Abstract
The OrthoMCL database (http://orthomcl.cbil.upenn.edu) houses ortholog group predictions for 55 species, including 16 bacterial and 4 archaeal genomes representing phylogenetically diverse lineages, and most currently available complete eukaryotic genomes: 24 unikonts (12 animals, 9 fungi, microsporidium, Dictyostelium, Entamoeba), 4 plants/algae and 7 apicomplexan parasites. OrthoMCL software was used to cluster proteins based on sequence similarity, using an all-against-all BLAST search of each species' proteome, followed by normalization of inter-species differences, and Markov clustering. A total of 511 797 proteins (81.6% of the total dataset) were clustered into 70 388 ortholog groups. The ortholog database may be queried based on protein or group accession numbers, keyword descriptions or BLAST similarity. Ortholog groups exhibiting specific phyletic patterns may also be identified, using either a graphical interface or a text-based Phyletic Pattern Expression grammar. Information for ortholog groups includes the phyletic profile, the list of member proteins and a multiple sequence alignment, a statistical summary and graphical view of similarities, and a graphical representation of domain architecture. OrthoMCL software, the entire FASTA dataset employed and clustering results are available for download. OrthoMCL-DB provides a centralized warehouse for orthology prediction among multiple species, and will be updated and expanded as additional genome sequence data become available.
INTRODUCTION
The ongoing sequencing of multiple genomes creates a growing need for functional annotation. Comparative approaches based on ortholog identification have been particularly useful, enabling protein function to be inferred based on information available from other species, and providing the raw material for evolutionary analysis (1). Homologous proteins share a common ancestry, and may be characterized as orthologs (which diverged from a common ancestral gene owing to speciation) or paralogs (which derive from a gene duplication event) (2). In general, orthologous genes are expected to retain similar (if not identical) function, while paralogs may more readily acquire novel functional roles.
OrthoMCL is a graph-clustering algorithm designed to identify homologous proteins based on sequence similarity, and distinguish orthologous from paralogous relationships without computationally intensive phylogenetic analysis (3). The algorithm first flags probable orthologous pairs identified by BLAST analysis as reciprocal best hits across two genomes (1), creating a graph in which edge weights connecting each protein pair are based on BLAST similarity scores. In addition, probable in-paralogs arising from duplication events subsequent to species divergence (2) are identified as sequences within the same genome that are (reciprocally) more similar to each other than either is to any sequence from other genomes, i.e. reciprocal better hits (3). Attaching these in-paralogous relationships, and incorporating edges connecting the resulting co-orthologs, overcomes the inability of simple reciprocal best hit approaches to detect many-to-many relationships (3,4). Edge weights are then adjusted to account for genome-to-genome similarity averages, and the resulting graph is clustered using the MCL algorithm (5), reducing large clusters containing weak single linkages into smaller clusters that are more robust in their representation of truly orthologous relationships (3). In contrast to TribeMCL (7), which clusters proteins based on all BLAST similarities, producing large protein families, OrthoMCL focuses on the identification of proteins whose similarity suggests true orthology. As a fully automated method, OrthoMCL is applicable to multiple species datasets by bypassing the labor-intensive manual curation involved in the construction of the NCBI KOG (euKaryotic Ortholog Groups) database (6). Preliminary results indicate that OrthoMCL groups exhibit higher levels of functional consistency than other ortholog identification algorithms (data not shown).
