Abstract
Protein structures are flexible, changing their shapes not only upon substrate binding, but also during evolution as a collective effect of mutations, deletions and insertions. A new generation of protein structure comparison algorithms allows for such flexibility; they go beyond identifying the largest common part between two proteins and find hinge regions and patterns of flexibility in protein families. Here we present a Flexible Structural Neighborhood (FSN), a database of structural neighbors of proteins deposited in PDB as seen by a flexible protein structure alignment program FATCAT, developed previously in our group. The database, searchable by a protein PDB code, provides lists of proteins with statistically significant structural similarity and on lower menu levels provides detailed alignments, interactive superposition of structures and positions of hinges that were identified in the comparison. While superficially similar to other structural protein alignment resources, FSN provides a unique resource to study not only protein structural similarity, but also how protein structures change. FSN is available from a server http://fatcat.burnham.org/fatcat/struct_neighbor and by direct links from the PDB database.
INTRODUCTION
Last years brought an exponential growth in the number of known protein structures and we can foresee even faster growth as a result of the NIH Structural Genomics Initiative and related development of high throughput structure determination techniques (1,2). Over 30 000 protein structures are now available from PDB (3) (see http://www.rcsb.org for the latest statistics), a single worldwide public depository of 3D biological macromolecular structure data. Of course among these thousands of structures many are similar and most structural classifications (described later) define 800–1300 basic types of structures, called folds. One of the most often asked questions in structural analysis of a protein is what fold it has, or in other words what other proteins are similar to it.
The question of comparing and classifying protein structures is of much interest and currently is one of the most active research areas in bioinformatics. Most of the existing approaches fall into two categories—classification or comparison. In the first category, resources such as SCOP or CATH provide hierarchical classification of all PDB deposited protein structures at different levels, including class (e.g. all alpha and mixed alpha/beta), fold (e.g. TIM, immunoglobulin and globin folds), superfamily (i.e. groups of homologous proteins), and family (i.e. groups of closely related proteins). Classification resources are either created or at least curated by hand and the ultimate decision about the position of a specific protein on the tree is made by a human curator based on some features of a protein, but often without an explicit alignment of the proteins being classified. In the second class, programs such as DALI (4), VAST (5) or CE (6) provide automated comparison of protein structures, together with a numerical score of each comparison, which allows for automated generation of lists of structural neighbors of each protein. There are dozens of other protein structure comparison programs, but their review is outside the scope of this publication.
Manually curated classification resources require significant effort to maintain and typically lag PDB by several months. Thus, users interested in structural classifications of newly deposited structures are often left without any help from resources such as SCOP or CATH. The second group is based on automated computer programs, thus these resources are easier to maintain and could be used even for new, user-supplied structures. However, structural comparisons are time consuming and if used in an interactive mode, obtaining results could take up to several hours. Therefore, in the three most popular protein comparison programs (DALI, VAST and CE), pre-calculated lists of structural neighbors were used to create databases where lists of protein structures similar to the query structure can be easily retrieved by a user from a relational database.
The Flexible Structural Neighborhood (FSN) database we present here falls squarely in the second category. The lists of structural neighbors are created from an all-by-all comparison of known protein structures using a specific protein structure comparison program, the FATCAT algorithm developed in our group (7,8). However, the similarity between the FSN and structural neighbors defined by DALI, VAST or CE is only superficial, as FSN is based on a protein structure comparison approach fundamentally different from that used in all the existing protein structure comparison databases. All protein structure comparison algorithms used previously to create structure classification databases are based on rigid body alignments, i.e. they try to answer a question about the largest common element between two (or more) protein structures. In contrast, FATCAT and other related approaches such as FlexProt (9) ask a different question: ‘What is the simplest reorganization of one of the proteins that would make one protein more similar to the other?’ This way, proteins that change their conformation upon ligand binding, regulatory proteins that change their structure upon activation (‘protein switches’) or distantly related proteins that underwent significant structural changes during evolution can be directly compared. The results of the comparison include not only the alignment and the score, but also positions of hinge points where the structure must be bent and rotated to make it more similar to the protein it is compared with.
