Abstract
Initiation of V(D)J recombination involves the synapsis and cleavage of a 12/23 pair of recombination signal sequences by RAG-1 and RAG-2. Ubiquitous nonspecific DNA-bending factors of the HMG box family, such as HMG-1, are known to assist in these processes. After cleavage, the RAG proteins remain bound to the cut signal ends and, at least in vitro, support the integration of these ends into unrelated target DNA via a transposition-like mechanism. To investigate whether the protein complex supporting synapsis, cleavage, and transposition of V(D)J recombination signals utilized the same complement of RAG and HMG proteins, I compared the RAG protein stoichiometries and activities of discrete protein-DNA complexes assembled on intact, prenicked, or precleaved recombination signal sequence (RSS) substrates in the absence and presence of HMG-1. In the absence of HMG-1, I found that two discrete RAG-1/RAG-2 complexes are detected by mobility shift assay on all RSS substrates tested. Both contain dimeric RAG-1 and either one or two RAG-2 subunits. The addition of HMG-1 supershifts both complexes without altering the RAG protein stoichiometry. I find that 12/23-regulated recombination signal synapsis and cleavage are only supported in a protein-DNA complex containing HMG-1 and a RAG-1/RAG-2 tetramer. Interestingly, the RAG-1/RAG-2 tetramer also supports transposition, but HMG-1 is dispensable for its activity.
The antigen binding regions of immunogobulin and T-cell receptors are encoded in arrays of noncontiguous coding segments, called variable (V), diversity (D), and joining (J), that are assembled during lymphocyte development to generate the variable-region exon of the mature antigen receptor gene (5). The assembly process, termed V(D)J recombination, involves a series of site-specific DNA rearrangements mediated by recombination signal sequences (RSSs) abutting the coding segments. Each RSS contains conserved heptamer and nonamer elements, separated by nominally conserved spacer DNA that is either 12 or 23 bp long (12-RSS and 23-RSS, respectively). The RSS facilitates the generation of functional antigen receptors via the “12/23 rule,” a restriction that limits recombination to a pair of coding segments whose flanking RSSs bear spacer DNAs of different lengths.
The products of recombination-activating genes 1 and 2, RAG-1 and RAG-2 (35, 42), initiate V(D)J recombination by introducing a DNA double-strand break (DSB) at a 12/23 pair of RSSs (at the heptamer-coding segment border). The cleavage reaction generates two distinct DNA intermediates: blunt, 5′-phosphorylated signal ends (SE) and coding ends terminating in covalently sealed DNA hairpin structures (38, 39, 43). DSB intermediates are generated in two biochemical steps: first-strand nicking at the 5′ end of the heptamer, followed by a direct transesterification reaction in which the 3′-OH exposed in the nicking step attacks the phosphodiester on the antiparallel DNA strand (26, 52). The RAG proteins also mediate other reactions in vitro, including a disintegration reaction similar to those catalyzed by retroviral integrases (28), the nicking of DNA hairpins (6, 45), the cleavage of 3′ flap structures (41), and the insertion of SE into double-stranded DNA via transposase-like strand transfer reactions (1, 19).
The RAG-RSS complex relevant to physiological V(D)J recombination is expected to contain two RSSs (one 12-RSS and one 23-RSS), which are brought together by the RAG proteins into a 12/23 paired signal complex (PC). In the context of the PC, also called a synaptic complex, DSBs are introduced at both RSSs in a coordinated or coupled cleavage reaction. Synaptic complex formation and coupled cleavage adhering to the 12/23 rule have been recapitulated in vitro (13, 40). Both are facilitated by certain nonspecific DNA-bending proteins of the HMG box family (e.g., HMG-1 and HMG-2), and both exhibit a divalent metal ion dependence. Ca2+ is sufficient to promote PC formation but not DNA cleavage. Coupled cleavage of 12/23 RSS pairs requires Mg2+; Mn2+ decouples the reaction, allowing cleavage to occur at an isolated RSS.
Both cleavage steps are minimally supported in vitro in a protein-DNA complex, termed a stable or single complex (SC), containing a RAG-1 dimer and one or two subunits of RAG-2 bound to an isolated RSS (4, 46, 48). HMG-1 and HMG-2 can be stably incorporated into the SC (30, 47, 51). The addition of HMG-1/HMG-2 does not significantly affect the cleavage of isolated 12-RSS substrates by the RAG proteins but stimulates both nicking and hairpin formation (the latter preferentially) on 23-RSS substrates (51), even in the absence of synapsis (47).
In contrast to the SC, whose RAG protein stoichiometry has been characterized, the number of RAG-1 and RAG-2 protomers present in the PC remains unclear. In one model of the PC, RAG-1 and RAG-2 are bound as a mixed tetramer containing two molecules of each protein, based on the following lines of evidence: (i) RAG-1 alone forms a stable dimer in solution (4) and retains this configuration when bound to DNA (37), regardless of whether RAG-2 is present (48); (ii) RAG-1 and RAG-2 associate as a tetramer in solution (in a 1:1 ratio) (4); and (iii) two active sites are present in each dimer of RAG-1, one contributed from each RAG-1 subunit (22, 46). However, recent studies provide indirect evidence suggesting that the PC contains a larger number of RAG-1 protomers, possibly a tetramer (22, 31). Discriminating between these models necessarily requires a direct approach that compares the RAG stoichiometries within the SC and PC side by side. A related issue, as yet unresolved, is whether signal end complexes (SEC) that support transposition require the same complement of RAG and HMG proteins as the PC for their assembly and activity.
Evidence has implicated both synaptic-complex assembly and the hairpin step of V(D)J cleavage as critical control points in the enforcement of the 12/23 rule (17, 53). However, the relative contribution of each factor, as well as the role of the HMG protein in these processes, remains uncertain, as no study has directly compared the catalytic activities of preformed paired RSS complexes.
To address these issues, I compared the stoichiometries and catalytic activities of the SC, PC, and SEC directly by using a combination of mobility shift and in-gel enzyme assays. I find that two distinct SC complexes are assembled on intact, prenicked, and precleaved RSS substrates (SC1 and SC2). Both SC complexes contain a stable RAG-1 dimer. One species contains a single RAG-2 monomer (SC1), whereas a slower-migrating species (SC2) contains two RAG-2 subunits. Both complexes are supershifted by HMG-1 (forming HSC1 and HSC2). The PC (and SEC) assembled with an RSS pair contains the same complement of RAG proteins as HSC2. The 12/23 rule is found to be imposed both during PC (and SEC) assembly and during catalysis of strand cleavage (or strand transfer). Interestingly, both the SC2 and SEC species assembled from precleaved substrates support transposition. Thus, HMG-1 is required for 12/23-regulated PC assembly and RSS substrate cleavage but is dispensable for RAG-mediated transposition.
MATERIALS AND METHODS
DNA constructs.
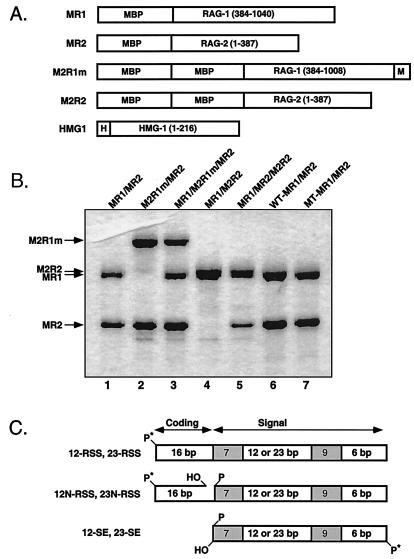
Eukaryotic expression constructs encoding core RAG-1 and RAG-2 fused at the amino terminus to one or two copies of maltose binding protein (MBP) have either been previously described or were assembled from previously published constructs by conventional techniques (M2R2) (47, 48). The prokaryotic expression construct pET11d-hHMG-1 has been described elsewhere (15). Fusion proteins used in this study are depicted in Fig. 1.
FIG. 1.
RAG proteins and RSS substrates used in this study. (A) Schematic diagrams of RAG-1, RAG-2, and HMG-1 fusion proteins (encoded residues are in parentheses) designated at the left. MBP, myc (M), and polyhistidine (H) sequences are also indicated. (B) Single and double MBP-RAG fusion proteins were coexpressed in the indicated combinations and purified by amylose affinity chromatography. Wild-type and active-site mutant (D600A) forms of RAG-1 (WT-MR1 and MT-MR1, respectively) were coexpressed with RAG-2 and similarly prepared. Protein samples were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and detected by silver staining. The positions of the RAG-1 and RAG-2 fusion proteins are shown at the left. (C) Schematic diagrams of radiolabeled intact (12-RSS, 23-RSS), prenicked (12N-RSS, 23N-RSS), and precleaved (12-SE, 23-SE) RSS substrates, along with the position of the 32P end label (∗P). Heptamer and nonamer sequences are shaded. The lengths of the flanking and spacer regions are also shown.
