Abstract
In the past year, DDBJ (http://www.ddbj.nig.ac.jp) collected and released 1 956 826 entries or 1 741 313 111 bases. The released data include ∼90 000 ESTs and cDNAs of Macaca fascicularis, and 280 million bases of mouse GSS. In addition to the data collection, we have indexed the submitted data to the International Nucleotide Sequence Database Collaboration (INSDC, http://www.insdc.org) to classify the entries into research projects behind data submissions. They are expected to be useful to the data submitters and users for enhancing the data submission, retrieval and systematic data analyses at INSDC. The results of indexing also allow one to grasp research projects in life sciences that promoted and produced the DNA sequences submitted to INSDC.
INTRODUCTION
After 18 years of the collaboration among EMBL Bank, GenBank and DDBJ, the three databases have more tightly been united as International Nucleotide Sequence Database Collaboration (INSDC). During this period of the collaboration, we at DDBJ have witnessed a dramatic growth and spread of DNA sequencing activity and considerable diversification in research projects behind it. Consequently, the entries submitted and served at INSDC have grown remarkably heterogeneous not only in size and quality of sequences but also in scale of research projects. In fact, after setting up 14 divisions for the submitted data mainly according to data sources, 2 divisions were excluded, and 8 divisions were created and added to accommodate those DNA sequences that were produced with new and divergent projects thereafter.
Among the eight divisions, STS (Sequence Tagged Site) (1), EST (Expressed Sequence Tag) (2), and GSS (Genome Survey Sequence) (3) were created first to deal with short fragments that were convenient identifiers or tags to discriminate among regions on a genomic DNA or among molecular species of tissue mRNAs. Second, with the advent of genome-sequencing projects, HTG (High Throughput Genome sequence) and HTC (High Throughput cDNA) were created to provide users with the preliminary sequence data that were not yet assembled or annotated (4). Third, ENV (Environment sequence) was created for DNA fragments from an unspecified mixture of organisms, because sequencing exploration was extended to various microbial floras inside and out of organisms (5). Fourth, TPA (Third Party Annotation) (6) was created to accept the submission of new annotations to the existing sequence at INSDC by researchers other than the submitters of the sequence. This division is expected to enrich the INSDC data in annotation. Finally, to accommodate massive fragments that were considered valuable for genome-scale annotation, MGA (Mass sequences for Genome Annotation) (7) was created.
Despite the notion that understanding of the contents of each division is sometimes essential for effective use of INSDC, the contents have not clearly been presented to users. Therefore, we at DDBJ have indexed the entries of the INSDC data to extract and represent such pieces of information to the data submitters and users. The information will provide substantial background knowledge not only to the submitters and users but also to the research community of life science in general.
In this paper, we first report on the data collection at DDBJ in the period from June 2004 to May 2005, and present and discuss the results obtained by indexing.
DATA COLLECTION AT DDBJ IN THE PAST YEAR
In the period mentioned above, we at DDBJ collected and released 1 956 826 entries or 1 741 313 111 bases including 179 684 entries or 129 790 492 bases from the Japan Patent Office. There are three noteworthy points for the collected data in this period. First, we collaborated with the National Institute of Biomedical Innovation and the University of Tokyo (Institute of Medical Science and Department of Medical Genome Sciences) to construct the Macaca fascicularis cDNA database (QFbase, http://genebank.nibio.go.jp/gbank/index_e.html). QFbase provides users with ∼85 000 EST sequences of the monkey derived from the brain, liver and testis. It also contains ∼4000 full-length cDNAs of which 1700 show the difference in protein coding region between the monkey and humans. The sequences are retrieved at DDBJ using getentry against the accession numbers, BB873801–BB894695 (20 895 3′ESTs) and CJ430287–CJ493524 (63 238 5′ESTs) or our keyword search tools. Second, the data contains ∼280 million bases of GSS data in Mus musculus molossinus (8). The mouse data submitted from RIKEN will certainly contribute to research in mouse genetics and the related areas. Finally, though not included in the above statistics we also collected and released 6 246 064 mouse entries and 2 559 204 human entries in MGA. The MGA data can be retrieved at DDBJ and downloaded at ftp://ftp.ddbj.nig.ac.jp/database/mga/ (MGA directory) and ftp://ftp.ddbj.nig.ac.jp/database/mga/project_index.html/ (project index). Among the mouse MGA data, 383 264 entries are also a part of the CIBEX gene-expression database (http://cibex.nig.ac.jp) (9) at DDBJ.