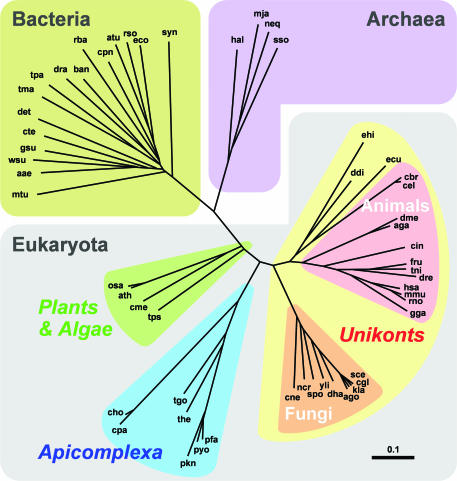
OrthoMCL was designed to address the difficulties inherent in identifying eukaryotic orthologs, focusing on the recognition of recent duplications, and the use of Markov clustering to separate groups linked by protein fusions (7). An initial report described clustering of six eukaryotic genomes and one reference prokaryotic species (Escherichia coli K12) (3). Many additional genome sequences have been released in the last 2 years, however, stimulating considerable demand for the identification of ortholog groups. This report describes clustering of the predicted proteomes for 35 eukaryotic and 20 diverse prokaryotic species (both bacteria and archaea), spanning the tree of life (Figure 1), and an online database for perusing, querying and retrieving of these clusters.
Figure 1.
A phylogeny was constructed for 55 sequenced genomes based on orthologous gene content. See Table 1 for species abbreviations. The tree was drawn using Phylodendron (http://iubio.bio.indiana.edu/treeapp/treeprint-form.html).
METHODS
Protein sequence data
Translated protein sequences for all eukaryotic genomes considered complete as on July 2005 were obtained from the following sources: bacterial and archaeal sequences from GenBank (8); many eukaryotic sequences (e.g. Drosophila melanogaster and Homo sapiens) from Ensembl (9); other sequences from the relevant sequencing centers or organism-specific databases (see Table 1). In some cases, this resulted in the inclusion of proteins derived from differentially spliced transcripts. Because various naming systems are used for protein identification at the different source sites, a unified sequence accession format (consisting of the genome abbreviation followed by a number) was used to provide each protein with a unique identifier. Original sequence identifiers were incorporated into the sequence description. A total of 627 098 protein sequences was obtained from 55 genomes (see Table 1).
Table 1.
The 55 genomes included in OrthoMCL-DB, with clustering statistics
| Lineage | Abbreviation | Full name | Data source | Sequences | Clustered | Groups |
|---|---|---|---|---|---|---|
| Archaea | ||||||
| Euryarchaeota | hal | Halobacterium sp. NRC-1 | GenBank | 2622 | 1878 | 1323 |
| mja | Methanococcus jannaschii DSSM 2661 | GenBank | 1786 | 1260 | 1054 | |
| Crenarchaeota | sso | Sulfolobus solfataricus P2 | GenBank | 2977 | 2220 | 1357 |
| Nanoarchaeota | neq | Nanoarchaeum equitans Kin4-M | GenBank | 536 | 351 | 336 |
| Bacteria | ||||||
| Proteobacteria | wsu | Wolinella succinogenes DSM 1740 | GenBank | 2044 | 1617 | 1338 |
| gsu | Geobacter sulfurreducens PCA | GenBank | 3446 | 2616 | 1987 | |
| atu | Agrobacterium tumefaciens C58 Uwash | GenBank | 5402 | 3826 | 2757 | |
| rso | Ralstonia solanacearum GMI1000 | GenBank | 5116 | 3856 | 2795 | |
| eco | Escherichia coli K12 | GenBank | 4242 | 3295 | 2536 | |
| Aquifex | aae | Aquifex aeolicus VF5 | GenBank | 1560 | 1294 | 1165 |
| Thermotoga | tma | Thermotoga maritima MSB8 | GenBank | 1858 | 1473 | 1297 |
| Green nonsulfur | det | Dehalocsoccoides ethenogenes 195 | GenBank | 1580 | 1237 | 963 |
| Deinococci | dra | Deinococcus radiodurans R1 | GenBank | 3182 | 2249 | 1848 |
| Spirochetes | tpa | Treponema pallidum subsp. pallidum str. Nichols | GenBank | 1036 | 703 | 621 |
| Green sulfur | cte | Chlorobium tepidum TLS | GenBank | 2252 | 1554 | 1361 |
| Planctomyces/Pirella | rba | Rhodopirellula baltica SH_1 | GenBank | 7325 | 3624 | 2261 |
| Chlamydia | cpn | Chlamydophila pneumoniae CWL029 | GenBank | 1052 | 722 | 599 |
