Abstract
Lateral gene transfer affects the evolutionary path of key genes involved in ancient metabolic traits, such as sulfate respiration, even more than previously expected. In this study, the phylogeny of the adenosine-5′-phosphosulfate (APS) reductase was analyzed. APS reductase is a key enzyme in sulfate respiration present in all sulfate-respiring prokaryotes. A newly developed PCR assay was used to amplify and sequence a fragment (∼900 bp) of the APS reductase gene, apsA, from a taxonomically wide range of sulfate-reducing prokaryotes (n = 60). Comparative phylogenetic analysis of all obtained and available ApsA sequences indicated a high degree of sequence conservation in the region analyzed. However, a comparison of ApsA- and 16S rRNA-based phylogenetic trees revealed topological incongruences affecting seven members of the Syntrophobacteraceae and three members of the Nitrospinaceae, which were clearly monophyletic with gram-positive sulfate-reducing bacteria (SRB). In addition, Thermodesulfovibrio islandicus and Thermodesulfobacterium thermophilum, Thermodesulfobacterium commune, and Thermodesulfobacterium hveragerdense clearly branched off between the radiation of the δ-proteobacterial gram-negative SRB and the gram-positive SRB and not close to the root of the tree as expected from 16S rRNA phylogeny. The most parsimonious explanation for these discrepancies in tree topologies is lateral transfer of apsA genes across bacterial divisions. Similar patterns of insertions and deletions in ApsA sequences of donor and recipient lineages provide additional evidence for lateral gene transfer. From a subset of reference strains (n = 25), a fragment of the dissimilatory sulfite reductase genes (dsrAB), which have recently been proposed to have undergone multiple lateral gene transfers (M. Klein et al., J. Bacteriol. 183:6028–6035, 2001), was also amplified and sequenced. Phylogenetic comparison of DsrAB- and ApsA-based trees suggests a frequent involvement of gram-positive and thermophilic SRB in lateral gene transfer events among SRB.
One of the oldest types of biological energy conservation on Earth is sulfate respiration, which developed ∼2.8 to 3.1 billion years ago (3, 37). The evolutionary diversification of sulfate-reducing prokaryotes (SRP) since then should, in principle, be reflected in the history of their genes. However, lateral gene transfer (LGT) (32), which appears to be an important and frequent event in evolution (26), even across domains (1), may blur the evolutionary conclusions about certain enzymes (8). There is mounting evidence that genes coding for key enzymes of sulfate respiration were transferred horizontally from gram-positive bacteria to archaeal Archaeoglobus spp. (14, 20, 41). More recently, Klein et al. (19) discovered the occurrence of multiple lateral transfers of genes coding for the dissimilatory sulfite reductase (dsrAB) among SRP. The phylogenetic tree based on comparative sequence analysis of DsrAB gene fragments of >30 reference strains was partially inconsistent with the corresponding 16S rRNA-based phylogenetic tree. It was concluded that dsrAB genes of several Desulfotomaculum spp. (low-G+C gram-positive division), Thermodesulfobacterium spp., and Desulfobacula toluolica had been laterally transferred from unidentified ancestors of sulfate-reducing bacteria (SRB) within the δ-proteobacteria.
Whereas dissimilatory sulfite reductase occurs also in non-sulfate-reducing, sulfite-respiring microorganisms, such as Pyrobacculum islandicum (29), Desulfitobacterium spp. (15, 19, 21), and Bilophila wadsworthia (24), other sulfate-respiring prokaryotes possess adenosine-5′-phosphosulfate (APS) reductase in addition to sulfite reductase (35). After activation of the chemically inert sulfate by ATP sulfurylase (9) the Fe-S flavoprotein APS reductase (EC 1.8.99.2) catalyzes the two-electron reduction of APS to sulfite and AMP (E0′ = −60 mV). It has been assumed that the same enzyme activity catalyzes also the inverse reaction in a variety of sulfur-oxidizing bacteria (for a review see reference 11); however, this “reverse” function has recently been questioned in connection with the phototroph Allochromatium vinosum (5). The genes for APS reductase, apsBA (Desulfovibrio vulgaris; GenBank accession no. Z69372) and aprBA (Archaeoglobus fulgidus and Allochromatium vinosum [14] and Desulfovibrio desulfuricans [12]) encode subunits that appear to form a 1:1 αβ heterodimer (12). Both subunits of the APS reductase are highly conserved (12), and the APS reductase genes have been proposed as a useful phylogenetic marker (14). However, it is still under debate whether the APS reductase genes of Archaeoglobus fulgidus were transferred from an ancestral donor within the domain Bacteria (14).
Recently, new assays for the PCR amplification of fragments from the apsA gene have been developed (7, 49) and utilized to study the diversity and distribution of SRB in gastrointestinal tracts. However, the lack of a thorough phylogenetic framework of APS reductase from cultivated sulfate reducers still prevents a reliable assignment of molecular, environmental sequences to known taxa of sulfate reducers and thus prevents the use of the apsA gene as a functional marker gene for molecular ecology studies.
This study analyzed the evolutionary relationship of a wide taxonomic range of SRP based on the α-subunit of the APS reductase (ApsA). A new PCR assay targeting the apsA gene was developed, and apsA PCR products were directly sequenced and comparatively analyzed. Incongruences between phylogenetic trees of ApsA and 16S rRNA genes revealed evidence for the intradomain lateral transfer of the apsA gene among distantly related gram-positive SRB and distinct groups of δ-proteobacteria, comprising members of the Syntrophobacteraceae and the Nitrospinaceae. A comparison of DsrAB- and ApsA-based phylogenetic trees revealed patterns of LGT for key enzymes of SRP.
MATERIALS AND METHODS
Microorganisms.
Reference strains of validly described sulfate-reducing microorganisms (see Table 2) were obtained from Kai Finster (Aarhus, Denmark), Alexander Galushko (Konstanz, Germany), Bernhard Schink (Konstanz, Germany), Hans Scholten (Marburg, Germany), and the Deutsche Sammlung von Mikroorganismen und Zellkulturen (Braunschweig, Germany) either as lyophilized cells or as actively growing cultures. Archaeoglobus veneficus (DSM11195) was isolated by K. O. Stetter (University of Regensburg, Regensburg, Germany).
TABLE 2.
PCR amplification of apsA gene fragments using genomic DNA of sulfate-reducing reference strains and selected characteristics
| Speciesa | Strainb | Oxida-tionc | Toptd (°C) | Genomic G+C content (mol %) | PCR product obtained with primer paire:
|
GenBank accession no.
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| APS-FW, APS-RVf (∼390 bp) | APS7-F, APS8-R (∼900 bp) | APS7a-F, APS8-R (∼900 bp) | APS7b-F, APS8-R (∼900 bp) | apsA | 16S rRNA | dsrAB | |||||
| Archaea, Euryarchaeota | |||||||||||
| Archaeoglobus fulgidus | DSM4304T | C | 83 | 46 | +g | +g | AE000988 | X05567, Y00275 | M95624 | ||
| Archaeoglobus profundus | DSM5631T | C | 82 | 41 | + | + (50) | AF418134 | AF297529 | AF071499 | ||
| Archaeoglobus veneficus | DSM11195T | C | 75–80 | 45.4 | + | + (50) | AF418132 | AF418181 | |||
| Bacteria | |||||||||||
| Nitrospira phylum | |||||||||||
| Thermodesulfovibrio islandicus | DSM12570T | I | 65 | 38 | − | + (48) | AF418113 | X96726 | AF334599 | ||
| Thermodesulfovibrio yellowstonii | DSM11347T | I | 65 | 29.5 | − | − (45) | − (45) | − (45) | |||
| Thermodesulfobacteria phylum | |||||||||||
| Thermodesulfobacterium commune | DSM2178T | I | 70 | 34 | − | + (48) | AF418114 | AF418169 | AF334596 | ||