Protein expression and purification.
Single and double MBP-RAG expression constructs (wild type or mutant) were cotransfected with pRSVT in 293 cells by calcium phosphate precipitation (49). Typically, 14 10-cm-diameter tissue culture dishes were transfected with 20 μg of expression construct(s) (20 μg for individually expressed proteins or 10 μg of both RAG-1 and RAG-2 constructs for coexpressed RAGs)/plate and 1 μg of pRSVT/plate and harvested at 48 h posttransfection. Fusion proteins were purified by amylose affinity chromatography as previously described (48, 49). Polyhistidine-tagged human HMG-1 was expressed in Escherichia coli and purified by a previously published procedure (47).
Oligonucleotide binding assays.
Intact and prenicked 12- and 23-RSS substrates were assembled and purified as described previously (46). Precleaved substrates containing only an RSS SE were similarly prepared by annealing cold 5′-phosphorylated DG10 (12-SE) or DG4 (23-SE) to their respective complements labeled at the 5′ end with 32P (26). Diagrams of the RSS substrates used in this study are shown in Fig. 1. Electrophoretic mobility shift assays (EMSA) were performed under the same conditions described previously (49), except that DAR 81/82 was omitted as a competitor. Briefly, binding reaction mixtures (10 μl) containing individually expressed (∼30 ng) or coexpressed RAG proteins (∼100 ng) and HMG-1 (5 μg/ml; when indicated in Fig. 2, 4, and 5) were preincubated with radiolabeled substrate DNA (∼0.02 pmol) in the presence of 5 mM Ca2+ for 1 min at 25°C. Subsequently, either 25-fold- (SE substrates) or 50-fold (intact and prenicked)-excess cold partner DNA (either intact, prenicked, or precleaved, as appropriate) was added, as determined by titration experiments (data not shown). Samples were incubated for an additional 10 min at 25°C and then fractionated at 4°C on native 4% gels. Protein-DNA complexes were visualized from dried gels by autoradiography with a Molecular Dynamics Storm 860 PhosphorImager.
FIG. 2.
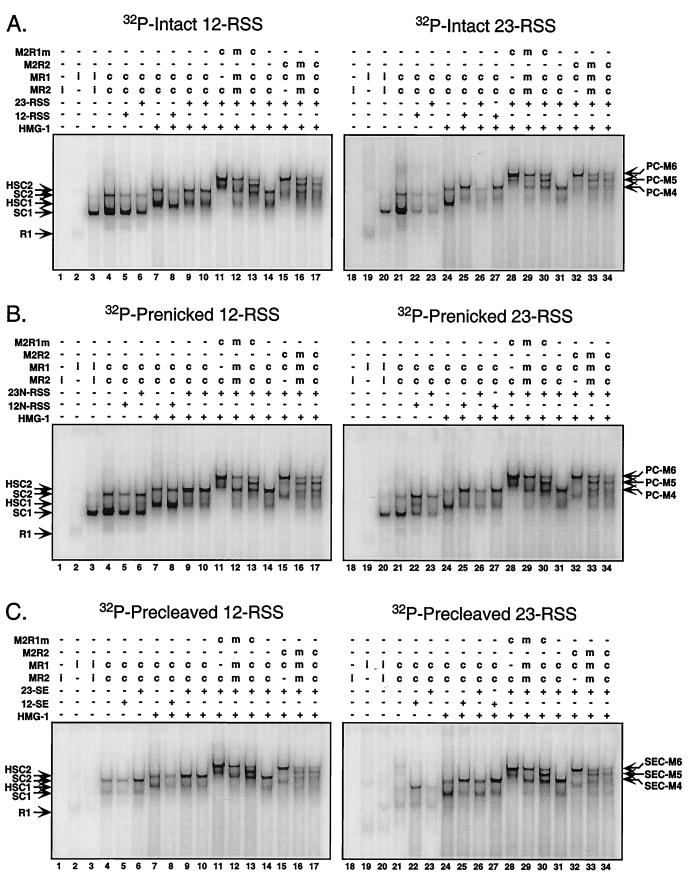
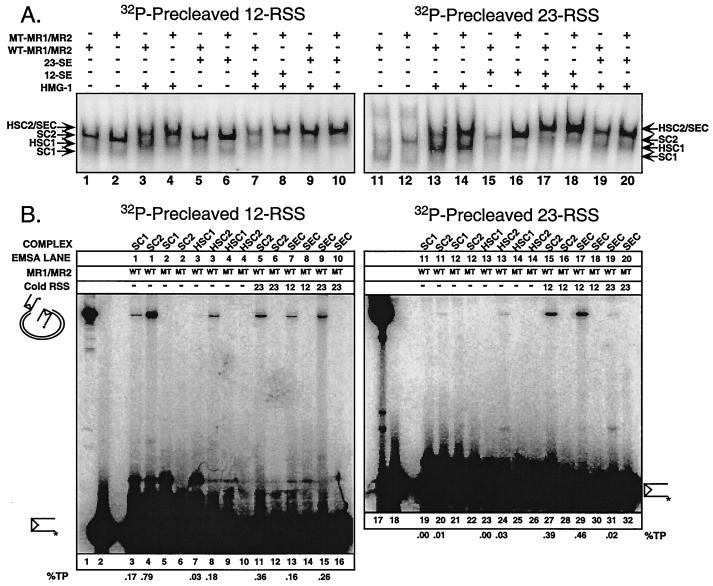
RAG synaptic complexes contain a RAG-1/RAG-2 tetramer core. EMSAs were performed by using 32P-end-labeled intact (A), prenicked (B), or precleaved (C) substrates containing either a canonical 12-RSS (left) or 23-RSS (right). Individually expressed (i) or coexpressed (c) single and/or double MBP-RAG proteins, defined in Fig. 1A, were incubated with radiolabeled substrate (Fig. 1C) in the absence or presence of HMG-1 and/or cold partner DNA as indicated above the gel. In some cases (e.g., lanes 12, 16, 29, and 33), samples contained an equal mixture (m) of the two coexpressed RAG protein preparations identified in the previous two lanes (e.g., lane 12 contains a mixture of coexpressed RAG proteins found in lanes 10 and 11). Protein-DNA complexes were fractionated by EMSA and visualized by autoradiography with a PhosphorImager. The positions of complexes containing dimeric RAG-1 alone (R1), dimeric RAG-1 and monomeric RAG-2 (SC1), and a RAG-1/RAG-2 tetramer (SC2) are shown at left. Complexes supershifted by the incorporation of HMG-1 are also indicated (HSC1 and HSC2). The positions of paired RSS complexes containing HMG-1 and a RAG-1/RAG-2 tetramer bound to intact (PC), prenicked (PC), or precleaved (SEC) substrates are noted at the right. Whether four, five, or six MBP moieties are distributed among the RAG subunits in these complexes is also indicated (M4, M5, and M6, respectively).
In-gel cleavage and transposition assays.
Preformed RAG complexes assembled on intact and prenicked substrates were assayed for cleavage activity by an in-gel cleavage assay described previously (46). Briefly, preparative-scale binding reaction mixtures (50 μl) were assembled in the presence of Ca2+ and fractionated by EMSA as described above. After disassembly, the gel was submerged in buffer containing 5 mM MgCl2 and incubated at 37°C for 1 h to initiate DNA cleavage within fractionated protein-DNA complexes. Following electrophoretic transfer of the DNA to DEAE-cellulose, reaction products attributed to complexes of interest (as visualized by autoradiography) were recovered, normalized, and fractionated by denaturing gel electrophoresis. A similar approach was used to determine transposition activity of RAG complexes assembled on SE substrates, except that samples were fractionated on native gels containing the target plasmid pcDNA1 (4.0 kb; Invitrogen), which was added to the acrylamide mixture before pouring the gel (2.5 μg/ml). After EMSA, the gel was incubated in transposition buffer (25 mM MOPS [morpholinepropanesulfonic acid]-KOH [pH 7.0], 60 mM potassium glutamate, 10 mM CaCl2, 10% glycerol) at 37°C for 1 h. Reaction products were recovered as described above and fractionated on a linear 4 to 20% gradient native polyacrylamide gel. PhosphorImager scans of dried gels were quantified with ImageQuant software.
