Abstract
Gene trapping is a method of generating murine embryonic stem (ES) cell lines containing insertional mutations in known and novel genes. A number of international groups have used this approach to create sizeable public cell line repositories available to the scientific community for the generation of mutant mouse strains. The major gene trapping groups worldwide have recently joined together to centralize access to all publicly available gene trap lines by developing a user-oriented Website for the International Gene Trap Consortium (IGTC). This collaboration provides an impressive public informatics resource comprising ∼45 000 well-characterized ES cell lines which currently represent ∼40% of known mouse genes, all freely available for the creation of knockout mice on a non-collaborative basis. To standardize annotation and provide high confidence data for gene trap lines, a rigorous identification and annotation pipeline has been developed combining genomic localization and transcript alignment of gene trap sequence tags to identify trapped loci. This information is stored in a new bioinformatics database accessible through the IGTC Website interface. The IGTC Website (www.genetrap.org) allows users to browse and search the database for trapped genes, BLAST sequences against gene trap sequence tags, and view trapped genes within biological pathways. In addition, IGTC data have been integrated into major genome browsers and bioinformatics sites to provide users with outside portals for viewing this data. The development of the IGTC Website marks a major advance by providing the research community with the data and tools necessary to effectively use public gene trap resources for the large-scale characterization of mammalian gene function.
INTRODUCTION
The large and continually growing number of genome sequencing projects provides an opportunity to greatly advance our understanding of genetics and disease. One of the keys to realizing this goal is the development of genomic resources to elucidate functional characteristics of these genes, especially in mammalian genomes. The mouse is an especially useful mammalian model system, providing an excellent subject for studies of gene function because of its short generational span, ease of handling, and the close structural and functional similarity of its genome to that of humans (1,2). Furthermore, using this organism, scientists have access to a wide range of procedures for genetic manipulation, including the use of embryonic stem (ES) cells to create mice with defined single-gene mutations using gene targeting and gene trapping techniques (3).
Gene trapping is a high-throughput method of creating mutagenized ES cells for use in generating knockout and other mutant mouse strains for research in functional genomics (4). Second generation gene trap vectors have recently enhanced the value of the method by offering the potential for creating conditional and other desired alleles using site-specific recombination (5–7). Major scientific initiatives are currently underway in North America and Europe to knock out every mouse gene in ES cells in order to characterize gene function and provide insight into systems associated with human disease (8,9).
A number of gene trap projects have already made notable progress toward this goal by generating resources of gene trap mouse ES cell lines harboring well-characterized insertional mutations (6,7,10–14), although until now the individual gene trap projects have been isolated, providing only details about their own cell lines. The International Gene Trap Consortium (IGTC) is a collaboration representing the major public gene trap resources worldwide, whose mission is to offer the scientific community access to all publicly available gene trap cell lines on a non-collaborative basis for nominal handling fees (15). The centralization of gene trap resources provides many advantages to the research community, allowing more effective utilization of the experimental opportunities offered by gene trap cell lines through standardized protocols for the identification and annotation of sequences from trapped loci, and the increased availability of experimental protocols. As reported here, the release of the IGTC Website (www.genetrap.org) marks a major advance, generating a standardized informatics pipeline and providing in one place both easy access to all publicly available gene trap cell lines and sophisticated tools for analysis of resource data. Gene trap centers currently involved in this effort are listed in Table 1.
Table 1.