| Thermodesulfobacterium thermophilum | DSM1276T | I | 65 | 31 | − | + (48) | AF418112 | AF334601 | |||
| Thermodesulfobacterium hveragerdense | DSM12571T | I | 70–74 | 40 | − | + (48) | AF418119 | X96725 | |||
| Firmicutes, “Clostridia,” Peptococcaceae | |||||||||||
| Desulfotomaculum kuznetsovii | DSM6115T | C | 60–65 | 49 | + | − (45) | + (59) | + (59) | AF418152 | Y11569 | AF273031 |
| Desulfotomaculum thermobenzoicum | DSM6193T | C | 62 | 53 | + | − (45) | + (59) | + (59) | AF418161 | L15628 | AF273030 |
| Desulfotomaculum geothermicum | DSM3669T | C | 54 | 50 | + | + (48) | AF418115 | X80789 | AF273029 | ||
| Desulfotomaculum sapomandens | DSM3223T | C | 38 | 48 | − | − | + (45) | − (59) | AF418150 | AF168365 | |
| Desulfotomaculum alkaliphilum | DSM12257T | C | 50–55 | 41 | − | + (48) | − | − (59) | AF418154 | AF097024 | AF418195 |
| Desulfotomaculum acetoxidans | DSM771T | C | 37 | 38 | − | + (48) | − (59) | − (59) | AF418153 | Y11566 | AF271768 |
| Desulfotomaculum putei | DSM12395T | I | 50–65 | 47 | + | − (45) | + (59) | + (59) | AF418147 | AF053929 | AF273032 |
| Desulfotomaculum nigrificans | DSM574T | I | 55 | 45 | − | − | + (45) | − (59) | AF418154 | U33455 | |
| Desulfotomaculum ruminis | DSM2154T | I | 28 | 49 | − | − (45) | l (45) | l (45) | AF418164 | Y11572 | |
| Desulfotomaculum aeronauticum | DSM10349T | I | 37 | 44 | − | + (45) | + (45) | + (45) | AF418156 | X98407 | AF273033 |
| Desulfotomaculum halophilum | DSM11559T | I | 35 | 56.3 | + | − (45) | − (45) | − (45) | AF418167 | ||
| Desulfosporosinus orientis | DSM8344 | I | 30 | 45 | − | − (45) | − (45) | − (45) | |||
| Proteobacteria delta subdivision | |||||||||||
| “Desulfobacteraceae”a | |||||||||||
| Desulfobacter curvatus | DSM3379T | C | 28–32 | 46 | − | + | AF418107 | AF418175 | AF418199 | ||
| Desulfobacter postgatei | DSM2034T | C | 28–33 | 46 | − | + (45) | l (45) | + (45) | AF418157 | AF418180 | AF418198 |
| Desulfospira joergensenii | DSM10085T | C | 26–30 | 49.9 | l | + | AF418116 | X99637 | |||
| Desulfotignum balticum | DSM7044 | C | 30 | 62.4 | − | + | AF418127 | AF418176 | |||
| Desulfobacula toluolica | DSM7467T | C | 28 | 42 | l | + | AF418128 | X70953 | |||
| Desulfobacterium autotrophicum | DSM3382T | C | 20–26 | 48 | − | + | AF418108 | AF418177 | AF418182 | ||
| “Desulfobacterium vacuolatum” | DSM3385T | C | 25–30 | 45 | − | + | AF418124 | AF418178 | AF418203 | ||
| Desulfocella halophila | DSM11763T | I | 34 | 35 | − | + | AF418117 | AF022936 | AF418200 | ||
| Desulfonema ishimotonii | DSM9680T | C | 30 | 55 | + | + (50) | AF418135 | U45992 | |||
| Desulfonema magnum | DSM2077T | C | 32 | 41.6 | − | + | AF418122 | U45989 | |||
| Desulfofrigus oceanense | DSM12341T | C | 10 | 52.8 | l | − (45) | + (55) | AF418145 | AF099064 | ||
| Desulfofaba gelida | DSM12344T | I | 7 | 52.5 | + | + | AF418118 | AF099063 | AF334593 | ||
| Desulfococcus biacutus | DSM5651T | C | 28–30 | 56.5 | l | l (50) | AF418151 | AJ277887 | |||
| Desulfococcus multivorans | DSM2059T | C | 35 | 57.4 | + | + (50) | AF418136 | AF418173 | U58126/7 | ||
| Desulfosarcina variabilis | DSM2060T | C | 33 | 51 | − | + | AF418121 | M34407 | AF191907 | ||
| Desulfobacterium indolicum | DSM3383T | C | 28 | 47 | + | + | AF418123 | AJ237607 | |||
| “Desulfobacterium oleovorans” | DSM6200T | C | n.a.i | n.a. | + | + | AF418126 | Y17698 | AF418201 | ||
| Desulfotalea psychrophila | DSM12343T | I | 10 | 46.8 | − | − (45) | − (45) | − (45) | |||
| “Desulfobulbaceae”a | |||||||||||
| Desulfobulbus rhabdoformis | DSM8777T | I | 31 | 50.6 | + | + | AF418110 | U12253 | AJ250473 | ||
| Desulfobulbus elongatus | DSM2908T | I | 35 | 59 | + | + | l (55) | AF418146 | X95180 | AF418202 | |
| Desulfocapsa sulfexigens | DSM10523T | A | 30 | 47.2 | + | + | AF418131 | Y13672 | |||
| Desulfocapsa thiozymogenes | DSM7269T | I | 30 | 50.7 | + | + (45) | + (45) | + (45) | AF418166 | ||
| Desulfofustis glycolicus | DSM9705T | I | 28 | 56 | l | + | AF418130 | X99707 | AF418191 | ||
| Desulforhopalus sp. strain LsV 20 | DSM13038 | n.a. | <20 | n.a. | − | l (45) | − (45) | AF418160 | AF099057 | ||
| Desulforhopalus singaporensis | DSM12130T | I | 31 | 50.6 | − | + (45) | − (45) | + (45) | AF418163 | AF118453 | AF418196 |
| “Desulfovibrionales”a | |||||||||||
| Desulfovibrio desulfuricans subsp. desulfuricans Essex 6 | DSM642T | I | 30 | 59 | + | + | AF226708 | AF192153 | AJ249777 | ||
| Desulfovibrio intestinalis | DSM11275T | I | 37 | 54.5–55.5 | + | + | AF418106 | Y12254 | AF418183 | ||
| Desulfomonas pigra | DSM749T | I | 37 | 66 | + | + | AF418129 | AF192152 | AF418184 | ||
| Desulfovibrio termitidis | DSM5308T | I | 35 | 67–68 | + | + | + (55) | AF418142 | X87409 | AF418184 | |
| Desulfovibrio vulgaris | DSM644T | I | 30–36 | 65 | +g | +g | Z69372 | M34399 | U16723 | ||
| Desulfovibrio burkinensis | DSM6830T | I | 37 | 67 | + | + | + (55) | AF418143 | AF053752 | AF418186 | |
| Desulfovibrio fructosovorans | DSM3604T | I | 35 | 64 | l | + | AF418109 | AF050101 | AF418187 | ||
| Desulfovibrio africanus | DSM2603T | I | 30–36 | 65 | + | + (50) | + (55) | AF418140 | X99236 | AF271772 | |
| Desulfovibrio giganteus | DSM4370T | I | 37 | 56 | + | + (50) | + (55) | AF418141 | AF418170 | ||
| Desulfovibrio profundus | DSM11384T | I | 25 | 53 | + | + (50) | AF418133 | AF418172 | |||
| Desulfomicrobium baculatum | DSM1743T | I | 28–37 | 57 | + | + | AF418120 | ||||
| Desulfomicrobium apsheronum | DSM5918T | I | 25–30 | 52 | + | + | AF420281 | U64865 | AF418188 | ||
| Desulfohalobium retbaense | DSM5692T | I | 37–40 | 57.1 | l | + (50) | AF418125 | X99235 | AF418190 | ||
| Desulfonatronum lacustre | DSM10312T | I | 37–40 | 57.3 | + | + (50) | AF418137 | AF418171 | AF418189 | ||
| Desulfonatronovibrio hydrogenovorans | DSM9292T | n.a. | 37 | 48.6 | + | + | AF418111 | X99234 | AF418197 | ||
| “Syntrophobacteraceae”a | |||||||||||
| Syntrophobacter wolinii | DSM2805MT | I | n.d.j | n.d. | + | + (45) | + (45) | + (45) | AF418165 | X70905 | AF418192 |
| Syntrophobacter pfennigii | DSM10092T | I | 37 | 57.3 | + | + (45) | + (45) | + (45) | AF418168 | X82875 | |
| Syntrophobacter fumaroxidans | DSM10017T | I | 37 | 60.6 | + | + (45) | + (59) | + (59) | AF418138 | X82874 | AF418193 |
| Desulforhabdus amnigena | DSM10338T | C | 37 | 52.5 | + | + (45) | + (59) | + (59) | AF418139 | X83274 | AF337901 |
| Thermodesulforhabdus norvegica | DSM9990T | C | 60 | 51 | + | + (45) | + (45) | + (59) | AF418159 | U25627 | AF334597 |
| Desulfacinum infernum | DSM9756T | n.a. | 60 | 64 | + | + (45) | + (59) | + (59) | AF418144 | L27426 | AF418194 |
| Desulfacinum hydrothermale | DSM13146T | C | 60 | 59.5 | + | + (45) | + (50) | AF418148 | AF170417 | ||
| “Nitrospinaceae”a | |||||||||||
| Desulfomonile tiedjei | DSM6799T | C | 37 | 49 | + | + (45) | + (45) | + (50) | AF418162 | M26635 | AF334595 |
| “Desulfarculus baarsii” | DSM2075T | C | 37 | 66 | + | + (45) | + (45) | + (55) | AF418149 | AF418174 | AF334600 |
| “Desulfobacterium anilini” | DSM4660T | C | 35 | 59.1 | − | + (45) | + (45) | + (45) | AF418158 | AJ237601 | |
| Desulfobacca acetoxidans | DSM11109T | C | 37 | 51.5 | − | − (48) | − (45) | − (45) | |||
Phylum and family names according to the taxonomic outline of Bergey’s Manual of Systematic Bacteriology (http://www.cme.msu.edu/Bergeys/april2001-genus.pdf).
DSMZ strain numbers. T, type strain.
C, complete oxidation of organic carbon substrates; I, incomplete oxidation; A, chemolithoautotrophic growth.
Topt, optimum growth temperature.
PCR annealing temperatures in degrees Celsius are in parentheses. Boldface, no PCR product obtained. l, low yield.
PCR annealing temperature 60°C according to Deplancke et al. (7).
Binding of the primer pair deduced from sequence.
Sequences from this study are in boldface.
n.a., not available.
n.d., not determined.
DNA isolation.
