprecipitation (IP) from 293 cells with anti-SLBP antibody in the absence (α-SLBP) or presence (α-SLBP + SLBP peptide) of the SLBP 13-amino-acid C-terminal peptide used for production of the anti-SLBP antiserum and detected by Western analysis. (B) Immunoprecipitation from 293 cells was performed with preimmune antiserum (Pre-Im) or anti-SLBP antiserum (α-SLBP) in the absence or presence of the 13-amino-acid C-terminal SLBP peptide. eIF4G was detected by Western analysis. The antibody (Ab) used for the immunoprecipitation is indicated. A nonspecific band is also present in all lanes. (C) eIF4G or eIF3 was immunoprecipitated from 293 cells with anti-eIF4G or anti-eIF3 antiserum, respectively, and eIF4G, eIF3, and SLBP were detected by Western analysis. Preimmune antiserum for each was also used. A nonspecific band detected during SLBP Western analysis was also present. (D) Recombinant SLBP was added to binding reaction mixtures containing recombinant human eIF4E (expressed in and purified from E. coli) and/or recombinant human eIF4G (expressed in insect cells) as indicated. eIF4E, eIF4G, and SLBP loaded onto m7GTP-Sepharose resin and that retained on the resin was detected by Western analysis. The faint eIF4E and eIF4G signals in lane 3 represent the insect homologs.
We next examined whether recombinant SLBP could interact with recombinant eIF4G. Recombinant SLBP was added to binding reaction mixtures containing recombinant human eIF4E (expressed in and purified from Escherichia coli) and/or recombinant human eIF4G (expressed in insect cells). Interaction of SLBP with either initiation factor was determined by Western analysis of the protein retained on m7GTP-Sepharose resin. SLBP was not retained on the resin in the absence or presence of eIF4E (Fig. 9D, lanes 1 and 2). A high level of SLBP was retained only when recombinant eIF4E and extract from insect cells expressing human eIF4G were added (Fig. 9D, lane 5), suggesting that eIF4G was required for SLBP to bind the resin. A low level of SLBP was retained in the absence or presence of eIF4E when control insect extract was added (Fig. 9D, lanes 3 and 4), suggesting that insect eIF4G can also interact with SLBP.
Together, the data from yeast and mammalian cells suggest that the physical interaction of SLBP with eIF4F requires eIF4G, and perhaps eIF3, but does not require eIF4E, eIF4A, or PABP.
DISCUSSION
The cell cycle-regulated histone mRNAs are unique in that they terminate in a highly conserved stem-loop that is bound by SLBP. Although the 3′-terminal stem-loop serves as the functional equivalent to a poly(A) tail in that it functionally interacts with the 5′ cap to enhance translation (17), there was no information on whether SLBP was necessary and sufficient to mediate this function. Because S. cerevisiae lacks a homolog to SLBP and yeast histone mRNAs are polyadenylated, this species could be used to examine the requirements for the histone stem-loop-mediated regulation. We found that histone stem-loop-mediated regulation could be recapitulated in yeast, with expression of SLBP necessary and sufficient to mediate the translation regulation associated with the histone stem-loop (Fig. 1). The stem-loop was functionally dependent on the presence of a cap structure at the 5′ terminus of the mRNA (Fig. 4A), a finding previously described for mammalian cells (17). Moreover, mutations to the conserved features of the stem-loop structure were deleterious to its translational function in yeast (Table 1) and mammalian cells (17). Additionally, deletion of the N-terminal or C-terminal domains, which alone do not affect SLBP binding to the histone stem-loop, reduced the ability of the protein to stimulate translation (Fig. 3). The ability of SLBP to function in the context of the yeast cellular environment suggests that at least some of its molecular interactions with the yeast translational machinery are similar to those of animals.
The observations presented in this study suggest that SLBP functionally and physically interacts with eIF4G and eIF3. This conclusion is supported by multiple independent lines of evidence. First, the function of SLBP in yeast was genetically dependent on eIF4G and eIF3 (Tables 3 and 4). Second, copurification of SLBP with the eIF4F/eIF3/PABP complex was observed when the latter was purified by m7GTP-Sepharose or poly(A)-agarose chromatography (Fig. 4C, 6, and 7). Third, eIF4G and eIF3 were required for the association of SLBP with the eIF4F/eIF3/PABP complex (Fig. 7). Fourth, copurification of eIF4G and eIF3 with SLBP was observed when the latter was affinity purified as a His-tagged protein (Fig. 8). Fifth, mammalian eIF4G coimmunoprecipitated with SLBP (Fig. 9B). Sixth, SLBP coimmunoprecipitated with eIF4G or with eIF3 from mammalian cells (Fig. 9C). Seventh, an interaction between recombinant mammalian eIF4G and SLBP was observed (Fig. 9D). The observation that recombinant human SLBP and eIF4G interact in vitro suggests that the interaction may be direct.