IGTC members
| IGTC members | Cell lines | Website |
|---|---|---|
| Baygenomics (USA) | 9848 | www.baygenomics.ucsf.edu/ |
| Centre for Modelling Human Disease (Toronto, Canada) | 4137 | www.cmhd.ca/genetrap/ |
| Embryonic Stem Cell Database (University of Manitoba, Canada) | 8559 | www.EScells.ca/ |
| Exchangeable Gene Trap Clones (Kumamoto University, Japan) | 49 | egtc.jp/show/index |
| German Gene Trap Consortium (Germany) | 13031 | www.genetrap.de/ |
| Sanger Institute Gene Trap Resource (Cambridge, UK) | 7354 | www.sanger.ac.uk/PostGenomics/genetrap/ |
| Soriano Lab Gene Trap Database (FHCRC, Seattle, USA) | 1627 | www.fhcrc.org/science/labs/soriano/trap.html |
| Telethon Institute of Genetics and Medicine–TIGEM (Naples, Italy) | 1435 | core.tigem.it/genetrap/public/ |
| TOTAL | 44605 |
IGTC: RESOURCE OVERVIEW
The IGTC Website centralizes access to all publicly available gene trap cell line data for the first time. This repository was created to address the needs of the international gene trap community by providing researchers with the data and informatics tools necessary to find gene trap cell lines with mutations in genes and loci of interest. IGTC member projects produce gene trap cell lines and directly submit gene trap sequence tags to the Genome Survey Sequences Database (dbGSS) division of GenBank at NCBI (16). Data from all publicly available cell lines are downloaded from dbGSS and subjected to the IGTC identification and annotation pipeline, which then automatically populates the MySQL database used to generate the annotation information presented on the Website. The IGTC Website has been designed to provide easy user access to the extensive array of assembled gene trap informatics data. Interface options include homology searches using BLAST, search and browse capabilities, and viewing trapped genes within biological pathways. The project also includes the integration of gene trap data at major genome browsers and other informatics data sites in order to offer a variety of outside portals to this data. Cell line requests from the IGTC site are forwarded to the originating gene trap resource, where the cell line is removed from cryogenic storage and sent to the user for experimental analysis. The IGTC site also provides useful documentation, on-line tutorials and scientific overviews on gene trapping and the use of gene trap cell lines.
GENE TRAP IDENTIFICATION AND ANNOTATION PIPELINE
Gene trap mutations are characterized through a process of sequencing, identification and annotation. This process involves obtaining cDNA or genomic sequence upstream or downstream of the insertion site and identifying and annotating the locus at which the insertion occurs. A full identification and annotation protocol has been developed for the IGTC that integrates genomic and transcript-based identification approaches and adds information from other major informatics resources, such as genome browsers and specialized informatics sites to annotate the identified loci (Figure 1). Use of both genomic and transcript-based annotation methods were incorporated to increase confidence in identification and localization on the genome of trapped loci, providing a significant improvement over identification strategies previously in use. This pipeline was designed to be both robust and flexible, allowing the incorporation of multiple methods of identification and mapping and the future integration of new genomic data resources that may be developed.
Figure 1.
IGTC identification and annotation pipeline. IGTC members submit gene trap cell line data to dbGSS. The first step of the IGTC pipeline is the download of all publicly available gene trap cell line data from dbGSS. Gene trap sequences are then processed through the dual identification protocol based on genomic localization and transcript alignment using MapTag and AutoIdent, respectively. Returned homologous genomic regions and transcripts are aligned to genomic sequence to generate confirmed overlapping sequence regions, which are used as the primary identification data. Confirmed genomic coordinates are queried against major informatics databases to obtain annotation data for the genomic locus identified, and the returned data are entered into the IGTC database.
Gene trap cell line sequences are submitted by each individual gene trap resource to dbGSS along with relevant ancillary information about the cell line, including its original source and the vector used in the trapping experiment. Within dbGSS, IGTC sequences are grouped using LinkOut (17) (http://www.dlib.org/dlib/march02/03inbrief.html KWAN), which links gene trap dbGSS entries to the IGTC Website. Requiring gene trap cell lines to be entered into dbGSS ensures that all IGTC lines are in the public domain, and that all data are consistent, transparent and freely accessible. Researchers can access additional information about methodology and protocols from the original trapping experiment via links to the individual gene trap project sites (Table 1).
The two complementary programs, MapTag (15) and AutoIdent (12), are used to locate the trapped gene on the mouse genome by analyzing the similarity between the unidentified gene trap sequence and genomic and transcript sequences, respectively. The minimum region of genomic overlap serves as the primary identification data, from which all subsequent annotation is derived. Finally, the identified map coordinates are annotated with gene features obtained from major genomic and informatics databases.