Cells of exponentially growing cultures (10 ml) were harvested by centrifugation and washed with 120 mM sodium phosphate buffer, pH 8.0, before DNA extraction. DNA from lyophilized cells of reference strains was directly extracted without further cultivation of the bacteria. Genomic DNA was extracted from reference strains using a direct-lysis protocol modified from that described by Moré et al. (30) as described previously (13). Briefly, cells were disrupted by bead beating (45 s at 6.5 m s−1) in a sodium dodecyl sulfate solution. DNA was purified from the supernatant with ammonium acetate, isopropanol, and ethanol precipitation steps. The DNA extracts were further purified using a silica matrix-based purification protocol (EasyPure; Biozym, Hess. Oldendorf, Germany). Aliquots of DNA extracts were analyzed by standard gel electrophoresis to verify extraction.
PCR amplification of apsA gene fragments.
The nomenclature of the APS reductase gene operon has not yet been resolved, and aps and apr have been used synonymously. In this paper, apsA is used to designate the APS reductase α-subunit gene.
An ∼390-bp apsA segment was amplified by PCR from genomic DNA of pure cultures as described by Deplancke et al. (7) using primers APS-FW and APS-RV (Table 1). Longer apsA fragments were amplified using primers APS7-F (and its derivatives) and APS8-R (∼900 bp; Table 1). The reaction mixture contained, in a total volume of 50 μl, 25 μl of 2× premix E (Epicentre Technologies, Madison, Wis.), a proprietary PCR premix (containing 400 μM deoxynucleoside triphosphates, 5 mM MgCl2, and 4× betaine as a PCR enhancer), 2 μM primer APS7-F, 0.5 μM primer APS8-R, and 1.25 U of AmpliTaq DNA polymerase (Applied Biosystems, Weiterstadt, Germany). DNA from pure cultures (∼20 ng of nucleic acids) was added as the template. All reaction mixtures were prepared at 4°C in 0.2-ml reaction tubes to avoid unspecific priming. Amplification was started by placing the reaction tubes into the preheated (94°C) block of a Gene Amp 9700 thermocycler (Applied Biosystems). The standard thermal profile for amplification was as follows: an initial denaturation step (3 min, 94°C) was followed by 30 to 35 cycles of denaturation (30 s, 94°C), annealing (55 s, 60°C), and extension (60 s, 72°C). After a terminal extension (7 min, 72°C), the samples were kept at 4°C until further analysis. For PCR screening of apsA gene fragments, the annealing temperature was altered in a range between 45 and 60°C as indicated in Table 2. Aliquots of the amplicons (5 μl) were analyzed by electrophoresis on 1% agarose gels and visualized after staining with ethidium bromide using a gel imaging system (MWG Biotech).
TABLE 1.
PCR primers utilized for the amplification of apsA gene fragments
| Primer | Sequence (5′→3′)e | Primer binding sitea | Reference |
|---|---|---|---|
| APS-FW | TGG CAG ATM ATG ATYMAC GG | 481–500 | 7b |
| APS-RV | GGG CCG TAA CCG TCC TTG AA | 847–866 | 7 |
| APS-uni-F | TGG CAG ATV ATG ATY MAC GG | 481–500 | This studyc |
| APS7-F | GGG YCT KTC CGC YAT CAA YAC | 206–236 | This study |
| APS7a-F | GGG YCT SAG CGC YAT CAA Y | 206–234 | This studyd |
| APS7b-F | GG YCT STC CGC YAT CAA Y | 205–234 | This studyd |
| APS8-R | GCA CAT GTC GAG GAA GTC TTC | 1139–1159 | This study |
Positions of the Desulfovibrio-vulgaris apsA open reading frame.
Published primer sequence contains an additional G at the 3′ end, which was a typing error (B. Deplancke, personal communication).
Primer is a modification of primer APS-FW.
Primer is a modification of primer APS7-F.
Degenerate positions are in boldface.
PCR amplification of dsrAB gene fragments.
An ∼1.9-kb fragment encompassing parts of the dissimilatory sulfite reductase genes dsrA and dsrB was amplified using primers DSR1-F and DSR4-R (41). Reaction mixtures contained, in a total volume of 50 μl, 25 μl of 2× premix E, 0.5 μM (each) primer, genomic DNA (∼20 ng of nucleic acids), and 2.0 U of AmpliTaq DNA polymerase. The thermal profile for amplification was as follows: an initial denaturation step (2 min, 94°C) was followed by 38 cycles of denaturation (45 s, 94°C), annealing (45 s, 54°C), and extension (90 s, 72°C) and one terminal extension step (5 min, 72°C).
PCR amplification of 16S rRNA gene fragments.
16S rRNA genes were amplified from genomic DNA of pure cultures using primers 27F and 1492R (22) or the primer pair 27F (22) and 1542R (18). Reaction mixtures contained, in a volume of 50 μl, 10 μl of 10× PCR buffer, 0.5 μM (each) primer, 50 μM deoxynucleoside triphosphates, 1.5 mM MgCl2, and 1.25 U of AmpliTaq DNA polymerase. DNA from pure cultures (∼20 ng of nucleic acids) was added as the template. The thermal profile for amplification was as follows: an initial denaturation step (2 min, 94°C) was followed by 30 to 35 cycles of denaturation (30 s, 94°C), annealing (45 s, 55°C), and extension (60 s, 72°C) and one terminal extension step (5 min, 72°C).
Sequencing.
PCR products of apsA genes were directly sequenced using primers APS-FW and APS-RV, APS-uni-F and APS-RV, APS7-F (and its derivatives), and APS8-R (Table 1), and dsrAB PCR products were sequenced using primers DSR1F and DSR4R and sequencing primers DSR6F (5′-ATC GGC ACM TGG AGA GAC-3′), DSR7F (5′-KCC ATC GCB CGT TCC GAC-3′), DSR8F (5′-GGC MAG AAC CGY GAG CGY-3′), DSR9F (5′-MCA ACC CST AYA TCT TCT-3′), and DSR10F (5′-GGA AGA RGG CAA RAA CCG-3′). 16S rDNA PCR products were sequenced using primers 27F, 533R, 907R (44), 1542R (18), 1114F, and 1368R (10). PCR products were purified with the QIAquick PCR purification kit (Qiagen, Hilden, Germany) and sequenced using the ABI BigDye terminator cycle sequencing kit (Applied Biosystems) with 100 ng of template DNA as specified by the manufacturer. Cycle sequencing products were purified from excess dye terminators and primers using Autoseq G-50 columns (Amersham-Pharmacia Biotech, Freiburg, Germany) and analyzed with an ABI 373 or 377 DNA sequencer (Applied Biosystems).
Sequence data analysis and phylogenetic placement.
Raw sequence data were assembled and checked with the Lasergene software package (DNASTAR, Madison, Wis.). The data were phylogenetically analyzed (i.e., alignment, treeing) using the ARB (version 2.5b; O. Strunk and W. Ludwig, Technische Universität München, Munich, Germany [http://www.biol.chemie.tu-muenchen.de/pub/ARB/]), PHYLIP (J. Felsenstein, PHYLIP [phylogeny inference package], version 3.6; Department of Genetics, University of Washington, Seattle), and PUZZLE (version 5.0) (39) software packages.
Briefly, deduced APS amino acid sequences were fitted manually into an alignment of APS sequences retrieved from public databases (2) using Genetic Data Environment (version 2.2) as implemented in the ARB software package. Regions of ambiguous homology and insertions or deletions (indels) not present in all sequences analyzed were excluded, yielding an amino acid data set with 252 positions. Trees were reconstructed from distance matrices using FITCH (PHYLIP), neighbor-joining (ARB and PHYLIP), parsimony (PROTPARS; PHYLIP), and maximum-likelihood (PROTML; Institute Pasteur [http://bioweb.pasteur.fr/seqanal/interfaces/molphy.html]; PUZZLE) methods. Distance matrices were calculated using PROTDIST with the Dayhoff PAM 001 matrix as the amino acid replacement model (6). FITCH trees were reconstructed using the global rearrangement and randomized species input order (“jumble,” random number seed 7, seven times) options. Bootstrap analyses were performed using parsimony analysis (500 resamplings; randomized input order, “jumble” three times) or neighbor joining (100 resamplings) as implemented in the PHYLIP package. PUZZLE analysis was performed using tree reconstruction by Quartet Puzzling (QP; 10,000 or 25,000 puzzling steps), approximation of parameters using a neighbor-joining tree, and either JTT (Jones, Taylor, Thornton [17]), variable-time (31), or the WAG matrix (45) as amino acid replacement models (amino acid frequency was estimated from the data set) and assuming either a uniform rate of evolution or a gamma distribution. All trees were reconstructed as “unrooted.”
Deduced dsrAB sequences were analyzed similarly. Regions of ambiguous homology and indels not present in all sequences analyzed were excluded, yielding an amino acid data set with 528 positions for the concatenated data set (19). Phylogenetic trees were reconstructed as described above.
16S rRNA gene sequences were phylogenetically analyzed using distance matrix (neighbor-joining and FITCH using the Kimura two-parameter model), parsimony (DNAPARS), and maximum-likelihood methods (fastDNAML [33]; PUZZLE) by including only nucleotide positions with >50 to 100% invariance. Statistical support for tree topologies was obtained by bootstrap resampling (parsimony, n = 100; neighbor joining, n = 500) or PUZZLE analysis (QP; nucleotide substitution model by Tamura and Nei [40]).
Nucleotide sequence accession numbers.
Sequences of reference strains were deposited in the GenBank database under accession no. AF418106 to AF418168 (apsA), AF418182 to AF418203 (dsrAB), and AF418169 to AF418181 (16S rRNA genes) as specified in Table 2.