| Cyanobacteria | syn | Synechococcus sp. WH8102 | GenBank | 2517 | 1782 | 1526 |
| Actinobacteria | mtu | Mycobacterium tuberculosis H37Rv | GenBank | 3991 | 2963 | 1983 |
| Gram-positive | ban | Bacillus anthracis Ames Ames | GenBank | 5311 | 3497 | 2361 |
| Eukaryota | ||||||
| Entamoeba | ehi | Entamoeba histolytica | TIGR | 9772 | 8149 | 2910 |
| Dictyostelium | ddi | Dictyostelium discoideum | dictyBase | 13 678 | 10 144 | 4974 |
| Plants/Algae | cme | Cyanodioschyzon merolae 10D | University of Tokyo | 5013 | 3802 | 3286 |
| tps | Thalassiosira pseudonana | JGI | 11 397 | 7767 | 5211 | |
| ath | Arabidopsis thaliana | TIGR | 28 952 | 25 546 | 11 390 | |
| osa | Oryza sativa | TIGR | 88 149 | 78 731 | 18 933 | |
| Apicomplexa | tgo | Toxoplasma gondii | ToxoDB | 7793 | 4522 | 3755 |
| cpa | Cryptosporidium parvum Iowa | CryptoDB | 3396 | 3287 | 3222 | |
| cho | Cryptosporidium hominis TU502 | CryptoDB | 3886 | 3532 | 3427 | |
| pfa | Plasmodium falciparum 3D7 | PlasmoDB | 5363 | 5054 | 4371 | |
| pyo | Plasmodium yoelii 17XNL | PlasmoDB | 7850 | 6056 | 4252 | |
| pkn | Plasmodium knowlesi | PlasmoDB | 6890 | 4692 | 3878 | |
| the | Theileria parva | TIGR | 4035 | 3003 | 2455 | |
| Fungi | sce | Saccharomyces cerevisiae S288C | SGD | 6702 | 5612 | 4633 |
| spo | Schizosaccharomyces pombe | Sanger | 4984 | 4328 | 3726 | |
| yli | Yarrowia lipolytica CLIB99 | Genolevures | 6666 | 5549 | 4464 | |
| kla | Kluyveromyces lactis CLIB210 | Genolevures | 5331 | 4957 | 4592 | |
| dha | Debaryomyces hansenii CBS767 | Genolevures | 6896 | 5602 | 4581 | |
| cgl | Candida glabrata CBS138 | Genolevures | 5272 | 4947 | 4342 | |
| cne | Cryptococcus neoformans | TIGR | 5882 | 4743 | 3845 | |
| ago | Ashbya gossypii | AGD | 4726 | 4565 | 4335 | |
| ncr | Neurospora crassa OR74A | Whitehead | 10 617 | 6298 | 5102 | |
| Microsporidium | ecu | Encephalitozoon cuniculi | GenBank | 1996 | 1348 | 1113 |
| Animals | cel | Caenorhabditis elegans | WORMBASE | 22 420 | 19 307 | 13 242 |
| cbr | Caenorhabditis briggsae | Sanger | 19 334 | 16 948 | 13 227 | |
| dme | Drosophila melanogaster | Ensembl | 19 177 | 16 251 | 8640 | |
| aga | Anopheles gambiae | Ensembl | 15 802 | 12 645 | 8662 | |
| cin | Ciona intestinalis | Ensembl | 15 851 | 11 460 | 8140 | |
| fru | Fugu rubripes | Ensembl | 33 003 | 28 145 | 14 277 | |
| tni | Tetraodon nigroviridis | Ensembl | 28 005 | 18 707 | 13 861 | |
| dre | Danio rerio | Ensembl | 32 062 | 26 692 | 12 738 | |
| gga | Gallus gallus | Ensembl | 28 416 | 22 826 | 12 420 | |
| mmu | Mus musculus | Ensembl | 31 535 | 27 299 | 17 917 | |
| rno | Rattus norvegicus | Ensembl | 32 543 | 28 318 | 17 445 | |
| hsa | Homo sapiens | Ensembl | 33 869 | 28 948 | 16 586 | |
OrthoMCL clustering
OrthoMCL was originally designed as a pipeline integrated with a GUS (Genomic Unified Schema) relational database (http://www.gusdb.org). In response to multiple requests from users, a stand-alone Perl script version of OrthoMCL is now available from the website, allowing this ortholog clustering algorithm to be run without a relational database. OrthoMCL accepts as input a tab-delimited summary of all-against-all sequence similarity search data, including estimates of statistical significance in the form of expectation values. For this dataset, a single FASTA file was compiled from all genomes, and a WU-BLASTP (10) search was performed using the following parameters: E = 1 × 10−5 wordmask = seg + xnu W = 3 T = 1000. BLAST results were fed into the stand-alone OrthoMCL program using a default MCL inflation parameter of 1.5.