Genomic localization: MapTag
MapTag was developed as an automated method of identifying homologous genomic regions for gene trap sequences using the Ensembl database (18) and SSAHA algorithm (19). MapTag identifies matches in genomic sequence and assembles individual related stretches of genomic similarity using a basic splice model that takes into account exon boundaries, allowing the processing of nucleotide sequences of cDNA or genomic origin. The program filters alignment results, applying simple heuristics using the overall match length, percent identity and exon coverage, and filtering to remove pseudogenes, to determine the best match. As gene trap sequence tags are typically short and imperfect, they can be difficult to identify to a unique locus. The protocol used by MapTag greatly improves the ability to differentiate between matches that show comparable levels of similarity by correctly selecting the result that exhibits a correspondence to the insertion site. The program returns the identified genomic region and an estimate of the match confidence.
Transcript identification: AutoIdent
The pipeline also uses transcript identification to provide an independent and orthogonal identification method. AutoIdent, an automated protocol developed by BayGenomics, uses the BLAST (20) algorithm to identify the most similar sequences in the GenBank non-redundant nucleotide database at NCBI. When the program identifies many high-scoring matches to very similar (synonymous) genes, AutoIdent adds steps to filter results and condense synonymous transcripts to obtain a single result. The program applies stringent criteria for acceptance of a high-quality gene identification but allows more relaxed criteria to identify multiple matching sequences or to a homologous sequence in another species. At the end of the process, AutoIdent returns the best match transcript along with alignment data.
Identification reconciliation and confirmation
Genomic and transcript identification data are stored in the IGTC database. The pipeline then compares results from the two identification protocols, using overlap between the genomic coordinates from MapTag and the genomic localization of transcripts returned by AutoIdent to confirm the identification. Gene trap sequences are assigned as localized when the genomic and transcript map coordinates overlap, or when only one protocol returns map coordinates. If the identified coordinates conflict or neither protocol returns map data, the gene trap sequence is classified as unlocalized and does not go through the rest of the annotation pipeline. This reconciliation step assures that each identified cell line is mapped to a genomic insertion locus with high confidence and in a manner that is fully documented.
Annotation of gene trap cell lines
Gene trap cell line annotation is based on the confirmed genomic coordinates as the primary identification data. Map coordinates are used to query the Ensembl and Entrez (21) databases to obtain gene features for the identified locus. These accessions are used as primary keys to further query Ensembl, Entrez and the Mouse Genome Informatics (MGI) resource (22) for secondary annotation data associated with the trapped genes, including major gene accession systems, Gene Ontology classifications (GO), protein domain, structure and function, PubMed and phenotype data, homology and orthology data, and microarray probesets. In addition to supporting an extensive array of informatics data, the IGTC site also includes tissue-specific expression data from the SymAtlas project (23) and biological pathway and GO hierarchy diagrams with trapped genes marked, produced using the GenMAPP program (24).
The IGTC database
The IGTC uses the open source MySQL database platform (www.mysql.com). The database is populated in an automated process using results from the MapTag and AutoIdent identification protocols and is structured to optimize information access via Web queries. New gene trap cell line entries in dbGSS are downloaded weekly and run through the IGTC pipeline. Identification results from MapTag are updated with each Ensembl build and AutoIdent is programmed to regularly BLAST sequences in the database to update accession numbers and other changes in the information provided by GenBank. Annotation data generation is synchronized with the Entrez, MGI and Ensembl databases, and the IGTC database is updated by downloading information from these sites as necessary. The information in the IGTC database is available upon request in a tab-delimited or database compatible format.