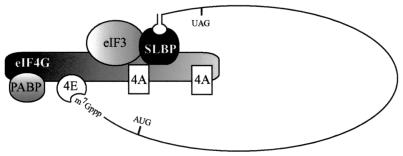
In addition to eIF4G and eIF3, eIF4E was necessary for SLBP translational function (Table 2). This finding is consistent with the observation that the translational enhancement conferred by the histone stem-loop structure is cap dependent (17) and that the translational function of SLBP requires a 5′ cap (Fig. 4A). Because no direct physical interaction between SLBP and eIF4E was observed (Fig. 9D), eIF4E likely contributes to SLBP function by facilitating the binding of eIF4F to the 5′ cap, which may be necessary for the interaction between SLBP and eIF4G in a manner analogous to the role of eIF4E in facilitating the interaction between eIF4G and PABP during the translation of capped and polyadenylated mRNAs (56, 60). Therefore, a model incorporating the present observations posits that the functional interaction of the 5′ cap with the 3′ histone stem-loop structure involves an interaction of the cap-bound eIF4E with eIF4G, which in turn interacts with the SLBP-bound 3′-terminal histone stem-loop structure (Fig. 10). The presence of other initiation factors, including eIF2 (which may be partially required for SLBP function) and eIF1, e1F1A, and eIF5, would be expected as part of the initiation complex.
FIG. 10.
Proposed physical and function model of the association of SLBP with the eIF4F/eIF3/PABP complex. SLBP is shown bound to the 3′-terminal stem-loop of a cell cycle-regulated histone mRNA. Association of SLBP with the eIF4F/eIF3/PABP complex requires eIF4G and perhaps eIF3. eIF4E is not required for the physical association of SLBP with the complex but is required for SLBP function in that it is necessary for the binding of the complex to the 5′ cap. Other initiation factors including eIF2 (which may be partially required), eIF5, eIF1, and eIF1A are not shown for simplicity but would be expected to be present.
Although yeast express two eIF4G proteins that are only 53% homologous (19), SLBP function was maintained as long as at least one eIF4G protein was expressed. Moreover, eIF4G1 or eIF4G2 were each found to copurify with SLBP when the latter was isolated from yeast strains solely expressing eIF4G1 or eIF4G2, confirming that SLBP can assemble with either eIF4G. The observed coimmunoprecipitation of eIF4G with SLBP and SLBP with eIF4G from mammalian cell extract as well as the interaction between recombinant mammalian SLBP and mammalian eIF4G (Fig. 9) supports the conclusion that the coassembly of SLBP and eIF4G in yeast may be representative of what occurs in mammalian cells. Further work will be needed to determine whether SLBP exhibits a functional preference for eIF4GI or eIF4GII (20) in mammalian cells.
Our results suggest that, from the perspective of translational regulation, SLBP has evolved as a functional mimic of PABP in that it binds to a 3′-terminal sequence, exhibits a functional requirement for the 5′ cap, copurifies with the eIF4F complex, and requires eIF4G to mediate its association with the complex. However, SLBP is distinct from PABP in that it may involve an interaction with eIF3 and regulates histone mRNA stability in a cell cycle fashion. Further investigation into the functional and physical parallels between SLBP and PABP in their interaction with the translational initiation machinery will provide valuable insight into the evolution of diverse molecular mechanisms that promote efficient translation.
Acknowledgments
Antibody raised against yeast eIF4G was generously provided by Michael Altmann (Berne, Switzerland). Yeast mutants were provided by Michael Altmann, Alan Sachs (University of California at Berkeley), Tom Dever and Alan Hinnebusch (NIH), Patrick Linder (Centre Médical Universitaire), and Nahum Sonenberg (McGill University).
This work was supported by a grant from the United States Department of Agriculture (NRICGP 99-35301-7866) and the National Science Foundation (MCB-9816657) to D.R.G and a grant from NIH (GM29832) to W.F.M. Work in the labs of S.J.M. and V.M.P. is supported by program and equipment grants from The Wellcome Trust (040800, 050703, 045619, and 056778), and S.J.M. is a Senior Research Fellow of The Wellcome Trust.