We also collaborated with Nara Institute of Science and Technology to refine the complete genome of Escherichia coli. The main aspect of the refinement was to resolve inconsistencies between W3110 strain sequenced by the Japanese team (http://ecoli.aist-nara.ac.jp/) and MG1655 strain sequenced by the US team (10). Since E.coli is considered reference to bacterial species in general, the refined genome sequence will contribute to many aspects in bacterial research. Incidentally, five complete genome sequences of E.coli strains are available among 246 complete bacterial sequences at the Genome Information Broker (GIB, http://gib.genes.nig.ac.jp) (11) of DDBJ as of August 2005. By getting access to this URL, users can explore any one of the 246 bacterial genomes by clone name, ORF name/number, function, gene name, product name, location, sequence (namely, homology search), and other features/qualifiers defined by INSDC. The result of retrieval is displayed either in graphics or in a table format.
NEW DEVELOPMENT
Indexing for breakdown statistics
We have indexed all entries of the INSDC data as of August 2005 with respect to the date of publication, nationality of submitters, and research projects for which sequences were produced and submitted, in addition to the original information in the entries. In particular, we identified the research project from ‘Reference’ line, and determined the nationality from the address of the primary submitter in the flat-file. By indexing across the 20 divisions mentioned above, we also elaborated various breakdown representations of the INSDC data that are periodically updated along with the regular DDBJ release four times a year.
Characteristics of the division
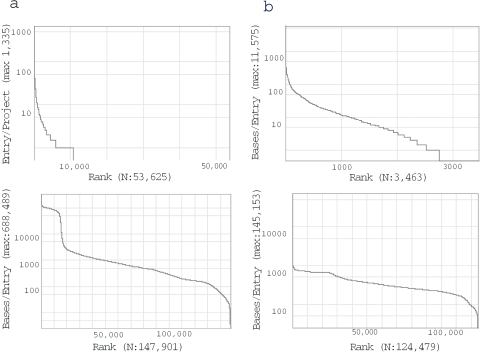
Although we have provided a concise definition of each division in our release notes, the definition may not necessarily be expressive of the contents. For providing more concrete ideas about the divisions, we represent the distribution of the number of projects and that of the sequence lengths for each of the 20 divisions. These pieces of information are available at ‘by division’ under ‘Breakdown Stats’ on the DDBJ home page mentioned in the abstract. To show typical examples in this subdivision, we show two sets of figures for ROD (rodents, one of the original 12 divisions) and ENV (one the 8 new divisions) in Figure 1a and b. The upper distributions in Figure 1a and b show that while most entries in ROD were individualized into independent research projects, those in ENV were grouped together into large research projects in which the top ten projects account for 20% of the total number of entries (see the web site mentioned above for details). Users also understand which flora has been intensively searched and analyzed from the list of projects given in the same web page. On the other hand, the lower distributions in the figures indicate that ∼20 000 entries include long genomic sequences and the remaining 120 000 contain short sequences representing exons and cDNAs, whereas those in ENV contain sequences similar to one another in length.
Figure 1.
(a) Distribution of research projects for ROD. Upper: x-axis indicates the research projects ranked with respect to the number of entries in them. The number in the parentheses is the total number of projects found. Lower: x-axis indicates the entries ranked with respect to the sequence length. The number in the parentheses is the total number of entries in the divisions. (b) Distribution of research projects for ENV; x-axis is the same as those in ROD. The figures for all 20 divisions are available via the DDBJ home page as mentioned in the text.
Temporal transition in the research projects
In addition to the research projects in each division, we provide the temporal transitions in the number of entries in a particular research project from 1992 to 2005 that would be useful for grasping temporal trends in the DNA sequencing activity in accordance with the advancement in life sciences. To see the transitions, users are invited to select ‘submission trends’ in ‘Breakdown Stats’. For instance, the figures given in the subdivision show a drastic shift in life sciences from the gene-oriented to genome-oriented research in the last several years.
Geographic distribution of data production
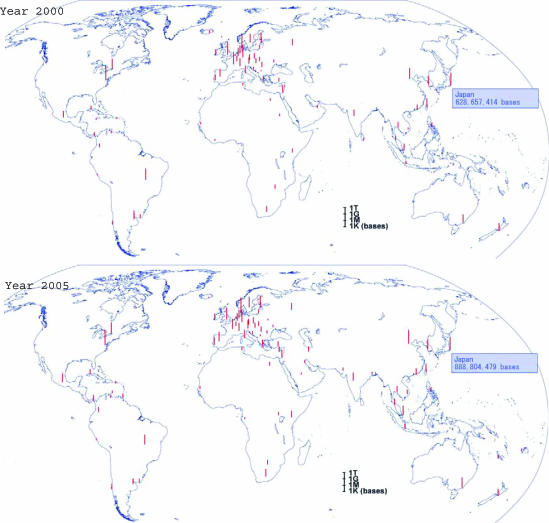
By indexing, we can also geographically differentiate the submitted data to INSDC as given in Figure 2. To obtain more information on this, users are invited this time to select ‘by geography’. The figure shows that the sequencing activity has been enhanced particularly in Asian and African countries in the last five years. For example, the number of Asian and African countries that submitted data to DDBJ is 36 in 2000 and 46 in 2005.