ACCESSING GENE TRAP DATA
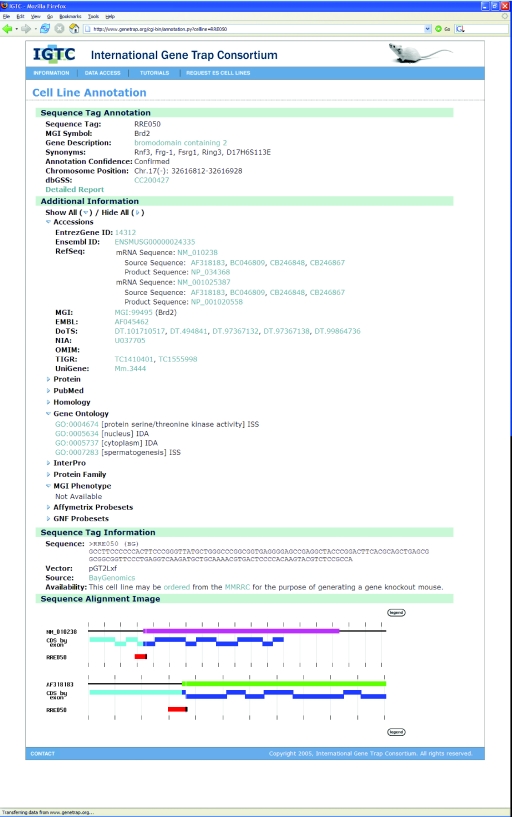
The IGTC site provides a user-friendly approach to gene trap data, allowing researchers to access the gene trap database from a sequence, accession number or ID, expression or pathway perspective using a variety of interfaces for searching and viewing gene trap data. The site is organized around the cell line annotation page, where the user can view all annotation data for a selected cell line (Figure 2). Primary identification and annotation data appear at the top of the page with a link to detailed identification results. This is followed by expandable lists of secondary annotation data, which provide information useful in the selection and analysis of cell lines of potential interest. Below this is a section containing details about the cell line and gene trap vector, including primer sequences and a description of the vector properties. To aid in the comparison of insertion sites of different cell lines that trap the same gene, the bottom of the cell line annotation page contains a diagram showing the gene trap sequence aligned to the transcripts returned by AutoIdent. The Website provides a similar annotation page for all trapped genes, organized by gene ID. Researchers can also access gene trap data via links from other major informatics resources, including the genome browsers and primary accession pages at NCBI (25), Ensembl (26) and UCSC (27).
Figure 2.
IGTC Website cell line annotation page. The image provides an example of the main data page for gene trap cell lines. Users can view primary identification and annotation data, with a link to detailed reports from the IGTC pipeline. An extensive amount of related annotation data is also presented to give researchers more information about the trapped locus. Finally, the page shows details about the cell line and vector and an image showing all gene trap cell line sequences aligned to the trapped gene. From this page, users are directed to the IGTC member site for cell line requests.
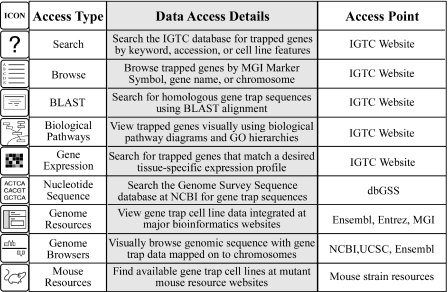
The IGTC Website offers users diverse ways to find gene trap cell lines, ranging from searches using protein data, microarray probesets or nucleotide sequences, to screening for traps placed in the context of biological pathways or in genes that demonstrate a particular expression profile. Figure 3 lists the ways in which gene trap data can be accessed, categorized by access type, details about available data type and data access point. Users can search the IGTC database by accession number, ID or keyword and chromosomal location. Results from database searches are displayed as a list of cell line IDs or gene symbols, which link to the individual cell line or gene annotation page. These lists can be exported as tab-delimited files for use in spreadsheet programs or custom databases. Users can also browse the database by MGI Marker Symbol, Gene Name or chromosome location. BLAST analysis can be performed using nucleotide sequence for a gene or locus of interest to search against gene trap sequences or genomic sequence of trapped genes. Trapped genes can be viewed with a pathway perspective using biological pathway diagrams and functional GO groupings, which are colored by the number of cell lines available for each gene. Users can also search for traps in genes with a designated expression profile by selecting a tissue of interest and choosing the expression level of the gene relative to the median tissue expression.
Figure 3.
Gene trap data access. Several methods for users to find gene trap cell lines of interest are illustrated.
Finally, users can browse gene trap cell lines displayed at major external informatics sites, such as genome browsers and gene pages. IGTC cell lines have been mapped to genomic sequence using the NCBI map viewer, UCSC genome browser and Ensembl genome browser. IGTC gene traps are maintained at these sites either as standard map tracks or as user-configured tracks. (Users should follow site-specific directions to view gene trap data using these browsers.) In addition, gene trap cell lines are listed on some of the major gene pages and in mouse strain resource databases, and the IGTC maintains a full list of partnering sites. By integrating gene trap data into the larger context provided by these bioinformatics resources, the IGTC can reach more potential users who are interested in genomic resources and functional genetics.