REFERENCES
- 1.Altmann, M., and H. Trachsel. 1994. The yeast S. cerevisiae system: a powerful tool to study the mechanism of protein synthesis in eukaryotes. Biochimie 76:853-861. [DOI] [PubMed] [Google Scholar]
- 2.Altmann, M., N. Schmitz, C. Berset, and H. Trachsel. 1997. A novel inhibitor of cap-dependent translation initiation in yeast: p20 competes with eIF4G for binding to eIF4E. EMBO J. 16:1114-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altmann, M., P. P. Muller, J. Pelletier, N. Sonenberg, and H. Trachsel. 1989. A mammalian translation initiation factor can substitute for its yeast homologue in vivo. J. Biol. Chem. 264:12145-12147. [PubMed] [Google Scholar]
- 4.Berset, C., H. Trachsel, and M. Altmann. 1998. The TOR (target of rapamycin) signal transduction pathway regulates the stability of translation initiation factor eIF4G in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 95:4264-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bond, U. M., T. A. Yario, and J. A. Steitz. 1991. Multiple processing-defective mutations in a mammalian histone pre-mRNA are suppressed by compensatory changes in U7 RNA both in vivo and in vitro. Genes Dev. 5:1709-1722. [DOI] [PubMed] [Google Scholar]
- 6.Borun, T. W., M. D. Scharff, and E. Robbins. 1967. Rapidly labeled, polyribosome-associated RNA having the properties of histone messenger. Proc. Natl. Acad. Sci. USA 58:1977-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bushell, M., W. Wood, G. Carpenter, V. M. Pain, S. J. Morley, and M. J. Clemens. 2001. Disruption of the interaction of mammalian protein synthesis eukaryotic initiation factor 4B with the poly(A)-binding protein by caspase- and viral protease-mediated cleavages. J. Biol. Chem. 276:23922-23928. [DOI] [PubMed] [Google Scholar]
- 8.Coppolecchia, R., P. Buser, A. Stotz, and P. Linder. 1993. A new yeast translation initiation factor suppresses a mutation in the eIF-4A RNA helicase. EMBO J. 12:4005-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cotten, M., O. Gick, A. Vasserot, G. Schaffner, and M. L. Birnstiel. 1988. Specific contacts between mammalian U7 snRNA and histone precursor RNA are indispensable for the in vitro RNA processing reaction. EMBO J. 7:801-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de la Cruz, J., I. Iost, D. Kressler, and P. Linder. 1997. The p20 and Ded1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94:5201-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dever, T. E., L. Feng, R. C. Wek, A. M. Cigan, T. F. Donahue, and A. G. Hinnebusch. 1992. Phosphorylation of initiation factor 2α by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell 68:585-596. [DOI] [PubMed] [Google Scholar]
- 12.Dominski, Z., J. A. Erkmann, J. A. Greenland, and W. F. Marzluff. 2001. Mutations in the RNA binding domain of stem-loop binding protein define separable requirements for RNA binding and for histone pre-mRNA processing. Mol. Cell. Biol. 21:2008-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dominski, Z., J. Sumerel, R. J. Hanson, and W. F. Marzluff. 1995. The polyribosomal protein bound to the 3′ end of histone mRNA can function in histone pre-mRNA processing. RNA 1:915-923. [PMC free article] [PubMed] [Google Scholar]
- 14.Dominski, Z., L. X. Zheng, R. Sanchez, and W. F. Marzluff. 1999. Stem-loop binding protein facilitates 3′-end formation by stabilizing U7 snRNP binding to histone pre-mRNA. Mol. Cell. Biol. 19:3561-3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Everett, J. G., and D. R. Gallie. 1992. RNA delivery in Saccharomyces cerevisiae using electroporation. Yeast 8:1007-1014. [DOI] [PubMed] [Google Scholar]
- 16.Gallie, D. R. 1991. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 5:2108-2116. [DOI] [PubMed] [Google Scholar]
- 17.Gallie, D. R., N. J. Lewis, and W. F. Marzluff. 1996. The histone 3′-terminal stem-loop is necessary for translation in Chinese hamster ovary cells. Nucleic Acids Res. 24:1954-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gingras, A. C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913-963. [DOI] [PubMed] [Google Scholar]