RESULTS
PCR amplification of apsA fragments from sulfate-reducing microorganisms.
PCR amplicons of the apsA gene with the expected sizes (390 to 400 bp) were obtained from a wide phylogenetic range of sulfate-reducing microorganisms using primer combination APS-FW and APS-RV (Table 2). These included mostly δ-proteobacteria of the order “Desulfovibrionales” and of the families “Desulfobulbaceae,” “Syntrophobacteraceae,” and “Nitrospinaceae.” However, we were unable to obtain an apsA amplicon using the standard PCR conditions (annealing temperature at 60°C [7]) for the thermophilic sulfate-reducing Thermodesulfobacterium spp. and Thermodesulfovibrio spp.; some gram-positive sulfate-reducing Desulfotomaculum spp.; some members of the Desulfobacteraceae; and Desulfobulbaceae, Desulfobacca acetoxidans, and Desulfobacterium anilini (Table 2). A new primer pair comprising APS7-F and APS8-R (Table 1) was developed based on the comparison of conserved sites in full-length apsA/aprA sequences of Desulfovibrio vulgaris, Desulfovibrio desulfuricans, Allochromatium vinosum (GenBank accession no. U84759), and Archaeoglobus fulgidus. Use of this primer pair allowed the amplification of an ∼900-bp fragment of the apsA gene from a wide range of sulfate-reducing microorganisms, thereby providing considerably more information for phylogenetic analyses. Notably, apsA PCR products were obtained from the thermophilic sulfate-reducing Thermodesulfobacterium spp. and Thermodesulfovibrio islandicus, as well as members of the “Desulfobacteraceae” (Table 2). For some species, amplification required a considerable reduction of the annealing temperature to 45°C using primers APS7-F and APS8-R, e.g., Desulfotomaculum spp. Even at the lower annealing temperatures, PCR amplification using these primers failed for most members of the “Syntrophobacteraceae.” Derivatives of primer APS7-F, e.g., APS7a-F and APS7b-F, with variations in degenerate codons (Table 1) were successfully used to obtain a PCR product also from members of the “Syntrophobacteraceae” and most Desulfotomaculum spp. We were unable to obtain amplification products using all primer pairs for only a few species, such as Desulfosporosinus orientis, Thermodesulfovibrio yellowstonii, Desulfotalea psychrophila, and Desulfobacca acetoxidans (Table 2); however, the 16S rRNA genes of these strains could be amplified, verifying the quality of the genomic DNA for amplification.
Phylogeny of APS reductase.
apsA PCR fragments (primer pair APS7-F and APS8-R) were directly sequenced, yielding sequences varying in length between 860 and 950 bp. The absence of ambiguous nucleotides in sequences of all strains tested indicated that probably only one apsA gene copy is present in each strain. The deduced ApsA amino acid sequences (n = 60) of fragments >860 bp and the published complete AspA sequences of Archaeoglobus fulgidus, Desulfovibrio vulgaris, and Desulfovibrio desulfuricans were aligned. Amino acid positions that could not be aligned unambiguously or that included indels were excluded from phylogenetic analysis by using filters, yielding a data set with 252 amino acid positions (Desulfovibrio vulgaris ApsA positions 82 to 375) for phylogenetic analysis.
ApsA amino acid sequences analyzed were >53% similar, and intrafamily similarities were >80% (Table 3). Notably, sequence similarities among Archaeoglobus spp. were low (71 to 83%), which contrasts with the close similarity of the known species on the 16S rDNA level (93 to 98%). The ApsA amino acid sequence of Archaeoglobus fulgidus differed the most from those of Archaeoglobus profundus and Archaeoglobus veneficus (73 and 71%, respectively). Unexpectedly, the ApsA amino acid sequences of members of the Syntrophobacteraceae and Nitrospinaceae were more similar to those of gram-positive Desulfotomaculum spp. than to those of gram-negative δ-proteobacterial SRB.
TABLE 3.
ApsA sequence similarities of selected sulfate-reducing microorganisms representing major lineages of sulfate-respiring prokaryotesa
| Sulfate-reducing microorganism | % Amino acid sequence similarity to:
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D. curvatus | D. postgatei | D. variabilis | D. rhabdoformis | D. glycolicus | D. vulgaris | D. fructosivorans | T. thermophilum | T. commune | T. islandicus | D. amnigena | S. wolinii | D. baarsii | D. ruminis | D. putei | D. kuznetsovii | A. veneficus | A. profundus | A. fulgidus | |
| Desulfobacter curvatus | |||||||||||||||||||
| Desulfobacter postgatei | 95.5 | ||||||||||||||||||
| Desulfosarcina variabilis | 83.3 | 82.3 | |||||||||||||||||
| Desulfobulbus rhabdoformis | 71.9 | 71.9 | 73.5 | ||||||||||||||||
| Desulfofustis glycolicus | 73.7 | 73.7 | 75.3 | 82.2 | |||||||||||||||
| Desulfovibrio vulgaris | 65.4 | 65.0 | 68.2 | 67.5 | 66.3 | ||||||||||||||
| Desulfovibrio fructosovorans | 65.5 | 65.5 | 68.3 | 70.0 | 68.9 | 87.7 | |||||||||||||
| Thermodesulfobacterium thermophilum | 67.3 | 66.5 | 66.2 | 62.4 | 63.7 | 64.5 | 66.1 | ||||||||||||
| Thermodesulfobacterium commune | 67.0 | 66.3 | 66.4 | 62.5 | 63.4 | 65.0 | 66.2 | 96.5 | |||||||||||
| Thermodesulfovibrio islandicus | 62.9 | 63.3 | 62.0 | 60.7 | 64.7 | 63.7 | 66.8 | 64.6 | 64.6 | ||||||||||
| Desulforhabdus amnigena | 60.1 | 60.5 | 58.6 | 57.0 | 57.1 | 56.5 | 59.7 | 59.5 | 60.0 | 59.6 | |||||||||
| Syntrophobacter wolinii | 62.0 | 62.4 | 60.5 | 57.2 | 58.0 | 57.1 | 61.0 | 62.0 | 62.9 | 59.8 | 90.4 | ||||||||
| “Desulfoarculus baarsii” | 57.7 | 58.5 | 55.2 | 54.4 | 53.6 | 55.3 | 58.9 | 57.0 | 58.0 | 58.5 | 78.7 | 79.5 | |||||||
| Desulfotomaculum ruminis | 59.1 | 59.5 | 56.6 | 56.3 | 55.2 | 56.3 | 59.1 | 55.1 | 55.6 | 57.7 | 77.4 | 77.9 | 76.9 | ||||||
| Desulfotomaculum putei | 59.8 | 60.6 | 58.1 | 57.7 | 57.0 | 56.6 | 59.8 | 56.1 | 56.6 | 58.8 | 78.9 | 77.1 | 76.4 | 93.6 | |||||
| Desulfotomaculum kuznetsovii | 60.5 | 60.1 | 59.4 | 57.3 | 57.9 | 59.7 | 62.5 | 62.0 | 62.1 | 60.2 | 79.2 | 79.3 | 74.4 | 74.3 | 75.8 | ||||
| Archaeoglobus veneficus | 60.2 | 62.2 | 57.0 | 58.0 | 58.2 | 57.8 | 60.2 | 59.5 | 59.2 | 57.3 | 61.8 | 60.4 | 60.3 | 60.9 | 61.9 | 63.5 | |||
| Archaeoglobus profundus | 60.6 | 61.0 | 58.6 | 57.0 | 58.4 | 57.7 | 57.7 | 62.6 | 62.7 | 57.7 | 59.2 | 59.5 | 58.9 | 59.2 | 59.4 | 64.5 | 83.3 | ||
| Archaeoglobus fulgidus | 61.0 | 61.4 | 59.3 | 56.4 | 57.8 | 54.6 | 57.0 | 58.7 | 59.6 | 56.5 | 61.8 | 61.2 | 59.1 | 59.4 | 60.9 | 61.6 | 71.0 | 73.4 | |
Microorganisms with inferred xenologous apsA are in boldface.
The deduced ApsA amino acid sequences were phylogenetically analyzed and compared using distance matrix (FITCH, neighbor-joining), maximum-parsimony, and maximum-likelihood methods (PROTML, PUZZLE). The phylogeny of SRP based on ApsA sequence analyses was compared to the phylogeny based on 16S rRNA gene and DsrAB analyses. All treeing methods used (distance matrix, parsimony, and maximum-likelihood) for ApsA- and 16S rRNA-based phylogenies indicated similar relative branching orders of most taxa (Fig. 1). Both trees were rooted with the archaeal Archaeoglobus spp. Certain branch points of the gram-positive SRB lineages and of the “Desulfobacteraceae” branch of the δ-proteobacteria were resolved poorly, as indicated by low bootstrap values. These branch points were indicated as “multifurcations” (27) (Fig. 1), which were also directly shown by PUZZLE analysis (not shown).