As compared with the rigid body alignments, flexible alignments often allow us to recognize similarities that were previously undetected, and in the cases when the structural similarity was known, they usually extend the length of the alignment to include structure fragments that seemed divergent. At the same time, because of the high penalty imposed for introducing flexibility, alignments between proteins that do not exhibit structural flexibility are left unchanged (7). FATCAT algorithm is available since 2004 as a public web server at http://fatcat.burnham.org for both pairwise structural alignments and structure database searches (8). FATCAT shares with its peers a common problem—it is relatively slow and it takes hours to perform a single structure against whole database comparison. Therefore we developed Flexible Structure Neighborhood, a database of structural comparisons that allow a user to quickly retrieve a previously calculated list of similar structures using a PDB coordinate as a query.
DATABASE IMPLEMENTATION
Results of an all-by-all comparison between protein domains as identified by SCOP and PDB representative set at 90% (see below) were performed by FATCAT (7) and the results were stored in a relational database, implemented using a MySQL package on a Linux server.
To take advantage of the existing protein classifications, protein chains are divided into domains following the SCOP classification. However, as SCOP typically lags several months behind PDB in processing new protein structures, proteins deposited to PDB that are still not processed and classified by SCOP require special protocol. In short, unclassified proteins are divided into chains and compared with SCOP domains and with each other. Therefore, two separate databases are available for searching—database of neighbors in proteins already processed by SCOP, and database of neighbors among more recent PDB depositions. Throughout the FSN website the SCOP identified domains are referred to as d1su4a2 (second domain in the chain A of PDB entry 1su4), while newly deposited PDB entries are referred to by their PDB four letter code + chain designation (e.g. 1su4a). At this point the FSN database is updated manually and efforts are under way to completely automate the update to keep pace with weekly PDB updates.
In April 2005, there were 30 527 protein structures deposited in PDB, among which 23 983 were annotated by SCOP, while 6544 were not. This translated into 61 775 SCOP annotated protein domains and 12 126 chains in new PDB entries. The 90% sequence similarity threshold lowered this numbers to 10 167 domains and 2393 new chains, which still resulted over 78 million pairwise structural comparisons. Detailed information for all of about 6 million (6 010, 391) similarities with P-value ≤ 0.1 are stored in the database, but by default, only those with P-value ≤ 0.01 are displayed for the user. The threshold P-value can be modified on the input form. Flexibility was found in ∼70% of all significantly similar [P-value < 0.05 (10)] comparisons.
USING FSN
Using a PDB code as a query, a user can retrieve a pre-computed list of structural neighbors by using the main FSN searching webpage at http://fatcat.burnham.org/fatcat/struct_neibor (Figure 1a) or by following the FATCAT link in the structural neighbor page at the PDB site at http://www.rcsb.org (Figure 1b). For a protein query that is not included in the 90% representative set, the server will refer the user to the structural neighbors of its closest homolog. In the FSN searching page, users can choose one of the non-redundant databases to be searched against, while the PDB initiated queries assume the default choice of the sum of SCOP domains and new PDB entries. Also the FSN webpage provides users an option to search against the whole PDB database instead of the 90% non-redundant databases to detect conformational changes among same/homologous proteins.
Figure 1.
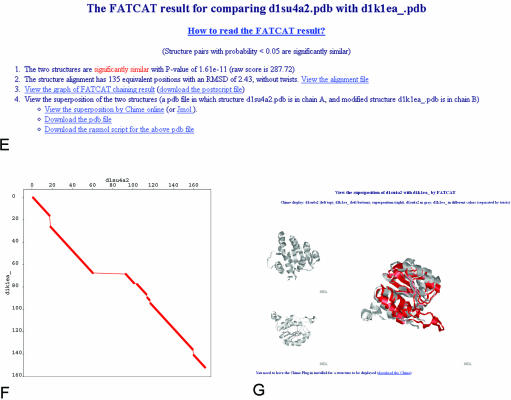
Workflow at the FSN database website and illustrations of the main result pages. An example of a calcium ATPase (PDB entry 1su4) is used throughout the figure. The query could be initiated directly from the FSN main interface (A) or by following the FATCAT link in PDB structural neighborhood page (B). Both ways lead to the protein summary page, where domain structure of the query protein is summarized and links to structural neighbors for each domain are provided (C). Clicking on each of the domains leads to a FSN page (D), where a list of similar structures is rank-ordered by the statistical significance of the structural similarity. This page provides links to an overall summary of hinge point positions (inset in D) and to FATCAT result pages (E) of each individual alignment. A FATCAT result page provides details of an alignment, and links to the graphical representation of the alignment (F), visualization page of the resulting superposition of input structures (G) and many others (not shown).