RESULTS
Paired RSS complexes contain a RAG-1/RAG-2 tetramer core.
As part of an ongoing effort to understand the molecular processes underlying V(D)J recombination, I have worked to characterize discrete protein-DNA complexes assembled on single RSS substrates containing various combinations of individually purified RAG-1, RAG-2, and HMG-1 (or HMG-2) (46-49). To extend these results, I wished to biochemically characterize RAG synaptic complexes containing a pair of RSS substrates in order to clarify their relationship to lower-order RAG-RSS complexes described previously (either lacking or containing HMG-1). For this study, forms of core RAG-1 and RAG-2 proteins, fused at the amino terminus to either one or two copies of MBP, were expressed individually or coexpressed in 293 cells and purified by amylose affinity chromatography (Fig. 1). A hexahistidine-tagged form of human HMG-1 was overexpressed in bacteria and purified from clarified lysates by Ni2+-chelation chromatography.
I first compared the requirements for assembly of single and paired RSS complexes by an EMSA, focusing particularly on the role of HMG-1 in protein-DNA complex formation. The DNA substrates used in these experiments contained a canonical 12- or 23-RSS that was either intact, prenicked, or precleaved (containing only an SE) (Fig. 1C and 2). Under these conditions, HMG-1 binds all three substrates tested in the absence of the RAG proteins (data not shown). RAG-2 shows no binding activity (Fig. 2, lanes 1 and 18), but RAG-1 alone binds all three substrates to low levels (R1; Fig. 2, lanes 2 and 19). When individually expressed RAG-1 and RAG-2 are mixed and analyzed, a protein-DNA complex of retarded mobility, termed SC1, is clearly observed in reactions containing intact or prenicked DNA substrates (Fig. 2A and B, lanes 3 and 20) but is only faintly visible in reactions containing precleaved substrates (Fig. 2C, lanes 3 and 20). A minor, slower-migrating species is also detected (at higher exposure) in samples containing intact or prenicked substrates (SC2). It is important to note at this point that SC1 and SC2 are designated based on their relative electrophoretic mobilities and their appearance in reactions containing only a single type of RSS substrate, rather than their intrinsic activities, which as shown below may be altered by the addition of cold partner DNA.
When both RAG-1 and RAG-2 are coexpressed, the abundances of SC1 and SC2 in all samples are increased (Fig. 2, lanes 4 and 21). Thus, coexpressed RAG proteins are used in the remaining samples. For intact substrates, the abundances of both the SC1 and SC2 complexes are reduced upon addition of cold intact partner DNA; the levels of reduction are comparable irrespective of whether the partner DNA contains a 12- or 23-RSS (Fig. 2A, lanes 5 and 6 and 22 and 23). In contrast, when cold prenicked or precleaved partner DNA is added to their respective samples, the formation of the SC1 complex is diminished, while SC2 complex formation is stimulated in a 12/23-dependent fashion, suggesting the presence of two RSS substrates in this complex (Fig. 2B and C; compare lanes 5 and 6 and 22 and 23). In all three cases, however, the mobility of the complex is not significantly altered by addition of cold partner DNA.
When binding reactions containing coexpressed RAG proteins are supplemented with HMG-1 in the absence of partner DNA, both the SC1 and SC2 complexes are supershifted (HSC1 and HSC2; Fig. 2, lanes 7 and 24). For intact substrates, the addition of HMG-1 and cold partner DNA stimulates the 12/23-regulated formation of a complex that comigrates with HSC2. The preference for the appropriate RSS partner over the inappropriate partner is about three- to fourfold. This species is considered to contain an RSS pair and is therefore designated PC (or SEC) at this point for consistency with previously published reports describing conditions for the detection of paired RSS complexes by EMSA (17). In contrast, the level of HSC1 complex formation is reduced (Fig. 2A, compare lanes 8 and 9 and 25 and 26). For prenicked and precleaved substrates, the addition of cold partner DNA has the same effect as in the absence of HMG-1 but the abundance of the PC (and SEC) is slightly greater than that of counterparts lacking HMG-1 (Fig. 2B and C, lanes 9 and 10 and 25 and 26).
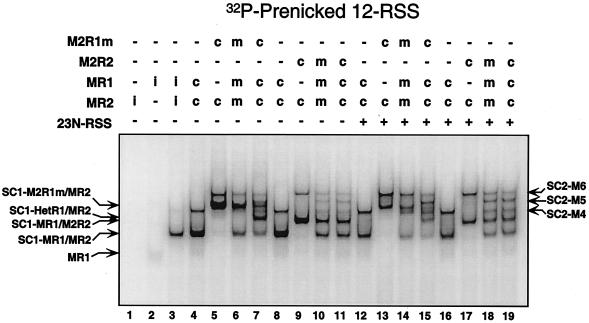
I next wished to examine the stoichiometry of the RAG proteins in the context of the PC (or SEC) for all of the DNA substrates listed above. To determine RAG protein stoichiometry, I coexpressed forms of RAG-1 and RAG-2 fused to one or two copies of MBP (thus altering their molecular weights) and analyzed protein-DNA complex formation by EMSA (48). The detection of one complex (or more) whose electrophoretic mobility is intermediate to those observed in binding reactions containing only one form of the RAG protein indicates heterodimer (or multimer) formation. As expected, the PC (or SEC) obtained from samples containing either MR1 or M2R1m coexpressed with MR2 displayed distinct mobilities, reflecting the difference in their relative molecular weights (PC [or SEC-]-M4 and PC [or SEC-]-M6, respectively; Fig. 2, compare lanes 10 and 11 and 27 and 28). When these two RAG protein preparations were mixed in the binding reaction, no species of intermediate mobility were detected by EMSA (Fig. 2, lanes 12 and 29). However, samples containing both forms of RAG-1 coexpressed with RAG-2 showed the presence of a single predominant species of intermediate mobility (PC [or SEC-]-M5; Fig. 2, lanes 13 and 30), consistent with a RAG-1 dimer configuration in the PC. The PCs assembled from reactions containing MR1 coexpressed with either MR2 or M2R2 also displayed distinct electrophoretic mobilities (Fig. 2, compare lanes 14 and 15 and 31 and 32). Interestingly, when both RAG protein preparations were mixed, an intermediate species was clearly evident (Fig. 2, lanes 16 and 33). Similar results were also obtained when MR1 was coexpressed with both forms of RAG-2 (Fig. 2, lanes 17 and 34). These data suggest that two RAG-1 and two RAG-2 subunits are present in the PC (and SEC) and that, unlike RAG-1, the RAG-2 subunits freely reassort in solution. Consistent with the conclusion that the PC (and SEC) contains a RAG-1/RAG-2 tetramer core, the PCs (and SECs) assembled from M2R1m or M2R2 comigrate (Fig. 2, compare lanes 11 and 15 and 28 and 32), as predicted if six MBP moieties are distributed among four RAG protomers. In contrast, the minor HSC1 species detected in the same two samples do not comigrate, an outcome expected in principle if HSC1 contains a RAG-1 dimer and monomeric RAG-2. Indeed, this stoichiometry was verified for HSC1 by using combinations of single and double MBP-RAG proteins in the absence of partner DNA (data not shown). By the same approach, SC2 was also found to possess the same RAG stoichiometry as the PC (and SEC) (Fig. 3). Thus, both SC1 and HSC1 contain a stable RAG-1 dimer and monomeric RAG-2, and SC2, HSC2, and PC (and SEC) contain a stable RAG-1 dimer and two subunits of RAG-2 capable of reassorting in solution.
FIG. 3.
The RAG protein stoichiometries of the SC2 and PC (or SEC) species are identical and differ from that of the SC1 species by the addition of a single RAG-2 subunit. Combinations of single and/or double MBP-RAG proteins were incubated with 32P-end-labeled prenicked 12-RSS substrate in the absence of HMG-1 with or without cold prenicked partner 23-RSS as shown above the gel (see the legend to Fig. 2 for more detail, including definitions of “i,” “c,” and“m”). Comparable results were obtained with intact and precleaved substrates as well.
The PC supports 12/23-dependent cleavage of intact and prenicked substrates.