Construction of the OrthoMCL database
Results from the OrthoMCL clustering were loaded into a custom MySQL relational database, along with additional computational analysis made available via the web interface. Pfam 17.0 domain assignments were generated for each sequence based on hmmpfam (http://hmmer.wustl.edu/), using the gathering cut-off (11). Summary statistics on sequence similarity for each group include percentage match pairs (fraction of protein pairs aligned in the initial all-against-all WU-BLASTP search), average E-value (based on log [E-value]), average percent coverage (fraction of aligned regions, based on the shorter sequence) and average percent identity. In addition, MUSCLE multiple sequence alignment (12) and BioLayout graphical visualization of sequence similarities (13) are provided for groups with ≤100 proteins. The OrthoMCL-DB web interface is run by Perl CGI scripts that implement a simple MVC (Model View Controller) architecture provided by the CGI::Application Perl module. The relational database schema and associated Perl scripts for data loading are available from the authors.
Species tree calculation
The unrooted species tree shown in Figure 1 was calculated using the PHYLIP program ‘neighbor’ for neighbor joining (14), where the distances between two species (dij) are calculated based on the number of ortholog groups shared between two species (nij), normalized to account for the number of ortholog groups observed in the two species considered separately (ni, nj):
Note that only ortholog groups containing proteins from at least two species were considered for this analysis.
RESULTS
OrthoMCL clustering
In this implementation of OrthoMCL, 511 797 of 627 098 protein sequences (81.6%) were clustered into 70 388 ortholog groups, as summarized for each species in Table 1. In some species—particularly those eukaryotes showing extensive gene duplications—the number of protein sequences is much higher than the number of ortholog groups identified. For example, while 3295 out of 4242 Escherichia coli sequences (78%) were clustered into 2536 groups (average of 1.3 E.coli sequences/group), 78 731 of 88 149 sequences from the Oryza sativa (rice) genome (89%) were represented by just 18 933 groups (average of 4.2 O.sativa sequences/group). An average of 7.3 sequences were identified per ortholog group (min. 2, max. 822), representing an average of 4.3 species (min. 1, max. 55). As a consequence of the conservative approach used for ortholog identification, OrthoMCL groups tend to be small, containing only a handful of sequences from a limited number of species. In some cases, ancient out-paralogs of these genes may be represented by other groups, and protein family clustering methods such as TribeMCL (7) could be helpful in identifying such relationships.
A relatively non-stringent E-value threshold (1 × 10−5) was used for inclusion of BLAST hits in the OrthoMCL graph, in order to ensure identification of distantly diverged orthologs. Although this might be expected to include many false positives, rules applied during group identification (reciprocal best/better hits, Markov clustering) eliminate most poorly alignable sequences. Considering the entire clustered dataset, 79% of all pairs within OrthoMCL groups were recognized in the initial BLAST search, and display an average E-value = 1 × 10−114, average percent identity = 53% and average percent coverage = 85%. The performance of this algorithm has been validated by comparison with other ortholog identification algorithms, and assessing consistency with EC number annotations (3).
