Abstract
We have determined the nucleotide sequence of a flagellin gene locus from the haloalkaliphilic archaeon Natrialba magadii, identified the gene products among proteins forming flagella, and demonstrated cotranscription of the genes. Based on the sequence analysis we suggest that different regions of the genes might have distinct evolutionary histories including possible genetic exchange with bacterial flagellin genes.
Representatives of the two kingdoms Archaea and Bacteria have analogous motility systems composed of long thin appendages (flagella), which rotate to produce thrust (1). The bacterial flagellum filament is mainly composed of a single type of protein. In contrast, the archaeal filament is formed from multiple types of flagellin, encoded by a few genes. Surprisingly, the archaeal flagellins (and related proteins) have homology with bacterial proteins from the type IV pilin family (4, 8) but not with bacterial flagellins (15). This fact, together with the presence of leader peptides (3, 21) and posttranslational modifications (15) in archaeal flagellins, suggests that archaeal flagella form in a manner similar to that in which bacterial pili form but not similar to that in which bacterial distal tip assembly occurs (8). The recent finding of a close relationship between the chemotaxis systems in gram-positive bacteria and archaea implies that the entire bacterial system was acquired by archaea (9). Since the chemotaxis system is involved in motility, this, in turn, raises a question about the origins and evolution of archaeal flagellin genes.
We sequenced flagellin genes from poorly understood haloalkaliphilic archaeon Natrialba magadii, analyzed their expression, and demonstrated their mosaic structure. We suggest that horizontal transmissions might be involved in the evolution of the genes.
Cloning, sequencing, and analysis of N. magadii flagellin genes.
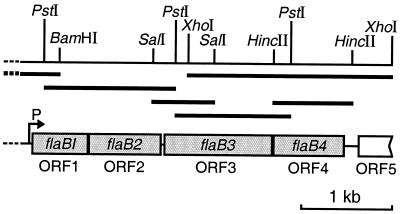
In the present study a 1.2-kbp DNA fragment containing Halobacterium halobium flgA1 and flgA2 genes (33) was used to search for flagellin genes in the genome of the haloalkaliphilic N. (formerly Natronobacterium) magadii. Using Southern blot hybridization (32) we identified 1.4- and 5.5-kbp fragments that gave positive signals with PstI-digested genomic DNA and 6.3- and 12.0-kbp fragments with BamHI-digested DNA. We cloned the 1.4- and 6.3-kbp fragments using plasmid-based libraries. The nucleotide sequences of the 1.4-kbp fragment and part of the 6.3-kbp fragment revealed the presence of two complete open reading frames (ORFs) and one partial ORF (Fig. 1). To sequence the distal part of the operon, we performed four rounds of inverse PCR using as templates SalI-, PstI-, HincII-, and XhoI-digested genomic DNA. The total 4,029-bp nucleotide sequence of the gene cluster contains four complete ORFs of 202, 260, 396, and 262 codons, located close to each other. In addition, we identified the 5′ end of the ORF (127 codons), 125 nucleotides (nt) away from the cluster comprising ORF1 to ORF4 (Fig. 1). ORF1, ORF2, and ORF4 are preceded by putative ribosome binding sites with the sequences 5′-GGGTG-3′, 5′-GGTGA-3′, and 5′-GTGA-3′ (25). The most favorable ribosome binding sites for ORF3 (5′-GGTGA-3′) and ORF5 (5′-GGAGT-3′) are located, however, far from initiation codons: 43 and 35 nt for ORF3 and ORF5, respectively. Thus, ribosomal subunits might bind trinucleotide sequences close to the translation initiation codon.
FIG. 1.
Organization of N. magadii flagellin operon. Restriction sites used for cloning and inverse PCR are indicated. Bent arrow, position of the transcription start point.
ORF1 to ORF4 were assigned to the flagellin gene family based on homology of the deduced amino acid sequences with archaeal flagellins (Fig. 2A) and were designated genes flaB1, flaB2, flaB3, and flaB4, respectively (Fig. 1). The amino acid sequence encoded by ORF5 has some similarity to putative flagellum-related protein FlaF from Methanococcus voltae and Methanococcus jannaschii (24% identity and 49% positives in 93 N-terminal amino acids) (4). Flagellins from N. magadii and other archaea, including archaea that inhabit very different niches, display high sequence similarity at the N termini (amino acids [aa] 13 to 60) (Fig. 2A). This indicates that the hydrophobic N termini in all flagellated archaea have an important role in flagellum core formation (30). The less well conserved nine N-terminal residues represent a signal peptide, which is most likely cleaved between glycine and glutamine in the RGQ sequence (Fig. 2A) (20). Although central regions of the N. magadii flagellins have limited similarity, the C-terminal 25 amino acids are very similar to each other (50% identity) but not to the corresponding flagellin regions from other archaeal species. FlaB3, the product of the flaB3 gene, contains an insertion in the middle region and, therefore, is significantly larger (395 aa) than other flagellins (mostly 195 to 260 aa). The only other example of a large flagellin (580 aa) was identified in Pyrococcus kodakaraensis (27). As an adaptation to living at high pH, the N. magadii flagellins contain a threefold-lower content of arginine and lysine residues (1.9 to 3.5%) than flagellins of halophile H. halobium (8.6 to 10.9%). Since the extracellular pH approaches the pK values of amide groups in Arg and Lys, it can affect the charge of these residues and may influence stability, folding, or solubility of the proteins (17).