I next evaluated the catalytic activity of the complexes observed by EMSA. To assess RSS cleavage activity, I used an in-gel cleavage assay to compare catalytic activities of multiple, discrete protein-DNA complexes in a single native polyacrylamide gel (46). In the first experiment, wild-type or active-site mutant (D600A) RAG-1 (14, 21, 23) coexpressed with RAG-2 was incubated under binding conditions (in the presence of Ca2+) with intact 12- or 23-RSS substrates in the absence or presence of HMG-1 and/or cold partner DNA in combinations comparable to those shown in Fig. 2. Samples were fractionated by EMSA, and the gel was soaked in buffer containing Mg2+ for 1 h at 37°C to initiate DNA cleavage within the protein-DNA complexes. Autoradiographs of the DNA following electrophoretic transfer to DEAE-cellulose show that wild-type and mutant RAG-1s assemble protein-DNA complexes to comparable levels (Fig. 4A). DNA derived from the SC1, SC2, HSC1, HSC2, and PC species was recovered, normalized, and fractionated by denaturing gel electrophoresis (Fig. 4B). To account for slight variations in the actual amount of DNA loaded on the gel (despite attempted normalization), the cleavage products were quantified and expressed as percentages of total DNA present on the gel.
FIG. 4.
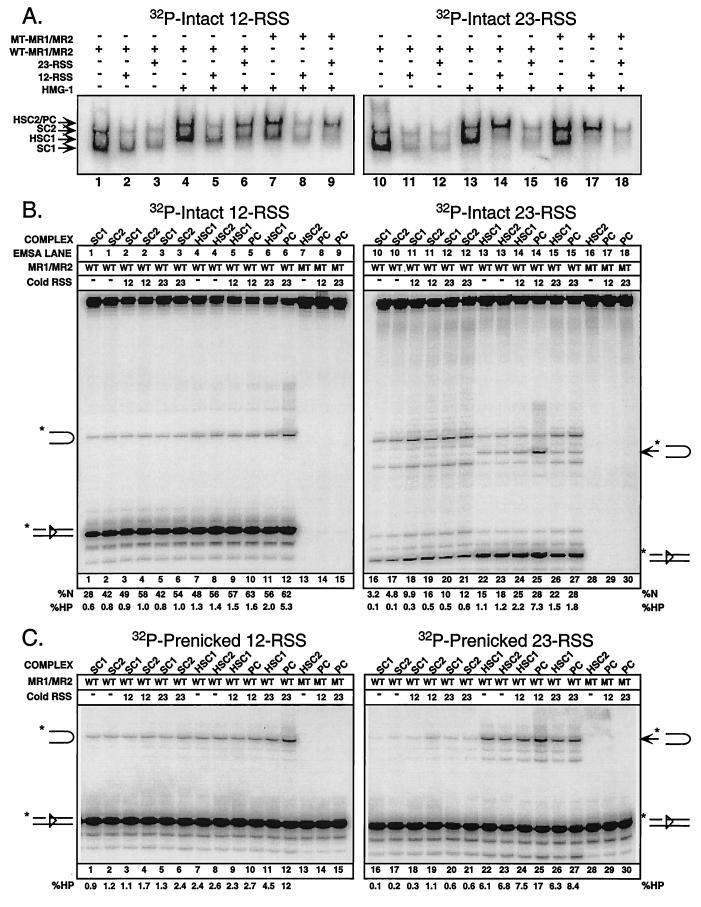
12/23-regulated RAG-mediated hairpin formation requires HMG-1. (A) Wild-type or active-site mutant (D600A) MR1 coexpressed with MR2 (WT-MR1/MR2 or MT-MR1/MR2, respectively) was incubated with intact 12-RSS (left) or 23-RSS (right) substrates with or without HMG-1 and/or cold partner DNA (12-RSS or 23-RSS) under binding conditions, as indicated above the gel, and subjected to an in-gel cleavage assay (see Materials and Methods). The autoradiographs of electrophoretically transferred DNA shown here were used to isolate reaction products from complexes of interest. The positions of SC1, HSC1, SC2, and HSC2/PC (comigrating) species, defined for Fig. 2, are shown at the left. (B) Recovered reaction products from panel A were normalized and fractionated by denaturing gel electrophoresis. Positions of nicked and hairpin products are shown at the left and right. The hairpin species shown on gels of 12- and 23-RSS substrates have previously been shown to comigrate. The percentages of nicked (%N) and hairpin (%HP) products in each lane are quantified below the gel and account for slight variations in the amount of DNA actually loaded. (C) In-gel cleavage experiments using prenicked 12-RSS (left) or 23-RSS (right) substrates were performed as described above and presented in the same order. Autoradiographs of DNA transferred from the gel resemble those shown in panel A and reveal a distribution of protein-DNA complexes similar to that observed in comparable lanes depicted in Fig. 2B (data not shown). Quantitative analysis of hairpin formation is shown below the gel. Note that all reaction products shown on both sides of panels B and C are derived from a single native gel subjected to the in-gel cleavage reaction. The abundance and distribution of the cleavage products observed are representative of independent experiments.
Consistent with previous results (46), significant nicking was detected in 12-RSS substrates recovered from wild-type SC1 formed in the absence of cold partner DNA, as well as low levels of the hairpin product (Fig. 4B, lane 1). The abundance of nicks, but not hairpin products, is slightly enhanced in the SC2 species obtained from the same lane (Fig. 4B, lane 2). The addition of intact 12- or 23-RSS partner DNA, while reducing complex formation, appears to modestly stimulate nicking independent of RSS composition (Fig. 4B, lanes 3 to 6). Supplementing HMG-1 in the absence of cold partner DNA supershifts the SC1 and SC2 species (HSC1 and HSC2, respectively) but does not appreciably alter the cleavage activity of either complex (Fig. 4B, lanes 7 and 8). Addition of cold 12-RSS partner DNA, while impairing HSC1 and PC formation, does not dramatically change the cleavage activity of either complex (Fig. 4B, lanes 9 and 10). However, while the addition of cold 23-RSS partner DNA has relatively little effect on HSC1 activity, PC cleavage activity is stimulated approximately fourfold relative to that observed in the PC formed in the absence of cold 12-RSS partner DNA (Fig. 4B, lanes 11 and 12; compare lane 10 to lane 12). Importantly, the PC formed by using a RAG-1 D600A mutant is essentially inactive, even if cold partner DNA is added (Fig. 4B, lane 13 to 15).
An in-gel cleavage assay using an intact 23-RSS substrate yielded results generally consistent with the data described above. However, the 23-RSS SC1 and SC2 species assembled in the absence of partner DNA generate lower levels of nicked products, have a propensity to nick aberrantly, and produce virtually no hairpin product compared to their 12-RSS SC counterparts (Fig. 4B, lanes 16 and 17). The addition of cold partner DNA enhances nicking activity appreciably (two- to threefold); nicking is preferentially stimulated in the SC2 species when 12-RSS partner DNA is added (Fig. 4B, lanes 18 to 21). The HSC1 species formed in the presence of HMG-1 exhibits enhanced nicking activity with reduced levels of aberrant nicking and supports hairpin formation to detectable levels (Fig. 4B, lane 22). The HSC2 species obtained from the same lane possesses the same activity as HSC1 with respect to nicking and hairpin formation (Fig. 4B, lane 23). However, when cold 12-RSS partner DNA is added, hairpin formation is not significantly altered in HSC1 but is preferentially stimulated in the PC obtained from the same lane (Fig. 4B, lanes 24 and 25). Such stimulation is not observed when cold 23-RSS partner DNA is added (Fig. 4B, lanes 26 and 27). As observed with the 12-RSS substrate, no cleavage activity is detected in control PCs containing a RAG-1 active-site mutant, regardless of whether cold partner RSS is present (Fig. 4B, lanes 28 to 30).
To determine whether the nicking step was rate limiting for the 12/23-regulated stimulation of hairpin formation observed in PCs assembled on intact RSS substrates, I next performed a similar experiment using a prenicked, rather than intact, RSS substrate (Fig. 4C). The results are largely consistent with those shown in Fig. 4B, except that a slight 12/23-dependent stimulation (approximately twofold) of hairpin formation in the 23-RSS SC2 complex is detectable, and the addition of cold prenicked 23-RSS partner DNA simulates hairpin formation in the PC, albeit to a lesser extent than prenicked 12-RSS partner DNA. Taken together, these results show that a PC containing a RAG-1/RAG-2 tetramer and HMG-1 supports 12/23-regulated synapsis and cleavage of RSS substrates in the presence of Mg2+.
HMG-1 promotes synapsis of SE substrates but is dispensable for RAG-mediated transposition.