Only six ortholog groups, representing ribosomal proteins and tRNA synthetases, contain proteins from all 55 genomes. It is not surprising that so few universal ortholog groups can be identified by similarity-based clustering alone, given the reduced gene content of some minimalist genomes, and the high degree of horizontal transfer and gene displacement observed in bacterial and archaeal species. A total of 20 583 ortholog groups contain only in-paralogs from a single species lineage, representing both organism-specific inventions, and ancient duplications retained in one lineage only (among those in the dataset).
Reconstructing the tree of life from phyletic data
The total number of shared ortholog groups for all pairwise species comparisons (available from the OrthoMCL-DB website as an Excel spreadsheet) can be used as an indication of phylogenetic distance between species (15), providing the basis for evolutionary reconstruction based on total proteomic evidence. The number of shared ortholog groups ranges from a low of 54 groups representing sequences from both Nanoarchaeum equitans and Chlamydophila pneumoniae, to a high of 15 954 groups with members from both Mus musculus and Rattus norvegicus. A tree of life constructed from these data closely reflects current understanding of organismal evolution (Figure 1), clustering the Archaea, Bacteria and Eukaryota in distinct groups, and clearly defining known eukaryotic assemblages, including the Plants/Algae, Apicomplexa and Unikonts [animals, fungi, microsporidia, slime molds (Dictyostelium) and amoebae (Entamoeba)] (16).
This total evidence tree reflects the evolutionary history of complete genomes, and it is interesting to note the relatively uniform branch lengths for all taxa, in contrast to the extreme variations in branch length often observed for trees based on individual genes. Differences between the topology of this tree and individual gene phylogenies, such as rRNA trees (17), include the grouping of Dictyostelium, Entamoeba and microsporidia with animals, and the deeper branching of Plants/Algae than Apicomplexa within the eukaryotic world. Some of these differences may be explained by events producing significant changes in gene content: gene loss, evolutionary convergence (especially in pathogen species), endosymbiosis and other cases of massive horizontal gene transfer. Despite the low resolution of prokaryotic phylogeny in this analysis (based on a limited taxonomic sampling), the observed topology resembles other analyses of prokaryotes (18).
OrthoMCL-DB web interface
The OrthoMCL-DB web interface provides a convenient means to search for sequences (and their corresponding ortholog groups) based on protein accession number or text keywords. In addition, a BLAST-based sequence similarity search function is provided, allowing users to find their favorite sequence or identify homologs that have been clustered into ortholog groups. Users are cautioned that identifying a homolog in a given ortholog group does not necessarily imply that the query sequence is in fact an ortholog to members of that group. Ortholog groups themselves can be searched by group accession number, or based on ortholog group summary statistics, including group size, average pairwise BLAST expectation value, average pairwise percent identity/coverage or percentage of matched pairs.
To further assist users in extracting biologically interesting ortholog groups, an interface permits queries based on phyletic patterns of conservation, using either a graphical form or text-based expressions. Both methods allow the user to identify ortholog groups by defining the desired pattern of the species representation. The graphical form lists all 55 species, organized by taxonomic clade, with toggle buttons that the user clicks to change status. A green check mark ‘√’ icon is used to represent required presence of a protein from a given species or clade, a red ‘x’ icon for required absence, or a gray circle icon ‘•’ meaning that the presence or absence of proteins from this species should not affect the result. This query form may be used, for example, to identify all groups containing proteins found in all eukaryotes but completely absent from the bacteria, regardless of their presence or absence in archaea.
For more intricate queries, such as the identification of genes that are specifically amplified in insects, a text-based form allows patterns to be expressed using a custom grammar called phyletic pattern expression (PPE). Individual grammatical units of PPE expressions are composed of two parts:
A species specification, composed of a three-letter species abbreviation (e.g. ‘tgo’), or a list of species abbreviations linked by plus sign ‘+’ (e.g. ‘tgo+pfa+hsa’). Several abbreviations are also permitted, such as ‘BAC’ for all bacterial genomes, ‘EUK’ for all eukaryotic genomes, ‘API’ for all apicomplexan genomes, ‘ALL’ to represent all 55 genomes and ‘OTHER’ to represent all other genomes not already specified anywhere in the composite expression (a complete list of clade abbreviations can be found in the website).
