Abstract
The miRBase database aims to provide integrated interfaces to comprehensive microRNA sequence data, annotation and predicted gene targets. miRBase takes over functionality from the microRNA Registry and fulfils three main roles: the miRBase Registry acts as an independent arbiter of microRNA gene nomenclature, assigning names prior to publication of novel miRNA sequences. miRBase Sequences is the primary online repository for miRNA sequence data and annotation. miRBase Targets is a comprehensive new database of predicted miRNA target genes. miRBase is available at http://microrna.sanger.ac.uk/.
INTRODUCTION
MicroRNAs (miRNAs) are a class of non-coding RNA gene whose final product is a ∼22 nt functional RNA molecule. They play important roles in the regulation of target genes by binding to complementary regions of messenger transcripts to repress their translation or regulate degradation (1–3). miRNAs have been implicated in cellular roles as diverse as developmental timing in worms, cell death and fat metabolism in flies, haematopoiesis in mammals, and leaf development and floral patterning in plants [reviewed in (4,5)]. Recent reports have suggested that miRNAs may play roles in human cancers (6–8).
The biogenesis of miRNA sequences has been largely elucidated [reviewed in (9)]. The mature miRNA (often designated miR) is processed from a characteristic stem–loop sequence (called a pre-mir), which in turn may be excised from a longer primary transcript (or pri-mir). Only a handful of primary transcripts have been fully described, but evidence suggests that miRNAs are transcribed by RNA polymerase II, and that the transcripts are capped and polyadenylated.
Since the discovery of the founding members of the miRNA class, lin-4 and let-7 in Caenorhabditis elegans [reviewed in (10)], over 2000 miRNA sequences have been described in vertebrates, flies, worms and plants, and even in viruses. However, the functions of only a handful of these miRNAs have been experimentally determined. In parallel with novel gene identification efforts, the miRNA community is therefore focused on predicting and validating miRNA gene targets.
The miRBase database brings together the gene naming and sequence database roles previously fulfilled by the microRNA Registry (11), with the first automated pipeline for predicting miRNA target genes in multiple animal genomes. These three functions are briefly discussed in turn.
miRBase REGISTRY
The rapid growth of the miRNA field has been facilitated by the adoption of a consistent gene naming scheme, which has been applied since the first large-scale miRNA discoveries (12–14). The miRNA Registry (11) has acted as an independent arbiter of gene names, and this function is continued by the miRBase Registry. Names are assigned by the Registry based on guidelines agreed by a number of prominent miRNA researchers and discussed elsewhere (15). In order to minimize the gaps in the naming scheme and to take advantage of the peer review process to assess the validity of submitted miRNAs, names are assigned after a manuscript describing their discovery is accepted for publication. Official gene names should be incorporated into the final version of a manuscript. The nomenclature guidelines require that novel miRNA genes are experimentally verified by cloning or with evidence of expression and processing. Homologous miRNAs from related organisms that are identified by sequence analysis methods may be named without the need for further experimental evidence.
miRNAs are assigned sequential numerical identifiers. The database uses abbreviated 3 or 4 letter prefixes to designate the species, such that identifiers take the form hsa-miR-101 (in Homo sapiens). The mature sequences are designated ‘miR’ in the database, whereas the precursor hairpins are labelled ‘mir’. The gene names are intended to convey limited information about functional relationships between mature miRNAs. For example, hsa-miR-101 in human and mmu-miR-101 in mouse are orthologous. Paralogous sequences whose mature miRNAs differ at only one or two positions are given lettered suffixes—for example, mmu-miR-10a and mmu-miR-10b in mouse. Distinct hairpin loci that give rise to identical mature miRNAs have numbered suffixes (e.g. dme-mir-281-1 and dme-mir-281-2 in Drosophila melanogaster). It should be noted that plant and viral naming schemes differ subtly.
However, miRNA names should not be relied upon to convey complex relationship information. Naming criteria may be subtly redefined over time, and opinion on the degree of conservation of mature sequence required for functional redundancy varies—some recent studies suggest that only the 5′ so-called seed region of the sequence forms a tight duplex with the target mRNA (16). Related hairpin precursor sequences may give rise to mature sequences with only marginal similarity and different miRNA numbers. The naming scheme is also complicated by instances where two different mature miRNA sequences appear to be excised from opposite arms of the same hairpin precursor. Such mature sequences are currently named of the form miR-17-5p (5′ arm) and miR-17-3p (3′ arm). Complex sequence relationships and names are discussed with the submitting author on a case by case basis.
miRBase SEQUENCES
In parallel with the miRNA community's need for a consistent naming scheme, miRNA research and informatics has benefited greatly from a dedicated database of miRNA sequences and annotation. The miRBase Sequence database takes over from the microRNA Registry database as the primary repository for miRNA data. We briefly describe recent growth and database improvements.
Rapid database growth
The miRBase Sequence database contains sequences of all published mature miRNA sequences, together with their predicted source hairpin precursors and annotation relating to their discovery, structure and function. The database has grown rapidly in the past 2 years, from 506 entries representing miRNA hairpin precursors in six species (release 2.0, June 2003) to 2909 entries in 36 species (release 7.0, June 2005).