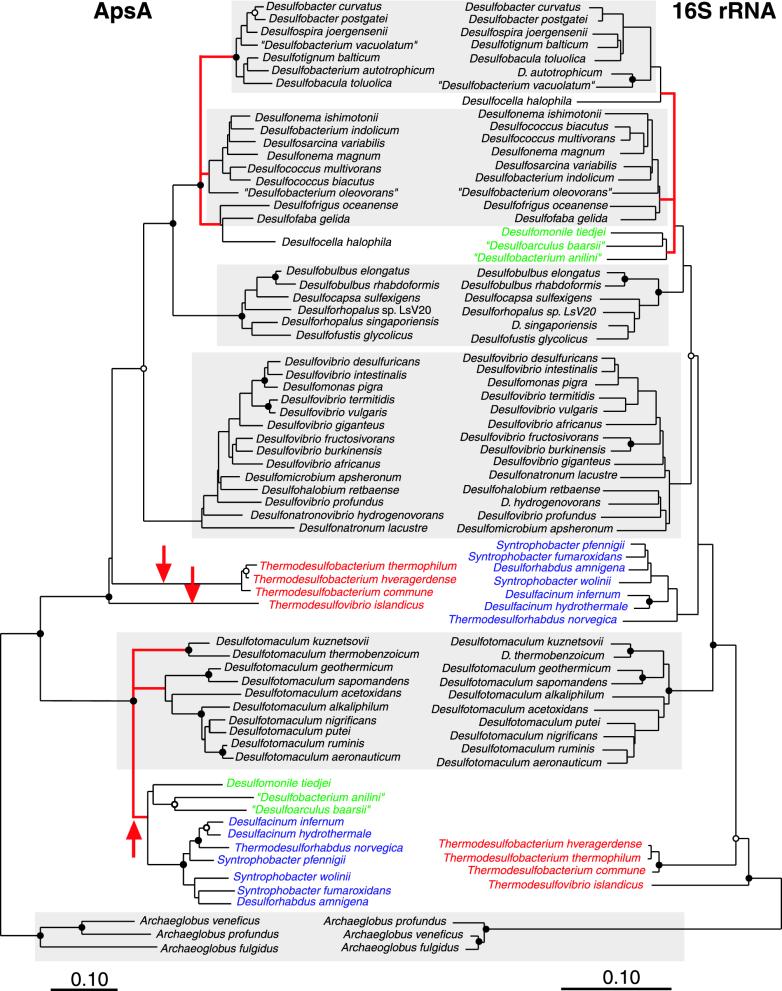
FIG. 1.
Comparison of ApsA- and 16S rDNA-based phylogenetic trees of members of major lineages of the sulfate-reducing microorganisms. Both trees were calculated using distance matrix-based FITCH analysis. Archaeoglobus spp. were used as the outgroup reference. Microorganisms with inferred laterally transferred apsA genes are color coded: red, Thermodesulfobacterium spp. and Thermodesulfovibrio islandicus; blue, “Syntrophobacteraceae”, green, “Nitrospinaceae.” Red lines, multifurcations; red arrow, lineage with inferred lateral gene transfer. Bootstrap support values were obtained from neighbor-joining (100 resamplings) and parsimony analyses (500 resamplings). Solid circles, nodes with a bootstrap support and QP support of >80% for all analyses; open circles, nodes with support of >60 and <80%. Groups monophyletic in the two trees are shaded in gray. Bars, 0.1 changes per nucleotide or amino acid position.
ApsA- and 16S rRNA-based trees both indicated that Archaeoglobus, gram-positive SRB, and δ-proteobacterial SRB form monophyletic lineages. SRB belonging to the order “Desulfovibrionales” and to the families “Desulfobacteraceae” and “Desulfobulbaceae” were recovered as sublineages within the δ-proteobacteria. However, there were major differences in the relative branching order between the 16S rRNA- and ApsA-based trees for the thermophilic Thermodesulfovibrio islandicus and Thermodesulfobacterium spp., δ-Proteobacteriabelonging to the “Syntrophobacteraceae,” the “Nitrospinaceae” (Desulfobacterium anilini, Desulfoarculus baarsii, Desulfomonile tiedjei), and the desulfobacteraceal Desulfocella halophila. All “Syntrophobacteraceae” and “Nitrospinaceae” analyzed (Table 2) grouped well within the phylogenetic radiation of low-G+C gram-positive genus Desulfotomaculum (Fig. 1). Desulfotomaculum halophilum, for which only a short apsA amplification fragment was recovered (GenBank accession no. AF418167), was found to group with the low-G+C gram-positive SRB (not shown in Fig. 1).
Thermodesulfovibrio islandicus and Thermodesulfobacterium spp. each formed monophyletic lineages and branched off between the gram-positive SRB and the δ-proteobacterial SRB with high support values (bootstrapping: 100 [neighbor-joining], 99 [parsimony], and 93% [QP]). PUZZLE analysis indicated a weak association of Thermodesulfovibrio islandicus and Thermodesulfobacterium spp., but only with a low support value (QP support of <70%). In contrast, 16S rRNA gene analysis clearly showed that the two species represented lines of descent close to the root of the Bacteria.
The most parsimonious explanation for these significant topological differences between the ApsA- and 16S rRNA-based trees is the occurrence of multiple lateral transfers of apsA genes between SRP of the “Syntrophobacteraceae,” the “Nitrospinaceae,” and the thermophilic Thermodesulfovibrio islandicus and Thermodesulfobacterium spp. Another topological difference between the two trees was observed: the ApsA-based tree indicated that Desulfocella halophila is closely related to Desulfofaba gelida and Desulfofrigus oceanense, although only with low bootstrap support, whereas the 16S rRNA-based trees indicated a closer relationship of Desulfocella halophila to the Desulfobacter branch of the “Desulfobacteraceae,” but again only with low bootstrap support.
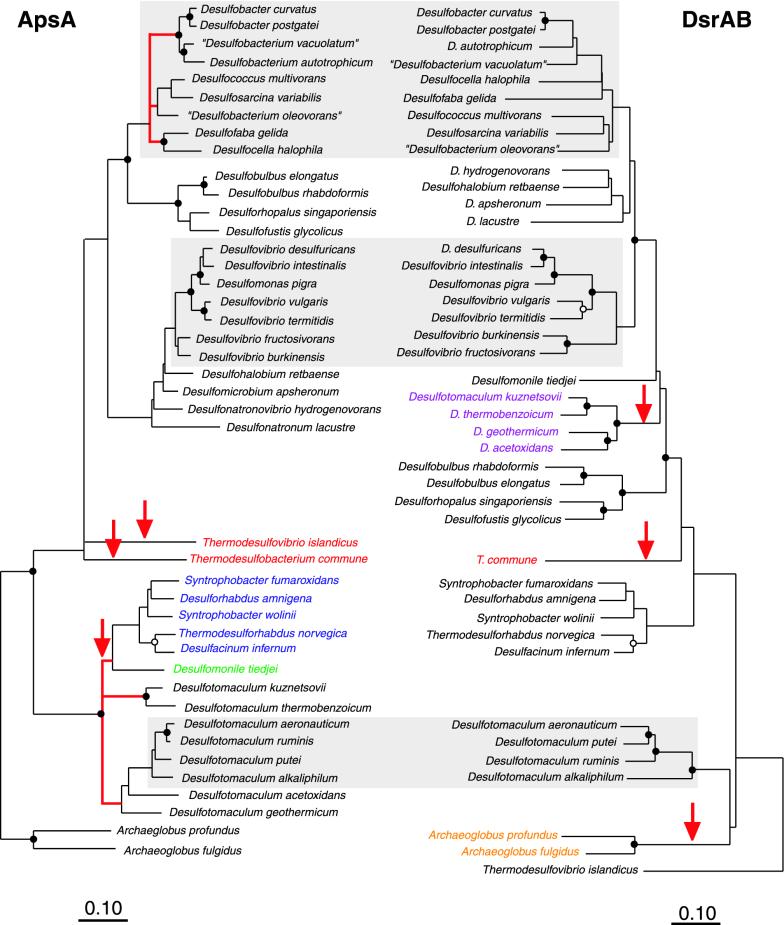
We also compared ApsA- and DsrAB-based phylogenies from a subset of 42 sulfate-reducing microorganisms. This comparison was of particular interest because of the recently described occurrence of lateral dsrAB gene transfer events (19). Fragments of the dsrAB genes were amplified from reference strains (n = 25) using primer pair DSR1-F and DSR4-R (41) and sequenced directly, or sequences available in public databases were used for comparison (as indicated in Table 2). The overall tree topologies of ApsA- and DsrAB-derived analyses were similar (Fig. 2). Some of the differences in topologies were due to different occurrences of lateral transfer events affecting mostly a subset of gram-positive SRB and Archaeoglobus spp. for the dsrAB genes and members of the “Syntrophobacteraceae” and “Nitrospinaceae” for the apsA genes. The only SRB affected by a dual lateral transfer of both genes were Thermodesulfobacterium spp. Other topological differences included the relative branching order of the “Desulfobulbaceae” clade, which was closer to the “Desulfobacteraceae” in the ApsA-based trees (>80% bootstrap support); in DsrAB-based trees, the “Desulfovibrionaceae” clade was closer to the “Desulfobacteraceae.” Although the “Desulfovibrionales” formed a consistent clade in 16S rRNA- and ApsA-based trees, they were recovered only in two separate branches: the “Desulfomicrobiaceae” and “Desulfohalobiaceae” grouped within the radiation of the “Desulfobacteraceae,” whereas the “Desulfovibrionaceae” did not (>80% bootstrap support).
FIG. 2.
Comparison of ApsA- and DsrAB-based phylogenetic trees of members of major lineages of the sulfate-respiring microorganisms. Both trees were calculated using distance matrix-based FITCH analysis. The DsrAB tree was rooted with Thermodesulfovibrio islandicus. Bootstrap analyses, node labeling, and color coding are as described in the legend of Fig. 1. Microorganisms with inferred laterally transferred dsrAB genes are color coded: purple, Desulfotomaculum spp.; orange, Archaeoglobus spp.; red, Thermodesulfobacterium commune. Bars, 0.1 changes per amino acid position.