A logical comparison operator, such as >, <, =, ≥ or ≤, and a number representing the number of sequences from these species that must be present in the ortholog group (e.g. ‘tgo>5’ specifies ortholog groups containing at least five in-paralogs from Toxoplasma gondii). Alternatively, when appended with the character ‘T’ (for Taxa), this number represents the number of species that must be represented in the ortholog group. For example, ‘EUK>=5T AND hsa>=10’ would generate all ortholog groups representing at least five eukaryotic species and containing at least 10 human proteins.
Multiple expression units may be combined using ‘AND’ or ‘OR', and may use parentheses to provide explicit execution ordering.
OrthoMCL-DB also provides a query history page, detailing all of the queries executed in the current session. Previous query results may be retrieved, and separate results can be further merged via intersection, union or subtraction operations, permitting very complicated queries to be generated by combining different ortholog group query methods. For example, the user may wish to identify ortholog groups that are well conserved (percent identity ≥ 70%), entirely absent in bacteria and archaea, present in at least five eukaryotic genomes, and expanded in Homo sapiens to include at least 10 recent paralogs.
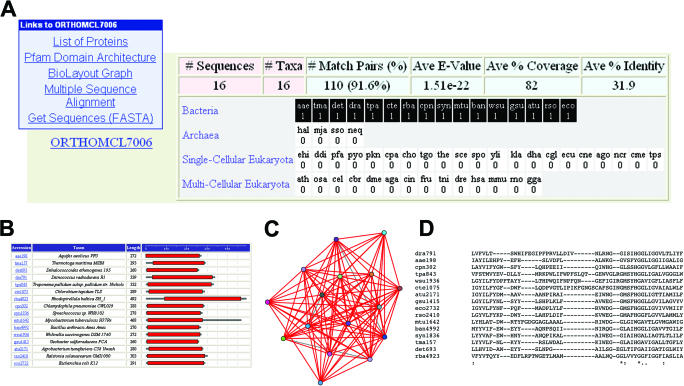
Ortholog groups are displayed to reflect patterns of phyletic conservation using a concise tabular form, along with summary statistics for the ortholog group and hyperlinks to view or download related sequence data (Figure 2). Precomputed information available for most ortholog groups includes Pfam domain architecture, visualizations of OrthoMCL similarity graphs generated using BioLayout software and multiple sequence alignments generated using MUSCLE. These resources provide useful insights into the evolution and organization of proteins within individual ortholog groups.
Figure 2.
An OrthoMCL group is a cluster of sequences from multiple species predicted to be orthologous to each other. (A) Ortholog group summary information, including group size (# Sequences, # Taxa), BLAST statistics (% Match Pairs, Average E-value, Average % Coverage, Average % Identity) and the phyletic pattern profile for all species in the dataset is shown. Rows in the phyletic pattern profile table represent bacteria, archaea, single-cellular eukaryotes and multi-cellular eukaryotes (plants and animals); each box represents a single species, with black or white background denoting presence or absence in the ortholog group, and the number of protein sequences found in the ortholog group listed. Mouse-over expands abbreviations to provide the full species name. Links at top left access a tabular list of information for each member of the ortholog group (including links to the reference database), a graphical representation of Pfam domain architecture (B), a BioLayout graph of pairwise similarity scores (C), a MUSCLE multiple sequence alignment (D) and a sequence retrieval option. The example shown illustrates a ‘prolipoprotein diacylglyceryl transferase’, whose distribution is restricted to the bacteria.
In summary, OrthoMCL-DB provides flexible web-based access to the results of a powerful algorithm for automated ortholog identification, applied to most of the currently available eukaryotic genomes and a representative selection of prokaryotic genomes. We anticipate reclustering and updating the database at least twice a year, as additional eukaryotic genomes become available; inclusion of additional prokaryotic genomes will also be considered.