To assess whether the SEC described in Fig. 2 supports transposition of SE, I used an in-gel assay similar to the one described above, except that plasmid DNA was added to the acrylamide solution used to pour the native gel for the EMSA. The plasmid DNA in this assay serves as the target of the strand transfer reaction catalyzed by the RAG proteins. In preliminary experiments, the plasmid pcDNA1 was shown to be large enough so that the protein-DNA complexes migrated faster than the plasmid DNA in a native 4% gel and small enough to allow sufficient electrophoretic transfer to DEAE-cellulose following the EMSA (data not shown). Wild-type or active-site mutant (D600A) RAG-1 coexpressed with RAG-2 was incubated under binding conditions (in the presence of Ca2+) with radiolabeled precleaved substrate in the absence or presence of HMG-1 and/or cold partner DNA in combinations comparable to those for Fig. 4. Protein-DNA complexes were separated by an EMSA, and the gel was soaked in buffer containing 10 mM Ca2+ for 1 h at 37°C, conditions known to support transposition in vitro (19). Autoradiographs of the DNA following electrophoretic transfer to DEAE-cellulose show that the wild-type and mutant forms of RAG-1 support protein-DNA complex formation to comparable levels (and to levels similar to the data shown in Fig. 2C), with the exception that the RAG-1 active-site mutant appears to assemble SC2 complexes more efficiently than wild-type RAG-1 (Fig. 5A). The basis for this phenomenon is unknown but is currently under investigation. The DNA recovered from the SC1, HSC1, SC2, HSC2, and SEC species was normalized and fractionated on a native 4 to 20% linear-gradient gel along with radiolabeled, linearized pcDNA1 and free precleaved substrate DNA (Fig. 5B, lanes 1 and 2). Double-ended insertion results in a DNA DSB of pcDNA1, with radiolabeled substrate DNA covalently linked to both ends of the linearized plasmid (19).
FIG. 5.
HMG-1 is dispensable for RAG-mediated transposition in a preformed SEC. (A) Precleaved 12-RSS (left) or 23-RSS (right) substrates were incubated with wild-type or active-site mutant (D600A) MR1 coexpressed with MR2 (WT-MR1/MR2 or MT-MR1/MR2, respectively) with or without HMG-1 and/or cold partner DNA (12-SE or 23-SE) under binding conditions, as indicated above the gel, and then subjected to an in-gel transposition assay (see Materials and Methods). The autoradiographs of the DEAE-cellulose paper to which the DNA was transferred are shown. The positions of SC1, HSC1, SC2, HSC2/SEC (comigrating) species, defined in Fig. 2, are shown at the left. (B) Reaction products were isolated from complexes of interest by using the autoradiographs in panel A and analyzed on a native linear 4 to 20% gradient gel. The position of pcDNA1, linearized by double-ended insertion, is indicated at the left. Linearized 5′-end-labeled pcDNA1 (lanes 1 and 17) and precleaved substrate (lanes 2 and 18; indicated at the left) serve as markers. The percentages of integrated substrate DNA are quantified below the gel (%TP).
Unexpectedly, a wild-type SC2 species assembled on a precleaved 12-RSS formed in the absence of cold partner DNA supports transposition, as a radiolabeled product that comigrates with linearized, radiolabeled pcDNA1 is clearly visible (Fig. 5B, lane 4). A smaller amount of this product is also observed in DNA recovered from the SC1 species present the same lane of the gel (Fig. 5B, lane 3). However, as this may be due to a low level of cross-contamination with SC2, whether the SC1 species truly supports some degree of transposition is unclear. As an important control, no double-ended insertion product is recovered from SC1 or SC2 complexes assembled by using a RAG-1 active-site mutant (Fig. 5B, lane 5 and 6), demonstrating that the radiolabeled substrate is covalently linked to the plasmid rather than associated via RAG-DNA interactions. Interestingly, HMG-1 suppresses transposition activity in HSC1 and HSC2 species formed in the absence of cold partner DNA (Fig. 5B, lanes 7 and 8). When cold 23-SE partner DNA is added in the absence of HMG-1, transposition activity in SC2 is reduced about twofold relative to the activity exhibited by its counterpart formed in the absence of partner DNA and HMG-1 (Fig. 5B, compare lane 11 to lane 4). This outcome is consistent with the possibility that two radiolabeled 12-SE substrates are present in SC2, one of which is exchanged for a cold 23-SE when this RSS partner is present. When both HMG-1 and cold 12-SE partner DNA are present, the SEC possesses no more activity than HSC2 (Fig. 5B, compare lane 13 to lane 8). However, when cold 23-SE partner DNA is present, transposition activity is stimulated to a level comparable to that for the 12/23 SC2 species (Fig. 5B, compare lane 15 to lane 11). Importantly, all complexes formed by using a RAG-1 active-site mutant failed to support transposition (Fig. 5B, lanes 4, 5, 9, 10, 12, 14, and 16).
Interestingly, the SC2 assembled on a radiolabeled 23-SE substrate in the absence of HMG-1 and cold partner DNA exhibits little transposition activity, but, in the presence of cold 12-SE partner DNA, transposition is stimulated ∼40-fold (Fig. 5B, compare lanes 20 and 27). In contrast to complexes assembled on precleaved 12-RSS substrates, HMG-1 did not suppress transposition activity when incorporated into the SEC formed in the absence of cold partner DNA (Fig. 5B, lane 24), nor did it greatly enhance activity (relative to the SC2 complex) when cold 12-SE partner DNA was present (Fig. 5B, compare lanes 27 and 29). However, when both HMG-1 and cold 23-SE partner DNA were present, the activity of the SEC dropped dramatically, to a level similar to that for HSC2 assembled in the absence of cold partner DNA (Fig. 5B, compare lane 31 to lane 13). Based on these data, I conclude that a RAG-1/RAG-2 tetramer supports transposition. In the absence of HMG-1, the RAG-1/RAG-2 tetramer supports both 12/12 and 12/23 transposition to similar levels; both reactions are intrinsically more favorable than 23/23 transposition. HMG-1 appears to actively inhibit 12/12 transposition, thereby making 12/23 transposition the most favorable. However, transposition activities in preformed 12/23 SC2 and SEC species are essentially identical, suggesting that HMG-1 is dispensable for RAG-mediated transposition.
DISCUSSION
A RAG-1/RAG-2 tetramer core supports 12/23-regulated synapsis, cleavage, and transposition of V(D)J recombination signals.
All site-specific recombination reactions require the orchestrated synapsis of recombination sites mediated (in every example to date) by a multisubunit protein complex. V(D)J recombination provides an important example of this process, in which two lymphoid cell-specific proteins, RAG-1 and RAG-2, collaborate with ubiquitous nonspecific DNA-bending factors (e.g., HMG-1 and -2) to facilitate the assembly of a nucleoprotein complex that supports the initiation of V(D)J rearrangement. As part of an ongoing effort to understand how RAG-1 and RAG-2, with assistance by HMG-1, restrict binding and cleavage to a 12/23 pair of recombination signals, I investigated the composition and assembly of RAG synaptic complexes with model V(D)J recombination signals. In this study, I examined the requirements for synapsis, cleavage, and transposition of RSS substrates by using a combination of mobility shift and in-gel enzyme assays. The results presented here extend previous work by demonstrating that (i) a RAG-1/RAG-2 tetramer supports 12/23-regulated synapsis, cleavage, and transposition of V(D)J recombination signals; (ii) the synaptic complex is assembled by association of a stable RAG-1 dimer with two RAG-2 subunits that freely reassort in solution; (iii) HMG-1 stimulates both PC and SEC formation, yet, once the complexes are formed, HMG-1 is required only for 12/23-dependent cleavage and is dispensable for RAG-mediated transposition activity; and (iv) the 12/23 rule is clearly imposed, both at the level of RSS binding and at the level of hairpin formation.
Implications for synaptic complex assembly.
The data presented here suggest that, in the absence of HMG-1, the RAG proteins together assemble at least two distinct protein-DNA complexes with isolated recombination signals, termed SC1 and SC2. SC1 contains a stable RAG-1 dimer and monomeric RAG-2, consistent with data published previously (48), and SC2 contains an additional subunit of RAG-2. This result is in agreement with a report by Sadofsky and colleagues concluding that RAG-1 and RAG-2 form a tetramer in solution and retain this configuration when bound to DNA (4). However, in that study, the authors did not examine whether the RAG-1/RAG-2 tetramer they observed supported 12/23-regulated synapsis and cleavage of V(D)J recombination signals. In accord with previous studies (30, 47, 51), the SC1 and SC2 species are supershifted by the incorporation of HMG-1 (forming HSC1 and HSC2, respectively). Data shown here suggest the RAG protein stoichiometry is not altered by the presence of HMG-1. Although HMG-1 stimulates 23-RSS cleavage within an HSC1 species containing monomeric RAG-2, as noted previously (47), 12/23-regulated synapsis and cleavage of intact RSS substrates are only supported when the RAG-1/RAG-2 tetramer and HMG-1 occur in the context of a PC.