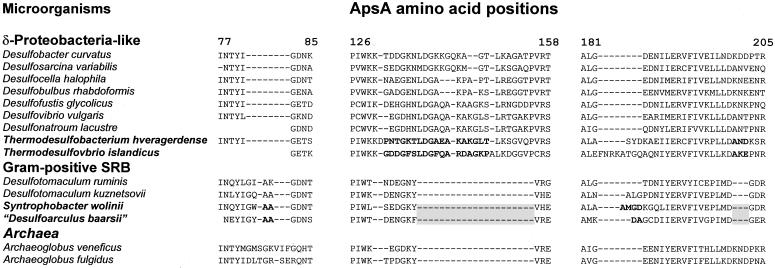
Indels present in ApsA.
Additional evidence for lateral transfer of apsA genes among SRB arises from six regions of the ApsA sequence with minor and major indels (excluded from phylogenetic analyses) present in different lineages (three regions shown in Fig. 3). Archaea, gram-positive SRB, and those gram-negative SRB with a presumed xenologous apsA gene (i.e., “Syntrophobacteraceae” and “Nitrospinaceae”) all have a major deletions between positions 137 and 156 of ApsA (numbering according to Desulfovibrio vulgaris; Fig. 3). On the other hand, certain insertions were not present in δ-proteobacterial SRB. For example, between positions 183 and 184 of the Desulfovibrio vulgaris ApsA are found two (Desulfomonile tiedjei, Desulfobacterium anilini [not shown]), three (Thermodesulfobacterium spp., Desulfotomaculum kuznetsovii, Desulfotomaculum thermobenzoicum [not shown]), four (“Syntrophobacteraceae”), or eight (Thermodesulfovibrio islandicus) additional amino acids.
FIG. 3.
Sequence alignments showing indels within ApsA among representatives of the major lineages of sulfate-respiring microorganisms. Note that δ-proteobacterial members of the Syntrophobacteraceae (i.e., Syntrophobacter spp.) and Nitrospinaceae (i.e., Desulfoarculus baarsii) carry insertions (boldface) and deletions (shaded in gray) characteristic of gram-positive Desulfotomaculum spp., whereas Thermodesulfobacterium spp. and Thermodesulfovibrio islandicus carry insertions (boldface) characteristic of δ-proteobacteria. Sulfate-reducing microorganisms with an inferred xenologous ApsA are in boldface. Amino acid positions are numbered according to ApsA of Desulfovibrio vulgaris.
DISCUSSION
This is the first comprehensive study to analyze the phylogeny of the APS reductase α-subunit (ApsA) from a taxonomically wide range of sulfate-reducing prokaryotes. Comparative analysis of ApsA- and 16S rRNA-based phylogenies revealed significant differences in tree topologies, which suggests that multiple lateral apsA gene transfers occurred among SRP.
Using the newly developed PCR primer set comprising APS7-F and APS8-R, apsA fragments with high sequence similarity to previously published apsA/aprA sequences were obtained from almost all SRP tested (Table 2). The apsA genes of certain strains were amplified only when a considerably lower annealing temperature was used, which indicates possible mismatches at the primer binding sites. Still, the new primer pair allowed us to obtain an amplified apsA DNA fragment considerably longer than those obtained by using the previously used primer pair APS-FW and APS-RV (7).
All ApsA sequences determined (including positions 73 to 386 of the Desulfovibrio vulgaris numbering) contained most of the core regions of the α-subunit of the APS reductase. Based on sequence comparisons with other flavin adenine dinucleotide-containing oxidoreductases, it has been suggested that this region contains the active site of APS reductase (14, 38). In general, the potential binding site of the substrate APS (residues 248 to 272) was highly conserved in all ApsA sequences analyzed. However, notably the gram-positive SRB and members of the “Syntrophobacteraceae” and of the “Nitrospinaceae” contained an additional aliphatic amino acid residue (between Desulfovibrio vulgaris positions 260 and 261) and a phenylalanine (between Desulfovibrio vulgaris positions 261 and 262) and had a deletion at position 265 (data not shown), which may affect APS binding. A more detailed comparison of structural features of APS reductase with the extensive AprA sequence data available will soon be possible since the three-dimensional structure of the APS reductase from Archaeoglobus fulgidus is being determined (36).
The universal phylogenetic tree based on rRNA, and the small-subunit (SSU) rRNA in particular, is still generally accepted to reflect the phylogeny of all organisms (8, 48). The SSU rRNA molecule is ubiquitous, exhibits functional constancy, and changes slowly in sequence, which makes it suitable as a phylogenetic marker. Most importantly, however, is the apparent lack of extensive LGTs affecting the rRNA genes (48). Only a few instances of a genus level transfer of 16S rRNA genes have been reported so far (43) but none across bacterial divisions. Based on the assumption that the 16S rRNA genes of SRP reflect their true evolutionary history, we compared both 16S rRNA- and ApsA-based trees to identify microorganisms which have been involved in LGT of apsA genes.
Major topological differences between the ApsA- and 16S rRNA-based trees suggest that seven species of the “Syntrophobacteraceae,” three members of the “Nitrospinaceae,” three Thermodesulfobacterium spp., and Thermodesulfovibrio islandicus carry xenologous apsA genes. The most parsimonious explanation for the discrepancies in tree topologies is the lateral transfer of apsA genes, which is supported by the following findings. (i) All treeing methods utilized for phylogenetic reconstruction agree on the tree topology, with strong statistical support for the decisive nodes (e.g., branching of gram-positive SRB relative to δ-proteobacterial gram-negative SRB; Fig. 1). The δ-proteobacterial members of the “Syntrophobacteraceae” and “Nitrospinaceae” were clearly recovered monophyletically with the gram-positive SRB. The thermophilic Thermodesulfovibrio islandicus and the Thermodesulfobacterium spp. branched off between the δ-proteobacterial SRB and the gram-positive SRB and not, as would be expected from 16S rRNA analysis, close to the root of the tree. (ii) The ApsA sequences are highly conserved (Table 2), which excludes treeing artifacts stemming from alignment errors. (iii) The patterns of indels of gram-positive SRB, the “Syntrophobacteraceae,” and the “Nitrospinaceae” were similar (Fig. 3).
An alternative interpretation of the discrepancy in tree topologies is a series of gene duplications and losses. However, it is unlikely that the convergent evolution of paralogous ApsA sequences in members of the δ-proteobacterial gram-negative “Syntrophobacteraceae” and “Nitrospinaceae” resulted in sequences closely related to the ApsA sequences of the gram-positive SRB rather than to those of δ-proteobacterial ApsA. Furthermore, and a “Syntrophobacteraceae” and “Nitrospinaceae” are distantly related at the 16S rRNA level (Fig. 1). A gene duplication and a loss event cannot be ruled out completely, however, for the apsA genes of the thermophilic Thermodesulfovibrio islandicus and Thermodesulfobacterium spp. since an orthologous apsA gene of the donor lineage of a putative LGT is not present. Finally, a putative gene duplication event would require in all cases either that the orthologous apsA gene was lost or that the orthologous apsA gene copy was overlooked. The PCR products were directly sequenced, and no evidence for the presence of two or more gene copies (i.e., ambiguous sequence data) was obtained. In addition, PCR fragments obtained by using two independent PCR assays targeting different regions of the apsA gene (i.e., using primer pairs APS-FW and APS-RV and APS7-F and APS8-R) were sequenced, and phylogenetic placement of ApsA sequences was the same regardless of whether the trees were based on the shorter or longer PCR fragment (i.e., using primer pairs APS-FW and APS-RV or APS7-F and APS8-R, respectively) (data not shown). Thus the presence of different apsA sequences in the reference strains studied was not indicated. This issue can only be resolved by extensive Southern hybridization experiments, which were beyond the scope of this study.
A donor lineage for the LGT event inferred for the thermophilic Thermodesulfovibrio islandicus and Thermodesulfobacterium spp. is not apparent; in contrast the gram-positive SRB clearly represent the donor lineage for the LGT affecting members of the “Syntrophobacteraceae” and of the “Nitrospinaceae.” Since the ApsA-based tree topology indicates only a weak association of the two families (Fig. 1 and 2), the possibility that both families received their xenologous apsA genes in two independent LGT events cannot be ruled out. A direct apsA gene donor, however, cannot be currently inferred since all analyzed gram-positive SRB form two separate clades (Fig. 1), which also indicates that the observed LGT was not a recent event. This is also supported by the absence of a conclusive difference in the G+C contents of xenologous apsA genes and orthologous dsrAB genes from within the same SRB (Fig. 2) (19) (G+C data not shown). Differences in G+C contents of host genomes and acquired genes have been used to detect recent LGT events (25). Thus, a recent LGT could have been detected by similar G+C contents and codon biases of the xenologous apsA gene of the recipient and the orthologous apsA of the putative donor SRB. However, codon biases and G+C contents of xenologous genes will be ameliorated to reflect the DNA composition the host genome over time since the same mutational processes affect all genes in the recipient genome.