Data availability
In addition to information available for browsing and querying via the web interface, the following data are available for bulk download as flat-files and/or SQL export files: all protein sequences from the current implementation of OrthoMCL-DB (in FASTA format), all clustering data (accession numbers for all proteins in each ortholog group), Pfam domain assignments for all proteins and summary statistics calculated for each group. An Excel spreadsheet lists the number of ortholog groups shared by all possible species pairs (data used to assemble the tree shown in Figure 1). The stand-alone version of OrthoMCL software is also downloadable.
Acknowledgments
This research was supported by NIH grant R01-AI058515, with website implementation covered by NIAID contract HHSN266200400037C, supporting the ApiDB Bioinformatics Resource Center. We thank Drs Li Li and Shailesh Date for helpful discussions, Lucia Peixoto for running MUSCLE software and Leon Goldovsky (European Bioinformatics Institute) for providing a special version of BioLayout Software. DSR is an Ellison Medical Foundation Scholar in Global Infectious Diseases. Funding to pay the Open Access publication charges for this article was provided by NIH grant R01-AI058515.
Conflict of interest statement. None declared.
REFERENCES
- 1.Tatusov R.L., Koonin E.V., Lipman D.J. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 2.Sonnhammer E.L., Koonin E.V. Orthology, paralogy and proposed classification for paralog subtypes. Trends Genet. 2002;18:619–620. doi: 10.1016/s0168-9525(02)02793-2. [DOI] [PubMed] [Google Scholar]
- 3.Li L., Stoeckert C.J., Jr, Roos D.S. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Remm M., Storm C.E., Sonnhammer E.L. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- 5.Van Dongen S. The Netherlands: University of Utrecht; 2000. Graph clustering by flow simulation. PhD Thesis. [Google Scholar]
- 6.Tatusov R.L., Fedorova N.D., Jackson J.D., Jacobs A.R., Kiryutin B., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N., et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enright A.J., Van Dongen S., Ouzounis C.A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L. GenBank. Nucleic Acids Res. 2005;33:D34–D38. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birney E., Andrews D., Bevan P., Caccamo M., Cameron G., Chen Y., Clarke L., Coates G., Cox T., Cuff J., et al. Ensembl 2004. Nucleic Acids Res. 2004;32:D468–D470. doi: 10.1093/nar/gkh038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altschul S.F., Gish W. Local alignment statistics. Methods Enzymol. 1996;266:460–480. doi: 10.1016/s0076-6879(96)66029-7. [DOI] [PubMed] [Google Scholar]
- 11.Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldovsky L., Cases I., Enright A.J., Ouzounis C.A. BioLayout(Java): versatile network visualisation of structural and functional relationships. Appl. Bioinformatics. 2005;4:71–74. doi: 10.2165/00822942-200504010-00009. [DOI] [PubMed] [Google Scholar]
- 14.Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 15.Snel B., Bork P., Huynen M.A. Genome phylogeny based on gene content. Nature Genet. 1999;21:108–110. doi: 10.1038/5052. [DOI] [PubMed] [Google Scholar]
- 16.Keeling P.J., Berger G., Durnford D.G., Lang B.F., Lee R.W., Pearlman R.E., Roger A.J., Gray M.W. The tree of eukaryotes. Trends Ecol. Evol. doi: 10.1016/j.tree.2005.09.005. in press. [DOI] [PubMed] [Google Scholar]
- 17.Pace N.R. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 18.Korbel J.O., Snel B., Huynen M.A., Bork P. SHOT: a web server for the construction of genome phylogenies. Trends Genet. 2002;18:158–162. doi: 10.1016/s0168-9525(01)02597-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
In addition to information available for browsing and querying via the web interface, the following data are available for bulk download as flat-files and/or SQL export files: all protein sequences from the current implementation of OrthoMCL-DB (in FASTA format), all clustering data (accession numbers for all proteins in each ortholog group), Pfam domain assignments for all proteins and summary statistics calculated for each group. An Excel spreadsheet lists the number of ortholog groups shared by all possible species pairs (data used to assemble the tree shown in Figure 1). The stand-alone version of OrthoMCL software is also downloadable.