Abstract
Pathguide: the Pathway Resource List (http://pathguide.org) is a meta-database that provides an overview of more than 190 web-accessible biological pathway and network databases. These include databases on metabolic pathways, signaling pathways, transcription factor targets, gene regulatory networks, genetic interactions, protein–compound interactions, and protein–protein interactions. The listed databases are maintained by diverse groups in different locations and the information in them is derived either from the scientific literature or from systematic experiments. Pathguide is useful as a starting point for biological pathway analysis and for content aggregation in integrated biological information systems.
MOTIVATION
Databases and methods for computational analysis and data mining are an increasingly integral part of biological research because they provide easy access to a wealth of biological information, which biologists require to support analyses and effectively answer research questions. While individual databases cover important information in certain areas of biological knowledge, integrated and reasonably comprehensive use of biological pathway and network datasets is severely hampered by the large number and fragmentation of available databases (1).
CREATING THE PATHWAY RESOURCE LIST
As a step toward effective navigation and selective pathway data integration, we have collected information on over 190 published online cellular pathway and network databases in Pathguide. By providing this community resource we aim to promote the development of an integrated view of the cell (the ‘cell map’) (1). While much cell map data are only found in the literature, the databases in the list already represent a significant amount of relatively well-organized pathway data at varying levels of accessibility that can eventually be integrated and comprehensively accessed.
TYPES OF DATA AND DATABASE CATEGORIES
Databases in the list are grouped into eight major categories based on the type of data made available, the data format and the biological focus (Table 1). The categories are approximate and a database can be in multiple categories if it contains multiple data types. Protein–protein interaction databases mainly store pairwise interactions or complexes between proteins and sometimes other molecular interaction types. Metabolic pathway databases generally store a series of biochemical reactions in pathways involved in metabolite conversions. Signaling pathway databases generally collect sets of molecular interactions and chemical modifications (such as post-translational protein modifications) as regulatory pathways. Gene regulation network databases capture transcription factors and the genes they regulate. Genetic pathway databases are composed of genetic interactions, such as epistasis and synthetic lethality, which occur when two mutations have a combined phenotypic effect that is not simply the sum of the effects caused by either mutation alone. Pathway diagram databases generally store hyperlinked pathway images; while it is difficult to extract computable information from these images, they are very useful for biologists as educational and quick reference tools. Essentially all databases listed focus on interactions or pathways/networks. However, we also include a number of protein-sequence databases that store pathway information as secondary information, e.g. the REBASE database of restriction enzymes contains information about catalytic events involving DNA.
Table 1.
Pathguide statistics
| Category | Number of databases |
|---|---|
| Protein–protein interactions | 79 |
| Metabolic pathways | 43 |
| Signaling pathways | 41 |
| Pathway diagrams | 22 |
| Transcription factors/gene regulatory networks | 20 |
| Protein–compound interactions | 14 |
| Genetic interaction networks | 5 |
| Protein sequence focused | 12 |
| Other | 11 |
| Unique total | 196 |
The number of pathway databases in each Path-guide database category is listed. Uncategorized databases are listed in “Other”. Databases can be in more than one category, so the unique total number of databases is shown.
COVERAGE
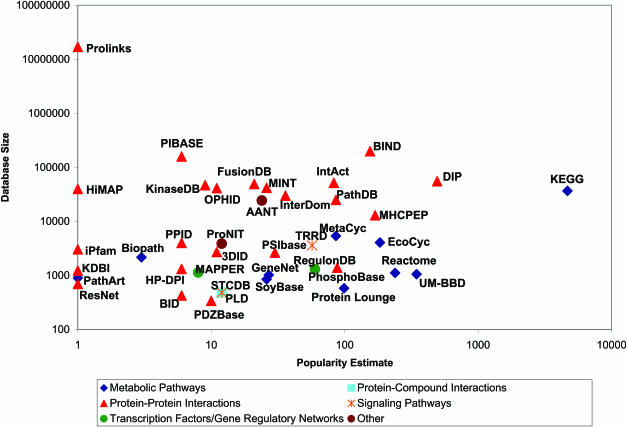
As one might expect, the categories of information captured in databases, so far, are biased by community biological interests and do not evenly cover the space of available pathway and interaction data (Figure 1). For instance, there are many protein–protein and protein–compound databases, plausibly because of technical developments in proteomics and interest in drug discovery, but there appear to be only two protein–RNA databases and none for RNA–compound interactions, although these categories are clearly of biological interest. Interactions and pathways define biological function at the molecular level. Therefore, pathway databases must grow to support the evolution of biological knowledge. There is still significant room for pathway database growth in underrepresented categories and areas of new biological discoveries, such as microRNA targets.
Figure 1.