The conclusion reached here that the SC2 and PC species both contain a RAG-1/RAG-2 tetramer contrasts with studies published recently by two other laboratories (22, 31). In a study by Oettinger and coworkers (31), RAG protein stoichiometry in SC and PC species assembled on intact RSS substrates was analyzed by using an approach similar to that presented here. Consistent with my results, the SC2 and PC were found to contain a pair of RAG-2 subunits and to possess the same complement of RAG-1 protomers, which do not reassort upon mixing. However, in contrast to data presented here, the authors could not could unambiguously determine RAG-1 stoichiometry in the SC or PC by using two different-size forms of RAG-1 coexpressed in bacteria, possibly due to limitations of that expression system. Instead, the relative content of RAG-1 and RAG-2 within the SC and PC species was evaluated indirectly. The authors showed by EMSA that the mobilities of the SC2 and the PC assembled with MBP-RAG-1 and core RAG-2 (expressed in HeLa cells) were slightly retarded relative to those of comparable complexes assembled with core RAG-1 and MBP-RAG-2 (expressed in insect cells), leading to the conclusion that RAG-1 is in excess over RAG-2 in both complexes. However, this interpretation may be confounded by differences in the sources and compositions of the RAG proteins used in the experiments, which could contribute to subtle changes in the mobilities of the complexes. Two factors to consider include differences in posttranslational modification of RAG-2 expressed in HeLa versus insect cells and differences in the composition and length of the adaptor used to fuse MBP to RAG-1 and RAG-2, as these expression constructs were prepared by different cloning strategies (21, 26). In this study, all RAG proteins were prepared from mammalian cells expressed by using similar plasmid constructs.
In another study by Roth and colleagues (22), a RAG-1 heterodimer containing an active-site mutation on one subunit was found to cleave a 12/23 plasmid substrate at both RSSs about fourfold less effectively than a wild-type RAG-1 heterodimer. These data led the authors to conclude, based on probability arguments, that the synaptic complex likely contains a pair of RAG-1 dimers. However, since the synaptic complex was not directly observed in this assay, one cannot determine from the data whether the cleavage events observed occurred within the context of a single complex or instead represented the combined activities of more than one complex. Because the RAG-1 dimer was present in large molar excess (∼100-fold) over the DNA substrate in these experiments, the cleavage events observed may reflect nearly simultaneous binding and cleavage at each RSS by separate RAG-1 heterodimers that do not physically interact, a possibility made more significant by the fact that cleavage can occur in vitro in the absence of synapsis (47). The probability of double cleavage in this scenario is the same as that in the two-dimer model, but the difference is that the two RAG-1 dimers do not associate in the same protein-DNA complex. If the PC were to contain a RAG-2 dimer (about which there is agreement between the study of Mundy et al. [31] and the data presented here) and a RAG-1 tetramer, then the PC assembled here from MR1/M2R2 would be unlikely to comigrate with the PC assembled from M2R1m/MR2, as the latter complex would contain an additional two MBP modules (representing ∼80 kDa of protein). The fact that all RAG-2 homo- and heterodimeric SC2 and PC species analyzed here comigrate with comparable species containing RAG-1 on the same gel argues strongly for the presence of a RAG-1 dimer in these complexes. The MBP moieties are unlikely to influence the RAG protein stoichiometry, as dimeric configurations for RAG-1 and RAG-2 have been established by using other approaches (4, 31, 37).
I considered the possibility that a pair of RAG tetramer complexes (one HSC2 containing a labeled RSS and another HSC2 containing an unlabeled partner RSS), rather than a single RAG tetramer complex (bound to a pair of RSS substrates), supports synapsis and cleavage through transient protein-protein interactions within the gel matrix. However, this possibility seems unlikely for three reasons. First, comparable HSC1 and SC2 species do not exhibit 12/23-regulated cleavage, which might reasonably be expected in this scenario since these complexes contain complements of protein similar to the PC. Second, diffusion of the protein-DNA complexes within the gel matrix would likely be impeded by their large size (≥500 kDa), impairing their ability to assemble an appropriate paired RSS complex. Third, the binding of two RSSs by a single RAG-1/RAG-2 tetramer is more plausible, given the following observations: (i) PC formation is stimulated in a 12/23-regulated fashion (which would not be expected if transient higher-order RAG complexes were involved in RSS synapsis), (ii) the nonamer binding domains on both RAG-1 subunits are intact and able to bind DNA (46), and (iii) RAG synaptic complexes evaluated for DNA stoichiometry by mobility shift assays demonstrate the presence of two RSSs within the PC (20).
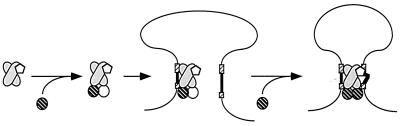
How is the synaptic complex thought to be assembled? Evidence presented here and elsewhere suggests the following sequence of events (Fig. 6). First, RAG-1 and RAG-2 associate in solution, forming a multimeric complex (3, 4, 27, 48). Next, the preassociated RAG-1/RAG-2 complex binds a single RSS (4, 18, 48). Finally, an appropriate partner RSS is integrated into a PC (17), likely through a direct-capture mechanism (31). Evidence presented here suggests that the RAG-1/RAG-2 tetramer and HMG-1 together mediate synapsis. However, the precise order of events that leads to the integration of HMG-1 and the second RAG-2 subunit into the PC remains unclear. Once the PC is assembled, the RAG proteins initiate DNA cleavage, retaining their tetramer configuration through both the nicking and transesterification steps of V(D)J recombination. The postsynaptic complex persisting after DNA cleavage is expected to retain the same RAG protein stoichiometry, as a RAG-1/RAG-2 tetramer complex supports SE synapsis. Although the SEC described here supports transposition, the physiological significance of this activity remains uncertain. The model proposed for the composition and assembly of the PC has several attractive features, including (i) its consistency with earlier observations that a RAG-1 dimer contains two active sites (46), assembles a 1:1 mixed-tetramer complex with RAG-2 in solution (4), and mediates both heptamer and nonamer contact in the presence of RAG-2 (49); (ii) its ability to accommodate variations in the accessibility of antigen receptor loci (5); and (iii) its sensitivity to intracellular RAG-2 levels, which are regulated in the cell cycle by a phosphorylation-dependent degradation mechanism (25).
FIG. 6.
A sequential-binding model of synaptic complex assembly, adapted from Sadofsky (40). In solution, RAG-1 (shaded ovals) exists as a dimer and associates with one or two RAG-2 subunits (hatched and open circles). A RAG complex minimally containing a RAG-1 dimer and monomeric RAG-2 associates with one RSS (a 12-RSS is shown here for simplicity) and then captures an appropriate RSS partner (31). RAG-1 interacts with both the nonamer (long hatched box) and the heptamer (short hatched box), and RAG-2 is localized at the heptamer-coding junction (12, 48, 49). The second RAG-2 subunit may associate with the complex either before or after acquisition of the second RSS (in this case, a 23-RSS). The HMG protein (open pentagon) selectively promotes binding to the 23-RSS through interactions with RAG-1 (2, 47, 51), although the number of protomers involved and the timing of their association with the RAG complex are unclear. Although not directly tested, a trans mode of RAG-1 association is depicted, consistent with the RAG-1 active-site organization observed in SC1 (46). Studies of the minimal intersignal distance required to support coupled cleavage and recombination suggest the heptamer-to-heptamer orientation of the RSSs shown here (11, 44). The RAG tetramer stoichiometry would be retained through both cleavage steps of V(D)J recombination and in the postcleavage complex as well (not shown). For additional details, see the text.