Figure 2.
Geographic distribution of data submissions in 2000 and 2005. The bar indicates the number of bases submitted to INSDC (Total) as a whole from a particular geographic region. The whole pictures for the latest release are available via the DDBJ home page as mentioned in the text. While you see it on a Windows PC, mouse-over to a bar shows the name of its country or geographic region and the number of bases submitted to it. Similar pictures [INSDC (Individual)] are also available for the three individual banks of INSDC by clicking INSDC (Total).
CONCLUDING REMARKS
The rapid advancements and diversification of life sciences have made it quite demanding to grasp the latest achievements qualitatively and quantitatively. In this situation, we may have to rigorously employ the information technology that facilitates the traffic of information across DNA sequence data, such as, ontology, natural language processing and others. As a member of INSDC, we at DDBJ can also contribute to facilitating information traffic by presenting the submitted DNA sequence data as mentioned above. We hope that such an effort may also enhance the accountability of the research community of life science to society.
Acknowledgments
DDBJ is funded by Ministry of Education, Culture, Sports, Science and Technology (MEXT) with Management expenses grants for national university corporations. Funding to pay the Open Access publication charges for this article was provided by Japan Society for the Promotion of Science Grant# 16255006.
Conflict of interest statement. None declared.
REFERENCES
- 1.Olson M., Hood L., Cantor C., Botstein D.A. Common language for physical mapping of the human genome. Science. 1989;245:1434–1435. doi: 10.1126/science.2781285. [DOI] [PubMed] [Google Scholar]
- 2.Adams M.D., Kelley J.M., Gocayne J.D., Dubnick M., Polymeropoulos M.H., Xiao H., Merril C.R., Wu A., Olde B., Moreno R.F., et al. Complementary DNA sequencing: expressed sequence tags and human genome project. Science. 1991;252:1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- 3.Smith M.W., Holmsen A.L., Wei Y.H., Peterson M., Evans G.A. Genomic sequence sampling: a strategy for high resolution sequence-based physical mapping of complex genomes. Nature Genet. 1994;7:40–47. doi: 10.1038/ng0594-40. [DOI] [PubMed] [Google Scholar]
- 4.Ouellette B.F., Boguski M.S. Database divisions and homology search files: a guide for the perplexed. Genome Res. 1997;7:952–955. doi: 10.1101/gr.7.10.952. [DOI] [PubMed] [Google Scholar]
- 5.Rondon M.R., August P.R., Bettermann A.D., Brady S.F., Grossman T.H., Liles M.R., Loiacono K.A., Lynch B.A., MacNeil I.A., Minor C., Tiong C.L., Gilman M., Osburne M.S., Clardy J., Handelsman J., Goodman R.M. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 2000;66:2541–2547. doi: 10.1128/aem.66.6.2541-2547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stoesser G., Baker W., van den Broek A., Garcia-Pastor M., Kanz C., Kulikova T., Leinonen R., Lin Q., Lombard V., Lopez R., Mancuso R., Nardone F., Stoehr P., Tuli M.A., Tzouvara K., Vaughan R. The EMBL Nucleotide Sequence Database: major new developments. Nucleic Acids Res. 2003;31:17–22. doi: 10.1093/nar/gkg021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tateno Y., Saitou N., Okubo K., Sugawara H., Gojobori T. DDBJ in collaboration with mass-sequencing teams on annotation. Nucleic Acids Res. 2005;33:D25–D28. doi: 10.1093/nar/gki020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abe K., Noguchi H., Tagawa K., Yuzuriha M., Toyoda A., Kojima T., Ezawa K., Saitou N., Hattori M., Sakaki Y., Moriwaki K., Shiroishi T. Contribution of Asian mouse subspecies Mus musculus molossinusto genomic constitution of strain C57BL/6J, as defined by BAC end sequence-SNP analysis. Genome Res. 2004;14:2439–2447. doi: 10.1101/gr.2899304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ikeo K., Ishi-I J., Tamura T., Gojobori T., Tateno Y. CIBEX: Center for Information Biology gene EXpression database. C. R. Biologies. 2003;326:1079–1082. doi: 10.1016/j.crvi.2003.09.034. [DOI] [PubMed] [Google Scholar]
- 10.Blattner F.R., Plunkett G., III, Bloch C.A., Perna N.T., Burland V., Riley M., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F., et al. The complete genome sequence of Escherichia coliK-12. Science. 1997;277:1432–1434. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 11.Fumoto M., Miyazaki S., Sugawara H. Genome Information Broker (GIB): data retrieval and comparative analysis system for completed microbial genomes and more. Nucleic Acids Res. 2002;30:66–68. doi: 10.1093/nar/30.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]