The 40 largest databases in Pathguide. Pathway databases are diverse, both in volume of data (Database size, vertical) and in the attention they appear to generate (Popularity Estimate, horizontal). Database size is the sum of interaction and pathway records (where available; some database statistics may be incomplete). Popularity Estimate is taken to be the number of web pages in Google that mention the database homepage URL. Five main database categories (see symbol legend) out of eight are represented in the top 40. Databases in multiple categories are only represented in one (arbitrarily chosen) category. See the Pathguide website for expanded database names.
META-DATA AND LINKS
Database names in Pathguide are linked to the database homepage and clicking on ‘more’ next to each database leads to a structured description of the database listing short name, full name, homepage Uniform Resource Locator (URL), last observed date, text description, sample data URL, availability (e.g. free to all users, license purchase required), PubMed links, Pathguide category, types of tools available, database statistics, organisms covered and a popularity measure. The popularity measure used is the number of web pages, as indexed by the Google Internet search engine, that mention a given pathway database homepage URL. A user can rank all databases by this measure by clicking ‘Order list by web popularity’ at the top of the Pathguide homepage. The measure is rough because not all websites mentioning a pathway database are relevant. Thus the exact ranking is likely not sound, but it is useful as an overview of the most popular set of databases in each category.
STANDARDS
Many computational pathway analysis methods gain power given a larger biological network. A single new link can lead to a significant new biological discovery. Collecting as many high-quality links as possible for network analysis requires well organized and convenient pathway database access. Databases that are freely accessible (open-access) and support standard languages facilitate their distribution and use. To encourage this, databases are highlighted if they are ‘Free to all users’ and can be downloaded in a standard format, such as the Proteomics Standards Initiative Molecular Interaction (2) and BioPAX (www.biopax.org) pathway data exchange standards and the Systems Biology Markup Language (SBML) (3) and CellML (4) pathway simulation model exchange standards. Biologists looking for data that can be analyzed in tools supporting these standards and computational biologists wishing to integrate available pathway data for global analyses may find this helpful.
IMPLEMENTATION
Pathguide is curated by the authors and regularly updated. Generation of web pages (HTML) is implemented using the scripting language PHP with a relational database (MySQL) backend, which stores all information in a structured manner. The Google ranking for a particular database is updated using a Perl script to query the Google SOAP Application Programming Interface for pages anywhere on the Internet that link to the database homepage address (URL), not counting links from the database site itself. We have designed Pathguide to facilitate research on biological pathways and to be complementary to existing database link resources, such as Michael Galperin's Molecular Biology Database Collection (5) and the UBiC Bioinformatics Links Directory (http://bioinformatics.ubc.ca/resources/links_directory).
FUTURE DEVELOPMENT
Future plans include adding search features (e.g. show me all databases with human protein–protein interaction information), automatically updated database content statistics (where available), graphical Pathguide content summaries, improved links to PubMed, differentiating primary (original content) and secondary (derived or predicted content) databases, automatic URL validation, homepage uptime statistics, an RSS feed to track updates and to include a section on pathway tools, a commonly requested feature. Comments, questions and information about missing pathway resources are most welcome.
Acknowledgments
Thanks to Robert Hoffmann for input on the Google ranking, Emek Demir for manuscript comments, users who have submitted new pathway resources and the editors of Nucleic Acids Research for supporting open-access publications. Funding to pay the Open Access publication charges for this article was provided by Memorial Sloan-Kettering Cancer Center.
Conflict of interest statement. None declared.
REFERENCES
- 1.Cary M.P., Bader G.D., Sander C. Pathway information for systems biology. FEBS Lett. 2005;579:1815–1820. doi: 10.1016/j.febslet.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Hermjakob H., Montecchi-Palazzi L., Bader G., Wojcik J., Salwinski L., Ceol A., Moore S., Orchard S., Sarkans U., von Mering C., et al. The HUPO PSI's molecular interaction format—a community standard for the representation of protein interaction data. Nat. Biotechnol. 2004;22:177–183. doi: 10.1038/nbt926. [DOI] [PubMed] [Google Scholar]
- 3.Hucka M., Finney A., Sauro H.M., Bolouri H., Doyle J.C., Kitano H., Arkin A.P., Bornstein B.J., Bray D., Cornish-Bowden A., et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 4.Lloyd C.M., Halstead M.D., Nielsen P.F. CellML: its future, present and past. Prog. Biophys. Mol. Biol. 2004;85:433–450. doi: 10.1016/j.pbiomolbio.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Galperin M.Y. The Molecular Biology Database Collection: 2005 update. Nucleic Acids Res. 2005;33:D5–D24. doi: 10.1093/nar/gki139. [DOI] [PMC free article] [PubMed] [Google Scholar]