Another lateral transfer of apsA genes probably affected the Archaeoglobales (Fig. 1); however, only limited support for this assumption is provided by our analyses. Such a LGT has been considered earlier, albeit one based on the phylogenetic analysis of a limited ApsA data set, including sequences of the sulfur-oxidizing phototroph Allochromatium vinosum, as well as Desulfovibrio vulgaris and Archaeoglobus fulgidus (14). Based on our analysis, the high degree of conservation of the ApsA sequences of Archaeoglobus spp. compared to those of the gram-positive Desulfotomaculum spp. and the relatively short length of the Archaeoglobales branch compared to the length in the 16S rRNA-based tree (Fig. 1) suggest that Archaeoglobus spp. possibly carry a xenologous apsA gene. Conversely, Archaeoglobus spp. appear to branch off deeply in the ApsA-based tree of the SRP analyzed, which justified the rooting of the tree with ApsA sequences of Archaeoglobus spp. Further support for xenologous apsA genes in Archaeoglobus spp. arises from phylogenetic analysis of DsrAB (Fig. 2) since there is increasing evidence that Archaeoglobus spp. carry xenologous dsrAB genes (19, 23, 29). Paralogous rooting of DsrA and DsrB indicated that Thermodesulfovibrio spp., and not Archaeoglobus spp. as expected from 16S rRNA-based trees (Fig. 1), are closest to the root (19). Thus, taking into consideration that Archaeoglobus spp. are currently the only recognized archaeal sulfate reducers, it appears likely that not only the dsrAB genes but also the apsA genes of Archaeoglobus spp. were laterally transferred. Interestingly, Archaeoglobus veneficus reduces only sulfite and thiosulfate, but not sulfate (16), even though this strain carries an apsA gene.
The high frequency of lateral transfers involving genes essential for sulfate reduction, such as the dsrAB genes (19) and the apsA genes, indicates that lateral transfer has been a frequent event affecting the evolutionary path of sulfate-respiring prokaryotes. As a possible explanation for the widespread lateral distribution of the dsr genes, it has been suggested that the dsr genes could be part of mobilizable metabolic islands (19). Several lines of evidence suggest that at least the APS reductase and the sulfite reductase genes do not form a genomic island in the genomes of SRP. (i) All lateral apsA transfers detected were not paralleled by a lateral transfer of the dsrAB genes, with the exception of those for Thermodesulfobacterium spp. and possibly the Archaeoglobus spp. (Fig. 2) (19). (ii) The only completed genome sequence of a sulfate-reducing microorganism, i.e., Archaeoglobus fulgidus (the sequencing of several others, i.e., those of Desulfovibrio vulgaris, Desulfovibrio desulfuricans, Desulfotalea psychrophila, and Desulfobacterium autotrophicum, is under way [http://wit.integratedgenomics.com/GOLD/prokaryagenomes.html]) reveals that dsrAB and aprBA genes are not present in one operon or physically close (nucleotide positions 381478 to 383834 and 1498455 to 1500855 of the Archaeoglobus fulgidus genome, respectively [20]). However, the ATP sulfurylase gene, sat, is adjacent to aprBA within the same operon (20). Although this linkage of genes remains to be shown for other SRP, it makes sense from an ecophysiological point of view. Whereas apsBA and sat are required for sulfate respiration, sulfite respiration is an independent metabolic trait, and indeed several microorganisms cannot reduce sulfate but reduce sulfite instead, e.g., Pyrobaculum islandicum (29), Desulfitobacterium spp. (15), Archaeoglobus veneficus (16), and Bilophila wadsworthia (24).
Although sulfate and sulfite respiration appears to be vital for growth of these microorganisms, this is not the case, and several types of sulfate reducers are capable of fermentative growth in the absence of sulfate or other inorganic electron acceptors (46). This versatility of energy conservation could have facilitated the integration of newly acquired xenologous aps or dsr genes into the genetic framework of the recipients without the recipients becoming dependent on these genes before the genes became fully functional in the cell with respect to codon usage or regulation. In fact, Syntrophobacter spp., which carry xenologous apsA genes, grow fermentatively as syntrophic propionate oxidizers, and had been isolated as such, and their ability to reduce sulfate was detected only later (42).
Further patterns of LGTs in SRP exist; these may also be linked to their ecophysiology. Most recipients of xenologous dsrAB and apsA genes are thermophilic (Table 2; Fig. 1 and 2), which suggests a thermophilic lifestyle of sulfate-reducing prokaryotes involved in LGTs (19). Moreover, some of the spore-forming gram-positive SRB are dsrAB recipients (Fig. 2) (19), and gram-positive SRB represent the donor lineage for xenologous apsA genes in members of the “Syntrophobacteraceae” and “Nitrospinaceae” (Fig. 1). Spore-forming SRB have a selective advantage in environments with fluctuating water availability and oxygen stress (47). It may be a further advantage to acquire new genes under these conditions, which could explain the frequent involvement of spore-forming gram-positive SRB in LGT.
Functional markers for physiologically coherent groups of microorganisms (guilds) such as the dsrAB and apsA genes have been used for the characterization of sulfate-reducing populations in a variety of habitats (4, 7, 28, 34, 49); however, a thorough phylogenetic framework has not yet been available. Besides the LGT events detected, we could show that the overall phylogeny of the ApsA-based tree is rather similar to the rRNA-based tree for most of the larger taxa of recognized SRB, including “Desulfovibrionales,” “Desulfobacteraceae,” and “Desulfobulbaceae.” In addition, we could add a substantial set of dsrAB reference sequences, including those of further members of the “Desulfovibrionales,” “Desulfobacteraceae,” “Desulfobulbaceae,” and the “Syntrophobacteraceae.” The phylogenetic analysis of DsrAB is in agreement with that established previously. Interestingly, some of the “Desulfovibrionales” (including Desulfomicrobium spp. D. lacustre, D. hydrogenovorans, and D. retbaense but not D. pigra) formed a cluster separate from members of the “Desulfovibrionaceae.”
With the phylogenetic framework of the two gene markers provided here and previously (19) it will now be possible to link environmental sequences at least to most of the recognized lineages of SRP. Since we could demonstrate that LGT is a frequent event in the evolution of SRP, phylogenetic inferences in environmental diversity studies should be interpreted cautiously.
Acknowledgments
This study was supported by the Max Planck Society, Munich.
I thank Ralf Conrad for continuing support and Thomas Gebhardt for Linux server maintenance, as well as Kai Finster, Alexander Galushko, Bernhard Schink, and Hans Scholten for providing sulfate-reducing strains, Werner Liesack for critically reading the manuscript, and Karen A. Brune for editing the manuscript. A special thank you goes to Bianca Wagner for her excellent technical assistance.
REFERENCES
- 1.Aravind, L., R. L. Tatusov, Y. I. Wolf, D. R. Walker, and E. V. Koonin. 1998. Evidence for massive gene exchange between archaeal and bacterial hyperthermophiles. Trends Genet. 14:442–444. (Erratum, 15:41, 1999.) [DOI] [PubMed] [Google Scholar]
- 2.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, B. A. Rapp, and D. L. Wheeler. 2000. GenBank. Nucleic Acids Res. 28:15–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cameron, E. M. 1982. Sulphate and sulphate reduction in early Precambrian oceans. Nature 296:145–148. [Google Scholar]
- 4.Cottrell, M. T., and S. C. Cary. 1999. Diversity of dissimilatory bisulfite reductase genes of bacteria associated with the deep-sea hydrothermal vent polychaete annelid Alvinella pompejana. Appl. Environ. Microbiol. 65:1127–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dahl, C. 1996. Insertional gene inactivation in a phototrophic sulphur bacterium: APS-reductase-deficient mutants of Chromatium vinosum. Microbiology 142:3363–3372. [DOI] [PubMed] [Google Scholar]
- 6.Dayhoff, M. O., R. M. Schwartz, and B. C. Orcutt. 1978. A model of evolutionary change in proteins, p.345–352. In M. O. Dayhoff (ed.), Atlas of protein sequence structure. National Biomedical Research Foundation, Washington, DC.
- 7.Deplancke, B., K. R. Hristova, H. A. Oakley, V. J. McCracken, R. Aminov, R. I. Mackie, and H. R. Gaskins. 2000. Molecular ecological analysis of the succession and diversity of sulfate-reducing bacteria in the mouse gastrointestinal tract. Appl. Environ. Microbiol. 66:2166–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doolittle, W. F. 1999. Phylogenetic classification and the universal tree. Science 284:2124–2128. [DOI] [PubMed] [Google Scholar]
- 9.Fauque, G., J. Legall, and L. L. Barton. 1991. Sulfate-reducing and sulfur-reducing bacteria, p.271–337. In J. M. Shively and L. L. Barton (ed.), Variations in autotrophic life. Academic Press, London, United Kingdom.