Stable accessions
miRNA names may change in time to reflect newly discovered relationships between sequences. Stable database accession numbers are therefore assigned to both hairpin (e.g. MI0000015) and mature (e.g. MIMAT0000029) sequences to enable tracking of sequence entities. A summary of the differences between releases is available. In addition, human and mouse gene symbols are provided in consultation with the Genome Nomenclature Committees (HGNC and MGNC).
Evidence tracking
The database contains miRNAs from two fundamentally different sources. Experimentally verified mature miRNAs are annotated with primary literature references and the experimental method used for discovery. The database also contains sequences that are predicted homologs of miRNAs verified in a related organism. For example, 223 of 313 distinct mature miRNA sequences from human (71%) have experimental evidence in human, while the remainder are clearly identifiable homologs of verified miRNAs from mouse, rat and zebrafish. Homologs are predicted based on sequence similarity and folding characteristics of the precursor hairpin, synteny analysis and conservation of the mature miRNA. The source of every miRNA is clearly annotated on the miRNA entry page (Figure 1) and distributed in the flat file downloads. The miRBase Sequence database does not currently contain predicted miRNAs that are without experimental evidence in any related organism.
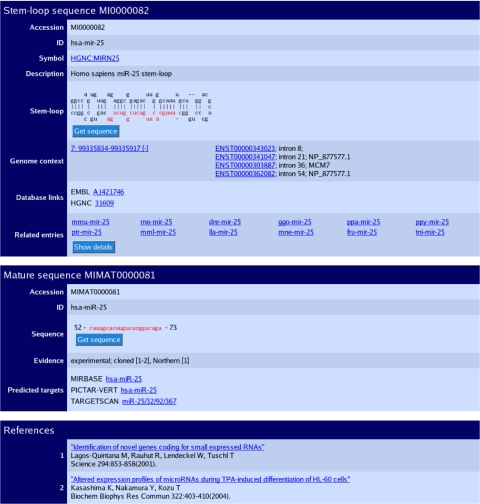
Figure 1.
The sequence database entry for hsa-mir-25. The three sections of the page describe the predicted stem–loop hairpin, mature sequences and primary references. The genomic coordinates and contextual information link to the Ensembl database. Each mature miRNA contains an evidence field, and links are provided to predicted target pages.
Genomic context
For organisms with an assembled genome sequence we provide coordinates of the genomic position of each miRNA sequence on the entry page (Figure 1) and also in GFF format on the FTP site. miRNA genes may be located within other genes, both protein-coding and non-coding (17,18), and the context of the genomic location with respect to Ensembl genes is also annotated (Figure 1). 35% of mammalian miRNA loci overlap annotated genes—over 90% of these are located in introns. In comparison, ∼14% of worm and fly miRNAs are intronic. Distributed annotation system (DAS) sources provide easy access to miRNA genomic locations, and the data are available for viewing within the Ensembl (19) and UCSC browsers (20).
miRBase TARGETS
As focus shifts from miRNA gene identification to functional characterization, miRBase includes not only miRNA sequence data but also information about their genomic targets. The function of a specific miRNA can be thought of as a product of the genes that it regulates. Although large-scale experimental detection of targets is currently difficult, a number of computational techniques exist for the prediction of miRNA targets in mRNA sequences (16,21–27). These methods can be used both to predict potential targets for miRNAs and for the selection of targets for experimental validation. For the most part, computational methods rely on first detecting potential binding sites (with a large degree of complementarity to the miRNA), followed by filtering out those sites that do not appear to be conserved in multiple species. This approach appears to work well, at least for species that have clearly defined orthologs in closely related species (e.g. human, mouse and rat). However, the conservation criterion is poor for those species for which we do not have closely neighbouring genome sequences.
The miRBase Targets database uses a novel fully automated pipeline (which will be described in detail elsewhere) to address some of these issues. All animal miRNA sequences from the miRBase Sequence database are scanned against 3′-untranslated regions (3′-UTRs) predicted from all available species in Ensembl (19) along with Caenorhabditis briggsae and Drosophila pseudoobscura. The core algorithm assigns P-values to individual miRNA–target binding sites, multiple sites in a single UTR, and sites that appear to be conserved in multiple species based on robust statistical models (22). The interface connects each miRNA to a list of predicted gene targets. The detailed target view page (Figure 2) illustrates individual binding sites for one or more miRNAs and their target in an orthologous 3′-UTR alignment. We are in the process of including annotation of experimentally validated miRNA targets.