- 19.Goyer, C., M. Altmann, H. S. Lee, A. Blanc, M. Deshmukh, J. L. Woolford, H. Trachsel, and N. Sonenberg. 1993. TIF4631 and TIF4632: two yeast genes encoding the high-molecular-weight subunits of the cap-binding protein complex (eukaryotic initiation factor 4F) contain an RNA recognition motif-like sequence and carry out an essential function. Mol. Cell. Biol. 13:4860-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gradi, A., H. Imataka, Y. V. Svitkin, E. Rom, B. Raught, S. Morino, and N. Sonenberg. 1998. A novel functional human eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 18:334-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graves, R. A., and W. F. Marzluff. 1984. Rapid reversible changes in the rate of histone gene transcription and histone mRNA levels in mouse myeloma cells. Mol. Cell. Biol. 4:351-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanic-Joyce, P. J., G. C. Johnston, and R. A. Singer. 1987. Regulated arrest of cell proliferation mediated by yeast prt1 mutations. Exp. Cell Res. 172:134-145. [DOI] [PubMed] [Google Scholar]
- 23.Hanson, R. J., J. Sun, D. G. Willis, and W. F. Marzluff. 1996. Efficient extraction and partial purification of the polyribosome-associated stem-loop binding protein bound to the 3′ end of histone mRNA. Biochemistry 35:2146-2156. [DOI] [PubMed] [Google Scholar]
- 24.Harris, M. E., R. Bohni, M. H. Schneiderman, L. Ramamurthy, D. Schumperli, and W. F. Marzluff. 1991. Regulation of histone mRNA in the unperturbed cell cycle: evidence suggesting control at two posttranscriptional steps. Mol. Cell. Biol. 11:2416-2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heintz, N., H. L. Sive, and R. G. Roeder. 1983. Regulation of human histone gene expression: kinetics of accumulation and changes in the rate of synthesis and in the half-lives of individual histone mRNAs during the HeLa cell cycle. Mol. Cell. Biol. 3:539-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hentschel, C. C., and M. L. Birnstiel. 1981. The organization and expression of histone gene families. Cell 25:301-313. [DOI] [PubMed] [Google Scholar]
- 27.Hershey, J. W. B., and W. C. Merrick. 2000. Pathway and mechanism of initiation of protein synthesis, p. 33-88. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Hinnebusch, A. G. 1996. Translational control of GCN4: gene-specific regulation by phosphorylation of eIF2, p. 199-244. In M. B. Mathews, N. Sonenberg, and J. W. B. Hershey (ed.), Translational control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Hinnebusch, A. G. 2000. Mechanism of regulation of initiator methionyl-tRNA binding to the ribosome, p. 185-244. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Imataka, H., A. Gradi, and N. Sonenberg. 1998. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 17:7480-7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamphear, B. J., R. Kirchweger, T. Skern, and R. E. Rhoads. 1995. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem. 270:21975-21983. [DOI] [PubMed] [Google Scholar]
- 32.Le, H., K. S. Browning, and D. R. Gallie. 2000. The phosphorylation state of poly(A)-binding protein specifies its binding to poly(A) RNA and its interaction with eukaryotic initiation factor (eIF) 4F, eIFiso4F, and eIF4B. J. Biol. Chem. 275:17452-17462. [DOI] [PubMed] [Google Scholar]
- 33.Le, H., R. L. Tanguay, M. L. Balasta, C.-C. Wei, K. S. Browning, A. M. Metz, D. J. Goss, and D. R. Gallie. 1997. Translation initiation factors eIF-iso4G and eIF-4B interact with the poly(A)-binding protein and increase its RNA binding activity. J. Biol. Chem. 272:16247-16255. [DOI] [PubMed] [Google Scholar]
- 34.Levine, B. J., N. Chodchoy, W. F. Marzluff, and A. I. Skoultchi. 1987. Coupling of replication type histone mRNA levels to DNA synthesis requires the stem-loop sequence at the 3′ end of the mRNA. Proc. Natl. Acad. Sci. USA 84:6189-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin, F., A. Schaller, S. Eglite, D. Schumperli, and B. Muller. 1997. The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J. 16:769-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marzluff, W. F. 1992. Histone 3′ ends: essential and regulatory functions. Gene Exp. 2:93-97. [PMC free article] [PubMed] [Google Scholar]
- 37.Morley, S. J., P. S. Curtis, and V. M. Pain. 1997. eIF4G: translation's mystery factor begins to yield its secrets. RNA 3:1085-1104. [PMC free article] [PubMed] [Google Scholar]
- 38.Mowry, K. L., and J. A. Steitz. 1987. Both conserved signals on mammalian histone pre-mRNAs associate with small nuclear ribonucleoproteins during 3′ end formation in vitro. Mol. Cell. Biol. 7:1663-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Naranda, T., S. E. MacMillan, and J. W. B. Hershey. 1994. Purified yeast translational initiation factor eIF-3 is an RNA-binding protein complex that contains the PRT1 protein. J. Biol. Chem. 269:32286-32292. [PubMed] [Google Scholar]