- 10.Ferris, M. J., and D. M. Ward. 1997. Seasonal distributions of dominant 16S rRNA-defined populations in a hot spring microbial mat examined by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 63:1375–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedrich, C. G. 1998. Physiology and genetics of sulfur-oxidizing bacteria. Adv. Microb. Physiol. 39:235–289. [DOI] [PubMed] [Google Scholar]
- 12.Fritz, G., T. Buchert, H. Huber, K. O. Stetter, and P. M. H. Kroneck. 2000. Adenylylsulfate reductases from archaea and bacteria are 1:1 alpha beta-heterodimeric iron-sulfur flavoenzymes—high similarity of molecular properties emphasizes their central role in sulfur metabolism. FEBS Lett. 473:63–66. [DOI] [PubMed] [Google Scholar]
- 13.Henckel, T., M. Friedrich, and R. Conrad. 1999. Molecular analyses of the methane-oxidizing microbial community in rice field soil by targeting the genes of the 16S rRNA, particulate methane monooxygenase, and methanol dehydrogenase. Appl. Environ. Microbiol. 65:1980–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hipp, W. M., A. S. Pott, S. Thum, I. Faath, C. Dahl, and H. G. Trüper. 1997. Towards the phylogeny of APS reductases and sirohaem sulfite reductases in sulfate-reducing and sulfur-oxidizing prokaryotes. Microbiology 143:2891–2902. [DOI] [PubMed] [Google Scholar]
- 15.Holliger, C., G. Wohlfarth, and G. Diekert. 1998. Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol. Rev. 22:383–398. [Google Scholar]
- 16.Huber, H., H. Jannasch, R. Rachel, T. Fuchs, and K. O. Stetter. 1997. Archaeoglobus veneficus sp. nov., a novel facultative chemolithoautotrophic hyperthermophilic sulfite reducer, isolated from abyssal black smokers. Syst. Appl. Microbiol. 20:374–380. [Google Scholar]
- 17.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8:275–282. [DOI] [PubMed] [Google Scholar]
- 18.Joulian, C., B. Ollivier, B. C. Patel, and P. A. Roger. 1998. Phenotypic and phylogenetic characterization of dominant culturable methanogens isolated from ricefield soils. FEMS Microbiol. Ecol. 25:135–145. [Google Scholar]
- 19.Klein, M., M. Friedrich, A. J. Roger, P. Hugenholtz, S. Fishbain, H. Abicht, L. L. Blackall, and D. A. Stahl. 2001. Multiple lateral transfers of dissimilatory sulfite reductase genes between major lineages of sulfate-reducing prokaryotes. J. Bacteriol. 183:6028–6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klenk, H. P., R. A. Clayton, J. F. Tomb, O. White, K. E. Nelson, K. A. Ketchum, R. J. Dodson, M. Gwinn, E. K. Hickey, J. D. Peterson, D. L. Richardson, A. R. Kerlavage, D. E. Graham, N. C. Kyrpides, R. D. Fleischmann, J. Quackenbush, N. H. Lee, G. G. Sutton, S. Gill, E. F. Kirkness, B. A. Dougherty, K. McKenny, M. D. Adams, B. Loftus, S. Peterson, C. Reich, L. K. McNeil, J. H. Badger, A. Glodek, L. Zhou, R. Overbeek, J. D. Gocayne, J. F. Weidman, L. Mcdonald, T. Utterback, M. D. Cotton, T. Spriggs, P. Artiach, B. Kaine, S. M. Sykes, P. W. Sadow, K. P. D’Andrea, C. Bowman, C. Fujii, S. Garland, T. M. Mason, G. J. Olsen, C. M. Fraser, H. O. Smith, C. R. Woese, and J. C. Venter. 1997. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 390:364–370. [DOI] [PubMed] [Google Scholar]
- 21.Lampreia, J., G. Fauque, N. Speich, C. Dahl, I. Moura, H. G. Trüper, and J. J. G. Moura. 1991. Spectroscopic studies on APS reductase isolated from the hyperthermophilic sulfate-reducing archaebacterium Archaeglobus fulgidus. Biochem. Biophys. Res. Commun. 181:342–347. [DOI] [PubMed] [Google Scholar]
- 22.Lane, D. J. 1991. 16S/23S rRNA sequencing, p.115–147. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acids techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom.
- 23.Larsen, Ø., T. Lien, and N.-K. Birkeland. 1999. Dissimilatory sulfite reductase from Archaeoglobus profundus and Desulfotomaculum thermocisternum: phylogenetic and structural implications from gene sequences. Extremophiles 3:63–70. [DOI] [PubMed] [Google Scholar]
- 24.Laue, H., M. Friedrich, J. Ruff, and A. M. Cook. 2001. Dissimilatory sulfite reductase (desulfoviridin) of the taurine-degrading, non-sulfate-reducing bacterium Bilophila wadsworthia RZATAU contains a fused DsrB-DsrD subunit. J. Bacteriol. 183:1727–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawrence, J. G., and H. Ochman. 1997. Amelioration of bacterial genomes: rates of change and exchange. J. Mol. Evol. 44:383–397. [DOI] [PubMed] [Google Scholar]
- 26.Lawrence, J. G., and H. Ochman. 1998. Molecular archaeology of the Escherichia coli genome. Proc. Natl. Acad. Sci. USA 95:9413–9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ludwig, W., O. Strunk, S. Klugbauer, N. Klugbauer, M. Weizenegger, J. Neumaier, M. Bachleitner, and K. H. Schleifer. 1998. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis 19:554–568. [DOI] [PubMed] [Google Scholar]
- 28.Minz, D., J. L. Flax, S. J. Green, G. Muyzer, Y. Cohen, M. Wagner, B. E. Rittmann, and D. A. Stahl. 1999. Diversity of sulfate-reducing bacteria in oxic and anoxic regions of a microbial mat characterized by comparative analysis of dissimilatory sulfite reductase genes. Appl. Environ. Microbiol. 65:4666–4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Molitor, M., C. Dahl, I. Molitor, U. Schaefer, N. Speich, R. Huber, R. Deutzmann, and H. G. Trüper. 1998. A dissimilatory sirohaem-sulfite-reductase-type protein from the hyperthermophilic archaeon Pyrobaculum islandicum. Microbiology 144:529–541. [DOI] [PubMed] [Google Scholar]
- 30.Moré, M. I., J. B. Herrick, M. C. Silva, W. C. Ghiorse, and E. L. Madsen. 1994. Quantitative cell lysis of indigenous microorganisms and rapid extraction of microbial DNA from sediment. Appl. Environ. Microbiol. 60:1572–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Müller, T., and M. Vingron. 2000. Modeling amino acid replacement. J. Comp. Biol. 7:761–776. [DOI] [PubMed] [Google Scholar]
- 32.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299–304. [DOI] [PubMed] [Google Scholar]
- 33.Olsen, G. J., H. Matsuda, R. Hagstrom, and R. Overbeek. 1994. FastDNAml: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput. Appl. Biosci. 10:41–48. [DOI] [PubMed] [Google Scholar]
- 34.Power, M. E., J. C. Araujo, J. R. Van Der Meer, H. Harms, and O. Wanner. 1999. Monitoring sulfate-reducing bacteria in heterotrophic biofilms. Water Sci. Technol. 39:49–56. [Google Scholar]
- 35.Rabus, R., T. A. Hansen, and F. Widdel. 1999. Dissimilatory sulfate- and sulfur-reducing prokaryotes, p.1–87. In M. Dworkin, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes: an evolving electronic database for the microbiological community. Springer, New York, N.Y.
- 36.Roth, A., G. Fritz, T. Buchert, H. Huber, K. O. Stetter, U. Ermler, and P. M. Kroneck. 2000. Crystallization and preliminary X-ray analysis of adenylylsulfate reductase from Archaeoglobus fulgidus. Acta Crystallogr. D 56:1673–1675. [DOI] [PubMed] [Google Scholar]
- 37.Schidlowski, M. 1979. Antiquity and evolutionary status of bacterial sulfate reduction: sulfur isotope evidence. Origins Life 9:299–311. [DOI] [PubMed] [Google Scholar]
- 38.Speich, N., C. Dahl, P. Heisig, A. Klein, F. Lottspeich, K. O. Stetter, and H. G. Trüper. 1994. Adenylylsulphate reductase from the sulphate-reducing archaeon Archaeoglobus fulgidus: cloning and characterization of the genes and comparison of the enzyme with other iron-sulphur flavoproteins. Microbiology 140:1273–1284. [DOI] [PubMed] [Google Scholar]
- 39.Strimmer, K., and A. von Haeseler. 1996. Quartet puzzling: a quartet maximum-likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13:964–969. [Google Scholar]
- 40.Tamura, K., and M. Nei. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10:512–526. [DOI] [PubMed] [Google Scholar]
- 41.Wagner, M., A. J. Roger, J. L. Flax, G. A. Brusseau, and D. A. Stahl. 1998. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J. Bacteriol. 180:2975–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wallrabenstein, C., E. Hauschild, and B. Schink. 1994. Pure culture and cytological properties of ‘Syntrophobacter wolinii.’ FEMS Microbiol. Lett. 123:249–254. [Google Scholar]
- 43.Wang, Y., and Z. Zhang. 2000. Comparative sequence analyses reveal frequent occurrence of short segments containing an abnormally high number of non-random base variations in bacterial rRNA genes. Microbiology 146:2845–2854. [DOI] [PubMed] [Google Scholar]
- 44.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whelan, S., and N. Goldman. 2001. A new empirical model of amino acid evolution. Mol. Biol. Evol. 18:691–699. [DOI] [PubMed] [Google Scholar]
- 46.Widdel, F. 1988. Microbiology and ecology of sulfate- and sulfur-reducing bacteria, p.469–585. In A. J. B. Zehnder (ed.), Biology of anaerobic microorganisms. Wiley, New York, N.Y.
- 47.Widdel, F. 1999. The genus Desulfotomaculum, p.1–6. In M. Dworkin, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes: an evolving electronic database for the microbiological community. Springer, New York, N.Y.
- 48.Woese, C. R. 2000. Interpreting the universal phylogenetic tree. Proc. Natl. Acad. Sci. USA 97:8392–8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zinkevich, V., and I. B. Beech. 2000. Screening of sulfate-reducing bacteria in colonoscopy samples from healthy and colitic human gut mucosa. FEMS Microbiol. Ecol. 34:147–155. [DOI] [PubMed] [Google Scholar]