Once a query is submitted, the server returns a summary page (Figure 1c) of information about the domain structure of the query protein (if query protein was processed by SCOP) and informs the user about substituting a close homolog in the search (if applicable). Each of the domains and/or chains in the query protein provides clickable links to its FSN page (Figure 1d).
The FSN page, based on the FATCAT database search page (8), provides a list of structurally similar proteins sorted by increasing P-values, as well as links to various statistics about this group of proteins. In particular, positions and distributions of twists (hinges) along the sequence of the query protein are shown as a graph in GIF (inset in Figure 1d) and PostScript format, as well as a plain text file. A table lists structurally similar proteins with each individual protein in a single row, providing detailed information about each comparison (SCOP classification of the hit, RMSD, length of the alignment, number of twists, etc.) and links to PDB and detailed FATCAT result pages. The FATCAT result page (Figure 1e) provides basic statistics of the comparison (RMSD, P-value) and links to alignment in different formats (Figure 1f shows a 2D representation of the alignment) and a visualization page for displaying the superposition of the query and target structures, either by CHIME plug-in (MDL equipped browsers required, http://www.mdl.com/products/framework/chime) or by Jmol (http://jmol.sourceforge.net/).
FUTURE PLANS
As mentioned earlier, at this point, FSN is updated manually. We are currently developing protocols for completely automated updates that could be performed weekly with every PDB update.
Acknowledgments
We gratefully acknowledge help from our colleagues from the Burnham Bioinformatics group and from PDB for being first beta testers of the FSN. This research was supported by the NSF grant DBI 0349600. Funding to pay the Open Access publication charges for this article was provided by NSF grant DBI 0349600.
Conflict of interest statement. None declared.
REFERENCES
- 1.Vitkup D., Melamud E., Moult J., Sander C. Completeness in structural genomics. Nat. Struct. Biol. 2001;8:559–566. doi: 10.1038/88640. [DOI] [PubMed] [Google Scholar]
- 2.Lesley S.A., Kuhn P., Godzik A., Deacon A.M., Mathews I., Kreusch A., Spraggon G., Klock H.E., McMullan D., Shin T., et al. Structural genomics of the Thermotoga maritima proteome implemented in a high-throughput structure determination pipeline. Proc. Natl Acad. Sci. USA. 2002;99:11664–11669. doi: 10.1073/pnas.142413399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holm L., Sander C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 5.Gibrat J.F., Madej T., Bryant S.H. Surprising similarities in structure comparison. Curr. Opin. Struct. Biol. 1996;6:377–385. doi: 10.1016/s0959-440x(96)80058-3. [DOI] [PubMed] [Google Scholar]
- 6.Shindyalov I.N., Bourne P.E. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Proteins Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- 7.Ye Y., Godzik A. Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics. 2003;19(Suppl 2):II246–II255. doi: 10.1093/bioinformatics/btg1086. [DOI] [PubMed] [Google Scholar]
- 8.Ye Y., Godzik A. FATCAT: a web server for flexible structure comparison and structure analog searching. Nucleic Acids Res. 2004;32:W582–W585. doi: 10.1093/nar/gkh430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shatsky M., Nussinov R., Wolfson H.J. Flexible protein alignment and hinge detection. Proteins. 2002;48:242–256. doi: 10.1002/prot.10100. [DOI] [PubMed] [Google Scholar]
- 10.Ye Y., Godzik A. Database searching by flexible protein structure alignment. Protein Sci. 2004;13:1841–1850. doi: 10.1110/ps.03602304. [DOI] [PMC free article] [PubMed] [Google Scholar]