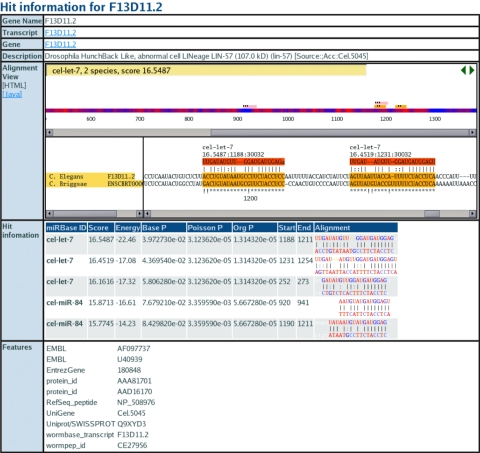
Figure 2.
miRBase Target view page for transcript F13D11.2. The alignment view shows the alignment of miRNA binding sites in orthologous 3′-UTRs. Bits scores, P-values, folding energies and alignments are shown for each miRNA match.
The miRBase Target database is designed with two main aims: to make available high-quality targets in a timely manner, and to remain as inclusive as possible with respect to the target prediction community. To this end, we provide a core set of predictions that are updated concurrently with the rest of the miRBase system. We also intend to provide a mechanism for viewing and comparing third-party target predictions contributed via DAS. The core predictions are generated in-house using the miRanda algorithm (v3.0) (21). The strengths of miRanda are that it is open source, scalable and incorporates robust statistical models. The provision of a P-value for each miRNA–target assignment allows the user to assess the confidence in the prediction. In addition, the method does not assume that the miRNA binding sites must be conserved, although in practice the most highly significant P-values tend to represent miRNA–target interactions that are conserved across multiple species. As new insights into miRNA–target binding mechanisms and improved prediction algorithms become available, they will be integrated into the system to provide the highest-quality target predictions to the user. In parallel with the miRBase Target pipeline, miRNA sequence entries also provide links to third-party target prediction websites (Figure 1).
AVAILABILITY
The miRBase database is freely available to all for online searching at http://microrna.sanger.ac.uk/. Sequences and annotation are also available for download from the FTP site in a number of formats, including FASTA format sequences and relational database dumps for easy upload to a MySQL or other database. Queries, feedback and data submissions and revisions are welcome by email to microrna@sanger.ac.uk.
Acknowledgments
We thank Mhairi Marshall and John Tate for website design, and are grateful to David Bartel and Tom Tuschl for ongoing nomenclature discussion. We also thank Michel Weber for assistance in providing data for viewing in the UCSC genome browser, Marc Rehmsmeier for discussion of P-value statistics and Antonio Giraldez for experimental work on target verification. Work at the Sanger Institute is supported by the Wellcome Trust. Funding to pay the Open Access publication charges for this article was provided by the Wellcome Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Filipowicz W., Jaskiewicz L., Kolb F.A., Pillai R.S. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr. Opin. Struct. Biol. 2005;15:331–341. doi: 10.1016/j.sbi.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 3.Sontheimer E.J., Carthew R.W. Silence from within: endogenous siRNAs and miRNAs. Cell. 2005;122:9–12. doi: 10.1016/j.cell.2005.06.030. [DOI] [PubMed] [Google Scholar]
- 4.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 5.Kidner C.A., Martienssen R.A. The developmental role of microRNA in plants. Curr. Opin. Plant Biol. 2005;8:38–44. doi: 10.1016/j.pbi.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 6.He L., Thomson J.M., Hemann M.T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., Hammond S.M. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 8.O'Donnell K.A., Wentzel E.A., Zeller K.I., Dang C.V., Mendell J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 9.Kim V.N. MicroRNA biogenesis: coordinated cropping and dicing. Nature Rev. Mol. Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 10.Pasquinelli A.E., Ruvkun G. Control of developmental timing by microRNAs and their targets. Annu. Rev. Cell Dev. Biol. 2002;18:495–513. doi: 10.1146/annurev.cellbio.18.012502.105832. [DOI] [PubMed] [Google Scholar]
- 11.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 13.Lau N.C., Lim L.P., Weinstein E.G., Bartel D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 14.Lee R.C., Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 15.Ambros V., Bartel B., Bartel D.P., Burge C.B., Carrington J.C., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez A., Griffiths-Jones S., Ashurst J.L., Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weber M.J. New human and mouse microRNA genes found by homology search. FEBS J. 2005;272:59–73. doi: 10.1111/j.1432-1033.2004.04389.x. [DOI] [PubMed] [Google Scholar]
- 19.Hubbard T., Andrews D., Caccamo M., Cameron G., Chen Y., Clamp M., Clarke L., Coates G., Cox T., Cunningham F., et al. Ensembl 2005. Nucleic Acids Res. 2005;33:D447–D453. doi: 10.1093/nar/gki138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karolchik D., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Enright A.J., John B., Gaul U., Tuschl T., Sander C., Marks D.S. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rehmsmeier M., Steffen P., Hochsmann M., Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stark A., Brennecke J., Russell R.B., Cohen S.M. Identification of Drosophila microRNA targets. PLoS Biol. 2003;1:E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajewsky N., Socci N.D. Computational identification of microRNA targets. Dev Biol. 2004;267:529–535. doi: 10.1016/j.ydbio.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 25.Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 26.Brennecke J., Stark A., Russell R.B., Cohen S.M. Principles of microRNA–target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krek A., Grun D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., Rajewsky N. Combinatorial microRNA target predictions. Nature Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]