- 40.Niepel, M., J. Ling, and D. R. Gallie. 1999. The effect of secondary structure on the functional interaction between the 5′-cap and the poly(A) tail during translation. FEBS Lett. 462:79-84. [DOI] [PubMed] [Google Scholar]
- 41.Pandey, N. B., and W. F. Marzluff. 1987. The stem-loop structure at the 3′ end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol. Cell. Biol. 7:4557-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pandey, N. B., A. S. Williams, J.-H. Sun, V. D. Brown, U. Bond, and W. F. Marzluff. 1994. Point mutations in the stem-loop at the 3′ end of mouse histone mRNA reduce expression by reducing the efficiency of 3′ end formation. Mol. Cell. Biol. 14:1709-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pandey, N. B., J.-H. Sun, and W. F. Marzluff. 1991. Different complexes are formed on the 3′ end of histone mRNA with nuclear and polyribosomal proteins. Nucleic Acids Res. 19:5653-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piron, P., P. Vende, J. Cohen, and D. Poncet. 1998. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J. 17:5811-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Powers, T., and P. Walter. 1999. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol. Biol. Cell 10:987-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Preiss, T., and M. W. Hentze. 1998. Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature 392:516-520. [DOI] [PubMed] [Google Scholar]
- 47.Rau, M., T. Ohlmann, S. J. Morley, and V. M. Pain. 1996. A reevaluation of the cap-binding protein, eIF4E, as a rate-limiting factor for initiation of translation in reticulocyte lysate. J. Biol. Chem. 271:8983-8990. [DOI] [PubMed] [Google Scholar]
- 48.Sachs, A. B. 2000. Physical and functional interactions between the mRNA cap and the poly(A) tail, p. 447-466. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 49.Sachs, A. B., and R. W. Davis. 1989. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell 58:857-867. [DOI] [PubMed] [Google Scholar]
- 50.Schmid, S. R., and P. Linder. 1991. Translation initiation factor 4A from Saccharomyces cerevisiae: analysis of residues conserved in the D-E-A-D family of RNA helicases. Mol. Cell. Biol. 11:3463-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sittman, D. B., R. A. Graves, and W. F. Marzluff. 1983. Histone mRNA concentrations are regulated at the level of transcription and mRNA degradation. Proc. Natl. Acad. Sci. USA 80:1849-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soldati, D., and D. Schümperli. 1988. Structural and functional characterization of mouse U7 small nuclear RNA active in 3′ processing of histone pre-mRNA. Mol. Cell. Biol. 8:1518-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun, J., D. R. Pilch, and W. F. Marzluff. 1992. The histone mRNA 3′ end is required for localization of histone mRNA to polyribosomes. Nucleic Acids Res. 20:6057-6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tarun, S. Z., and A. B. Sachs. 1995. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 9:2997-3007. [DOI] [PubMed] [Google Scholar]
- 55.Tarun, S. Z., and A. B. Sachs. 1996. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 15:7168-7177. [PMC free article] [PubMed] [Google Scholar]
- 56.Tarun, S. Z., S. E. Wells, J. A. Deardorff, and A. B. Sachs. 1997. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc. Natl. Acad. Sci. USA 94:9046-9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vasserot, A. P., F. J. Schaufele, and M. L. Birnstiel. 1989. Conserved terminal hairpin sequences of histone mRNA precursors are not involved in duplex formation with the U7 RNA but act as a target site for a distinct processing factor. Proc. Natl. Acad. Sci. USA 86:4345-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, Z. F., M. L. Whitfield, T. C. Ingledue, I. I. I., Z. Dominski, W. F. Marzluff. 1996. The protein that binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 10:3028-3040. [DOI] [PubMed] [Google Scholar]
- 59.Wei, C.-C., M. L. Balasta, J. Ren, and D. J. Goss. 1998. Wheat germ poly(A) binding protein enhances the binding affinity of eukaryotic initiation factor 4F and (iso)4F for cap analogues. Biochemistry 37:1910-1916. [DOI] [PubMed] [Google Scholar]
- 60.Wells, S. E., P. E. Hillner, R. D. Vale, and A. B. Sachs. 1998. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell 2:135-140. [DOI] [PubMed] [Google Scholar]
- 61.Williams, A. S., and W. F. Marzluff. 1995. The sequence of the stem and flanking sequences at the 3′ end of histone mRNA are critical determinants for the binding of the stem-loop binding protein. Nucleic Acids Res. 23:654-662. [DOI] [PMC free article] [PubMed] [Google Scholar]