The RAG-1/RAG-2 tetramer configuration proposed here also has precedents in studies of transposases. Bacteriophage Mu transposition, for example, is mediated by a protein-DNA complex, termed a transpososome, containing two Mu end DNA segments bound to a MuA tetramer (24). All four subunits interact specifically with DNA, occupying two of three potential binding sites at each Mu end (29), yet only two of four potential active sites are utilized for the chemical steps involved in Mu transposition (32, 54). However, aside from a superficial resemblance in the overall number of protomers in the synaptic complex, V(D)J recombination bears few other similarities to Mu transposition, especially relative to Tn5 (and Tn10) transposition (36), with which it shares more features. These include (i) the generation of DNA hairpin intermediates (7), (ii) the presence of a single enzyme binding site at each DNA end undergoing cleavage (36), and (iii) the structural resemblance between RAG-1 and the Tn5 transposase with respect to the spacing and organization of critical active-site carboxylate residues (the DDE motif) (33, 46). The similarity between RAG-1 and the Tn5 transposase is shown here to extend to the dimeric configuration of both proteins within synaptic complexes (8, 10). However, an important distinction between the two recombination systems is that only one protein, the transposase, is necessary to catalyze all reactions underlying Tn5 transposition, whereas RAG-1 requires RAG-2 for V(D)J recombinase activity. Despite this distinction, the protein complexes mediating synapsis in both cases apparently contain two potential active sites, both of which are used to catalyze sequential strand cleavage and strand transfer reactions.
Distinct roles for HMG-1 in synapsis, cleavage, and transposition.
HMG-1 (or HMG-2) is known to play a critical role in promoting RAG synaptic complex assembly and activity (13, 40). One way HMG-1 assists in this process is by facilitating RAG binding to and bending of the 23-RSS via interactions with the RAG-1 homeodomain and the RSS nonamer (2, 47). Two lines of evidence presented here suggest that HMG-1 additionally promotes association with the coding sequence to help organize a “competent” synaptic complex capable of selectively cleaving 12/23 RSS pairs. First, 12/23-regulated synapsis of intact RSS substrates requires the presence of HMG-1, but HMG-1 is not stringently required for synapsis of prenicked or precleaved substrates, indicating that the intact coding end itself acts as a barrier to synaptic complex assembly. HMG-1 may help the RAG proteins overcome this barrier, possibly by bending the DNA and/or stabilizing conformational changes or other structural intermediates integral to synapsis (9, 50). Second, 12/23-regulated cleavage of prenicked RSS substrates (transesterification) requires HMG-1, despite the less stringent requirement for HMG-1 in the assembly of paired complexes using these substrates. Therefore, in addition to promoting the synapsis of intact substrates, HMG-1 may assist the RAGs during the transesterification step by stabilizing a cleavage-competent complex capable of supporting hairpin formation in the context of a PC.
There remains some controversy over where the 12/23 rule is imposed, with both synapsis and transesterification being implicated as key control points in the enforcement of the 12/23 rule (17, 53). The data presented here provide direct evidence for a middle ground, as speculated in a recent review (13), in which the 12/23 rule is enforced both at the level of RSS binding (at least three- to fourfold) and at the level of hairpin formation (approximately fourfold). Although both factors individually are relatively modest under the conditions tested here, they could combine to provide about a 15-fold preference for 12/23 substrates, about half the ≥30-fold preference observed in vivo (16). The three- to fourfold preference for 12/23 synapsis found here likely underestimates the RAGs' true selectivity for 12/23 RSS pairs in vivo, as high local concentrations of chromosomal DNA would act as an effective nonspecific competitor for RAG binding in vivo. Moreover, chromosomal accessibility of antigen receptor loci is known to be tightly regulated and may impose temporal ordering to RAG-RSS association (5).
Like 12/23-regulated cleavage of intact and prenicked substrates, transposition in vitro also occurs in the context of a complex containing a RAG-1/RAG-2 tetramer core and exhibits dependence on the 12/23 rule. Remarkably, while HMG-1 is required for synapsis of intact substrates and promotes 12/23-regulated cleavage of intact and prenicked substrates, HMG-1 is dispensable for transposition activity once the SEC is formed, as the SC2 and SEC species assembled with a 12/23 pair of RSSs have the same transposition activity. This observation is consistent with recent evidence indicating that HMG-1 is not required for capture of target DNA (34) and that SEC formation occurs at low (but detectable) levels in the absence of the HMG protein (14). Interestingly, incorporation of HMG-1 is found to suppress 12/12 transposition, which otherwise occurs quite readily under these conditions, but has little effect on 23/23 transposition, which is not efficient in any case. Thus, in addition to being required for synapsis and cleavage of RSS substrates, HMG-1 may have an unexpected role in suppressing 12/12 transposition.
Acknowledgments
I appreciate the technical assistance of Dustin Volkmer and Lei Wang.
This work was supported by a grant to P.C.S. from the American Cancer Society (RSG-01-020-01-CCE) and the Health Future Foundation.
REFERENCES
- 1.Agrawal, A., Q. M. Eastman, and D. G. Schatz. 1998. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature 394:744-751. [DOI] [PubMed] [Google Scholar]
- 2.Aidinis, V., T. Bonaldi, M. Beltrame, S. Santagata, M. E. Bianchi, and E. Spanopoulou. 1999. The RAG1 homeodomain recruits HMG1 and HMG2 to facilitate recombination signal sequence binding and to enhance the intrinsic DNA-bending activity of RAG1-RAG2. Mol. Cell. Biol. 19:6532-6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aidinis, V., D. C. Dias, C. A. Gomez, D. Bhattacharyya, E. Spanopoulou, and S. Santagata. 2000. Definition of minimal domains of interaction within the recombination-activating genes 1 and 2 recombinase complex. J. Immunol. 164:5826-5832. [DOI] [PubMed] [Google Scholar]
- 4.Bailin, T., X. Mo, and M. J. Sadofsky. 1999. A RAG1 and RAG2 tetramer complex is active in cleavage in V(D)J recombination. Mol. Cell. Biol. 19:4664-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bassing, C. H., W. Swat, and F. W. Alt. 2002. The mechanism and regulation of chromosomal V(D)J recombination. Cell 109(Suppl.):S45-S55. [DOI] [PubMed] [Google Scholar]
- 6.Besmer, E., J. Mansilla-Soto, S. Cassard, D. J. Sawchuk, G. Brown, M. Sadofsky, S. M. Lewis, M. C. Nussenzweig, and P. Cortes. 1998. Hairpin coding end opening is mediated by RAG1 and RAG2 proteins. Mol. Cell 2:817-828. [DOI] [PubMed] [Google Scholar]
- 7.Bhasin, A., I. Y. Goryshin, and W. S. Reznikoff. 1999. Hairpin formation in Tn5 transposition. J. Biol. Chem. 274:37021-37029. [DOI] [PubMed] [Google Scholar]
- 8.Bhasin, A., I. Y. Goryshin, M. Steiniger-White, D. York, and W. S. Reznikoff. 2000. Characterization of a Tn5 pre-cleavage synaptic complex. J. Mol. Biol. 302:49-63. [DOI] [PubMed] [Google Scholar]
- 9.Bianchi, M. E., and M. Beltrame. 1998. Flexing DNA: HMG-box proteins and their partners. Am. J. Hum. Genet. 63:1573-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies, D. R., I. Y. Goryshin, W. S. Reznikoff, and I. Rayment. 2000. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science 289:77-85. [DOI] [PubMed] [Google Scholar]
- 11.Eastman, Q. M., T. M. Leu, and D. G. Schatz. 1996. Initiation of V(D)J recombination in vitro obeying the 12/23 rule. Nature 380:85-88. [DOI] [PubMed] [Google Scholar]
- 12.Eastman, Q. M., I. J. Villey, and D. G. Schatz. 1999. Detection of RAG protein-V(D)J recombination signal interactions near the site of DNA cleavage by UV cross-linking. Mol. Cell. Biol. 19:3788-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fugmann, S. D., A. I. Lee, P. E. Shockett, I. J. Villey, and D. G. Schatz. 2000. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu. Rev. Immunol. 18:495-527. [DOI] [PubMed] [Google Scholar]