SUMMARY
The establishment of the IGTC database and Website marks a major advance in making large-scale mouse knockout resources available to the scientific community. Through the collaboration of gene trap projects worldwide, a standardized identification and annotation pipeline has been developed to analyze gene trap data and offer the data access tools necessary for maximum resource utility. For the first time, researchers investigating gene function in the mouse can query all publicly available gene trap ES cell lines at a single site. The IGTC Website contains approximately 45 000 cell lines harboring mutations in nearly 40% of mouse genes (15), including many genes with gene trap cell lines representing multiple mutant alleles. Researchers can search and browse the resource based on accession numbers or IDs, keywords, sequence data, tissue expression and biological pathways, and can also access the IGTC site from other major informatics sites. Furthermore, they can easily request IGTC cell lines for functional characterization of gene function and disease models. For example, BayGenomics cell lines are available from the Mutant Mouse Regional Resource Center (MMRRC) at the University of California Davis (www.mmrrc.org). The MMRRC-UC Davis also offers to microinject ES cells to derive knockout mice for investigators. Soon, the Soriano and TIGEM collections will also be available from the MMRRC. The IGTC Website will grow and continue to develop new tools and features as the diversity of trapped genes and experimental options for using gene trap cell lines expand.
Acknowledgments
This work has been supported by NIH grants U01 HL66600 and P41 RR01081, Canadian Institutes of Health grant GOP-36055 and by The Wellcome Trust Sanger Institute. The authors thank Carol Bult at Mouse Genome Informatics, Deanna Church at the National Center for Biotechnology Information and Bob Kuhn at University of California at Santa Cruz for their helpful suggestions. Funding to pay the Open Access publication charges for this article was provided by the University of California, San Francisco.
Conflict of interest statement. None declared.
REFERENCES
- 1.Waterston R.H., Chinwalla A.T., Cook L.L., Delehaunty K.D., Fewell G.A., Fulton L.A., Fulton R.S., Graves T.A., Hillier L.W., Lindblad-Toh K., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 2.Sung Y.H., Lee H.-W., Song J. Functional genomics approach using mice. J. Biochem. Mol. Biol. 2004;37:122–132. doi: 10.5483/bmbrep.2004.37.1.122. [DOI] [PubMed] [Google Scholar]
- 3.Nolan P.M. Generation of mouse mutants as a tool for functional genomics. Pharmacogenomics. 2000;1:243–255. doi: 10.1517/14622416.1.3.243. [DOI] [PubMed] [Google Scholar]
- 4.Stanford W.L., Cohn J.B., Cordes S.P. Gene-trap mutagenesis: past, present and beyond. Nature Rev. Genet. 2001;2:756–768. doi: 10.1038/35093548. [DOI] [PubMed] [Google Scholar]
- 5.Branda C.S., Dymecki S.M. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev. Cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- 6.Schnutgen F., De-Zolt S., Van Sloun P., Hollatz M., Floss T., Hansen J., Altschmied J., Seisenberger C., Ghyselinck N.B., Ruiz P., et al. Genomewide production of multipurpose alleles for the functional analysis of the mouse genome. Proc. Natl Acad. Sci. USA. 2005;102:7221–7226. doi: 10.1073/pnas.0502273102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cobellis G., Nicolaus G., Iovino M., Romito A., Marra E., Barbarisi M., Sardiello M., Di Giorgio F.P., Iovino N., Zollo M., et al. Tagging genes with cassette-exchange sites. Nucleic Acids Res. 2005;33:e44. doi: 10.1093/nar/gni045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Austin C.P., Battey J.F., Bradley A., Bucan M., Capecchi M., Collins F.S., Dove W.F., Duyk G., Dymecki S., Eppig J.T., et al. The knockout mouse project. Nature Genet. 2004;36:921–924. doi: 10.1038/ng0904-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Auwerx J., Avner P., Baldock R., Ballabio A., Balling R., Barbacid M., Berns A., Bradley A., Brown S., Carmeliet P., et al. The European dimension for the mouse genome mutagenesis program. Nature Genet. 2004;36:925–927. doi: 10.1038/ng0904-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hicks G.G., Shi E.-G., Li X.-M., Li C.-H., Pawlak M., Ruley H.E. Functional genomics in mice by tagged sequence mutagenesis. Nature Genet. 1997;16:338–344. doi: 10.1038/ng0897-338. [DOI] [PubMed] [Google Scholar]
- 11.Wiles M.V., Vauti F., Otte J., Fuchtbauer E.-M., Ruiz P., Fuchtbauer A., Arnold H.-H., Lehrach H., Metz T., Von Melchner H., et al. Establishment of a gene-trap sequence tag library to generate mutant mice from embryonic stem cells. Nature Genet. 2000;24:13–14. doi: 10.1038/71622. [DOI] [PubMed] [Google Scholar]
- 12.Stryke D., Kawamoto M., Huang C.C., Johns S.J., King L.A., Harper C.A., Meng E.C., Lee R.E., Babbit P.C., Ferrin T.E., et al. BayGenomics: a resource of insertional mutations in mouse embryonic stem cells. Nucleic Acids Res. 2003;31:278–281. doi: 10.1093/nar/gkg064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansen J., Floss T., Wurst W., Van Sloun P., Schnütgen F., Von Melchnert H., Füchtbauer E.-M., Vauti F., Arnold H.-H., Ruiz P. A large-scale, gene-driven mutagenesis approach for the functional analysis of the mouse genome. Proc. Natl Acad. Sci. USA. 2003;100:9918–9922. doi: 10.1073/pnas.1633296100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.To C., Epp T., Reid T., Lan Q., Yu M., Li C.Y., Ohishi M., Hant P., Tsao N., Casallo G., et al. The Centre for Modeling Human Disease Gene Trap resource. Nucleic Acids Res. 2004;32:D557–559. doi: 10.1093/nar/gkh106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skarnes W.C., Nord A.S., Cox T., von Melchner H., Wurst W., Hicks G., Young S.G., Conklin B.R., Ruiz P., Soriano P., et al. A public gene trap resource for mouse functional genomics. Nature Genet. 2004;36:543–544. doi: 10.1038/ng0604-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L. GenBank. Nucleic Acids Res. 2005;33:D34–D38. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwan Y.K. LinkOut: Explore beyond PubMed and Entrez. D-Lib Mag. 2002;8 [Google Scholar]
- 18.Hubbard T., Andrews D., Caccamo M., Cameron G., Chen Y., Clamp M., Clarke L., Coates G., Cox T., Cunningham F., et al. Ensembl 2005. Nucleic Acids Res. 2005;33:D447–D453. doi: 10.1093/nar/gki138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ning Z., Cox A.J., Mullikin J.C. SSAHA: a fast search method for large DNA databases. Genome Res. 2001;11:1725–1729. doi: 10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Maglott D., Ostell J., Pruitt K.D., Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005;33:D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eppig J.T., Bult C.J., Kadin J.A., Richardson J.E., Blake J.A., Anagnostopoulos A., Baldarelli R.M., Baya M., Beal J.S., Bello S.M., et al. The Mouse Genome Database (MGD): from genes to mice—a community resource for mouse biology. Nucleic Acids Res. 2005;33:D471–475. doi: 10.1093/nar/gki113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su A.I., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl Acad. Sci. USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dahlquist K.D., Salomonis N., Vranizan K., Lawlor S.C., Conklin B.R. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nature Genet. 2002;31:19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- 25.Wheeler D.L., Barrett T., Benson D.A., Bryant S.H., Canese K., Church D.M., DiCuccio M., Edgar R., Federhen S., Helmberg W., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2005;33:D39–D45. doi: 10.1093/nar/gki062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stalker J., Gibbins B., Meidl P., Smith J., Spooner W., Hotz H.-R., Cox A.V. The Ensembl Web site: mechanics of a genome browser. Genome Res. 2004;14:951–955. doi: 10.1101/gr.1863004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karolchik D., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]