- 14.Fugmann, S. D., I. J. Villey, L. M. Ptaszek, and D. G. Schatz. 2000. Identification of two catalytic residues in RAG1 that define a single active site within the RAG1/RAG2 protein complex. Mol. Cell 5:97-107. [DOI] [PubMed] [Google Scholar]
- 15.Ge, H., and R. G. Roeder. 1994. The high mobility group protein HMG1 can reversibly inhibit class II gene transcription by interaction with the TATA-binding protein. J. Biol. Chem. 269:17136-17140. [PubMed] [Google Scholar]
- 16.Hesse, J. E., M. R. Lieber, K. Mizuuchi, and M. Gellert. 1989. V(D)J recombination: a functional definition of the joining signals. Genes Dev. 3:1053-1061. [DOI] [PubMed] [Google Scholar]
- 17.Hiom, K., and M. Gellert. 1998. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol. Cell 1:1011-1019. [DOI] [PubMed] [Google Scholar]
- 18.Hiom, K., and M. Gellert. 1997. A stable RAG1-RAG2-DNA complex that is active in V(D)J cleavage. Cell 88:65-72. [DOI] [PubMed] [Google Scholar]
- 19.Hiom, K., M. Melek, and M. Gellert. 1998. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell 94:463-470. [DOI] [PubMed] [Google Scholar]
- 20.Jones, J. M., and M. Gellert. 2001. Intermediates in V(D)J recombination: a stable RAG1/2 complex sequesters cleaved RSS ends. Proc. Natl. Acad. Sci. USA 98:12926-12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, D. R., Y. Dai, C. L. Mundy, W. Yang, and M. A. Oettinger. 1999. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 13:3070-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Landree, M. A., S. B. Kale, and D. B. Roth. 2001. Functional organization of single and paired V(D)J cleavage complexes. Mol. Cell. Biol. 21:4256-4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landree, M. A., J. A. Wibbenmeyer, and D. B. Roth. 1999. Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 13:3059-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lavoie, B. D., and G. Chaconas. 1996. Transposition of phage Mu DNA. Curr. Top. Microbiol. Immunol. 204:83-102. [DOI] [PubMed] [Google Scholar]
- 25.Lin, W. C., and S. Desiderio. 1995. V(D)J recombination and the cell cycle. Immunol. Today 16:279-289. [DOI] [PubMed] [Google Scholar]
- 26.McBlane, J. F., D. C. van Gent, D. A. Ramsden, C. Romeo, C. A. Cuomo, M. Gellert, and M. A. Oettinger. 1995. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell 83:387-395. [DOI] [PubMed] [Google Scholar]
- 27.McMahan, C. J., M. J. Sadofsky, and D. G. Schatz. 1997. Definition of a large region of RAG1 that is important for coimmunoprecipitation of RAG2. J. Immunol. 158:2202-2210. [PubMed] [Google Scholar]
- 28.Melek, M., M. Gellert, and D. C. van Gent. 1998. Rejoining of DNA by the RAG1 and RAG2 proteins. Science 280:301-303. [DOI] [PubMed] [Google Scholar]
- 29.Mizuuchi, M., T. A. Baker, and K. Mizuuchi. 1991. DNase protection analysis of the stable synaptic complexes involved in Mu transposition. Proc. Natl. Acad. Sci. USA 88:9031-9035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mo, X., T. Bailin, S. Noggle, and M. J. Sadofsky. 2000. A highly ordered structure in V(D)J recombination cleavage complexes is facilitated by HMG1. Nucleic Acids Res. 28:1228-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mundy, C. L., N. Patenge, A. G. Matthews, and M. A. Oettinger. 2002. Assembly of the RAG1/RAG2 synaptic complex. Mol. Cell. Biol. 22:69-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Namgoong, S. Y., and R. M. Harshey. 1998. The same two monomers within a MuA tetramer provide the DDE domains for the strand cleavage and strand transfer steps of transposition. EMBO J. 17:3775-3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naumann, T. A., and W. S. Reznikoff. 2000. Trans catalysis in Tn5 transposition. Proc. Natl. Acad. Sci. USA 97:8944-8949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neiditch, M. B., G. S. Lee, M. A. Landree, and D. B. Roth. 2001. RAG transposase can capture and commit to target DNA before or after donor cleavage. Mol. Cell. Biol. 21:4302-4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oettinger, M. A., D. G. Schatz, C. Gorka, and D. Baltimore. 1990. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science 248:1517-1523. [DOI] [PubMed] [Google Scholar]
- 36.Reznikoff, W. S., A. Bhasin, D. R. Davies, I. Y. Goryshin, L. A. Mahnke, T. Naumann, I. Rayment, M. Steiniger-White, and S. S. Twining. 1999. Tn5: a molecular window on transposition. Biochem. Biophys. Res. Commun. 266:729-734. [DOI] [PubMed] [Google Scholar]
- 37.Rodgers, K. K., I. J. Villey, L. Ptaszek, E. Corbett, D. G. Schatz, and J. E. Coleman. 1999. A dimer of the lymphoid protein RAG1 recognizes the recombination signal sequence and the complex stably incorporates the high-mobility group protein HMG2. Nucleic Acids Res. 27:2938-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roth, D. B., J. P. Menetski, P. B. Nakajima, M. J. Bosma, and M. Gellert. 1992. V(D)J recombination: broken DNA molecules with covalently sealed (hairpin) coding ends in scid mouse thymocytes. Cell 70:983-991. [DOI] [PubMed] [Google Scholar]
- 39.Roth, D. B., C. Zhu, and M. Gellert. 1993. Characterization of broken DNA molecules associated with V(D)J recombination. Proc. Natl. Acad. Sci. USA 90:10788-10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sadofsky, M. J. 2001. The RAG proteins in V(D)J recombination: more than just a nuclease. Nucleic Acids Res. 29:1399-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Santagata, S., E. Besmer, A. Villa, F. Bozzi, J. S. Allingham, C. Sobacchi, D. B. Haniford, P. Vezzoni, M. C. Nussenzweig, Z. Q. Pan, and P. Cortes. 1999. The RAG1/RAG2 complex constitutes a 3′ flap endonuclease: implications for junctional diversity in V(D)J and transpositional recombination. Mol. Cell 4:935-947. [DOI] [PubMed] [Google Scholar]
- 42.Schatz, D. G., M. A. Oettinger, and D. Baltimore. 1989. The V(D)J recombination activating gene, RAG-1. Cell 59:1035-1048. [DOI] [PubMed] [Google Scholar]
- 43.Schlissel, M., A. Constantinescu, T. Morrow, M. Baxter, and A. Peng. 1993. Double-strand signal sequence breaks in V(D)J recombination are blunt, 5′-phosphorylated, RAG-dependent, and cell cycle regulated. Genes Dev. 7:2520-2532. [DOI] [PubMed] [Google Scholar]
- 44.Sheehan, K. M., and M. R. Lieber. 1993. V(D)J recombination: signal and coding joint resolution are uncoupled and depend on parallel synapsis of the sites. Mol. Cell. Biol. 13:1363-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shockett, P. E., and D. G. Schatz. 1999. DNA hairpin opening mediated by the RAG1 and RAG2 proteins. Mol. Cell. Biol. 19:4159-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swanson, P. C. 2001. The DDE motif in RAG-1 is contributed in trans to a single active site that catalyzes the nicking and transesterification steps of V(D)J recombination. Mol. Cell. Biol. 21:449-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swanson, P. C. 2002. Fine structure and activity of discrete RAG-HMG complexes on V(D)J recombination signals. Mol. Cell. Biol. 22:1340-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swanson, P. C., and S. Desiderio. 1999. RAG-2 promotes heptamer occupancy by RAG-1 in the assembly of a V(D)J initiation complex. Mol. Cell. Biol. 19:3674-3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swanson, P. C., and S. Desiderio. 1998. V(D)J recombination signal recognition: distinct, overlapping DNA-protein contacts in complexes containing RAG1 with and without RAG2. Immunity 9:115-125. [DOI] [PubMed] [Google Scholar]
- 50.Thomas, J. O., and A. A. Travers. 2001. HMG1 and 2, and related ‘architectural' DNA-binding proteins. Trends Biochem. Sci. 26:167-174. [DOI] [PubMed] [Google Scholar]
- 51.van Gent, D. C., K. Hiom, T. T. Paull, and M. Gellert. 1997. Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J. 16:2665-2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Gent, D. C., K. Mizuuchi, and M. Gellert. 1996. Similarities between initiation of V(D)J recombination and retroviral integration. Science 271:1592-1594. [DOI] [PubMed] [Google Scholar]
- 53.West, R. B., and M. R. Lieber. 1998. The RAG-HMG1 complex enforces the 12/23 rule of V(D)J recombination specifically at the double-hairpin formation step. Mol. Cell. Biol. 18:6408-6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams, T. L., E. L. Jackson, A. Carritte, and T. A. Baker. 1999. Organization and dynamics of the Mu transpososome: recombination by communication between two active sites. Genes Dev. 13:2725-2737. [DOI] [PMC free article] [PubMed] [Google Scholar]