FIG. 2.
Homology between N. magadii and archaeal flagellins. (A) Multiple sequence alignment between N. magadii and representative archaeal flagellins was performed using ClustalW (39). Identical amino acids along with conserved substitutions were shaded with GeneDoc software (K. B. Nicholas and H. B. Nicholas, Jr., unpublished data). Amino acids similar in all proteins are white on a black background, and similar residues represented in >50% of the sequences are shaded in gray. The origins of the sequences were as follows: B1Nm to B4Nm, N. magadii; B1Hh, H. halobium (15); B2Mj, M. jannaschii (6); B2Ph, Pyrococcus horikoshii (22); B1Arf, Archaeoglobus fulgidus (24); B1Aerp, Aeropyrum pernix (23). (B) Comparison of the central parts of N. magadii FlaB3 and E. coli DEC 2a flagellins performed using BLAST (2).
Identification of flaB gene transcript and mapping of its 5′ terminus.
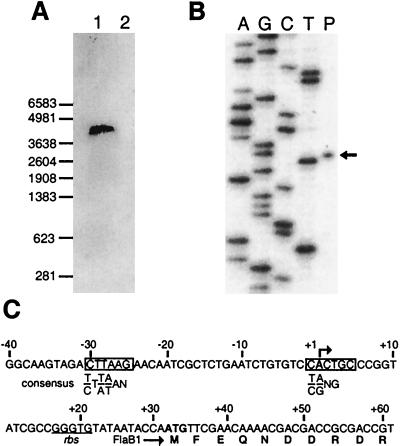
To identify the transcript(s) for the flagellin gene cluster, we performed Northern blot hybridization (32, 37) with the total RNA isolated from N. magadii cells and the random 32P-labeled probe synthesized with the HexaLabel DNA labeling kit (Fermentas AB, Vilnius, Lithuania). The probe carries the majority of the flaB3 gene and the conserved 5′ end of the flaB4 gene. Only one RNA transcript of about 4.5 kb in size was detected, indicating cotranscription of all flagellin genes and possibly ORF5 as described earlier for M. voltae (4, 21) (Fig. 3A). Extension of primer 5′-CCGTATTGATCAGCACGCCCGCGGCAATCG-3′ by reverse transcriptase mapped the transcription start point at position 102 of the sequence (Fig. 3B), 31 nt upstream of the translation start codon of the first flagellin gene, flaB1 (Fig. 3C). Sequence 5′-CTTAAG-3′, identified 23 nt upstream of the transcription initiation site, resembles TATA box consensus sequences of halophiles (7, 28), methanogenes, and Crenarchaeota (35) (Fig. 3C). On the other hand, the second transcription signal, the initiator element −1 CACTGC +5 is distinct from the +1 GGG +3 sequence from methanogenes but is similar to the −1 YRNG +3 and −1 TRSSSC +5 consensus sequences of halophiles and Crenarchaeota (35). This emphasizes the absence of a strict sequence requirement in this box for promoter activity. It is probably a general rule that promoters of archaeal flagellin genes do not significantly differ from transcription signals found in other archaeal genes (16, 21). The transcription of N. magadii flagellin genes appears to be similar to the “single-RNA” transcription start mechanism in H. halobium (15), which contrasts with the initiation of a few transcripts of different lengths from the same transcription start point found in P. kodakaraensis (27) and M. voltae (21). Although H. halobium flagellins have the strongest homology to N. magadii proteins, we must point out that the flagellin gene organization is different in H. halobium, where two transcriptional units comprise two and three genes (15).
FIG. 3.
Transcription of the flagellin gene cluster in N. magadii. (A) Northern blot analysis of flagellin mRNA. The positions of RNA markers (Promega Corporation, Madison, Wis.) are indicated on the left. (B) Mapping of the 5′ terminus of the flaB transcript by primer extension. The primer extension product (arrow [lane P]) was separated by electrophoresis in a denaturing 6% polyacrylamide gel along with the sequence ladders generated with the same oligonucleotide (lanes A, G, C, and T). (C) Nucleotide sequence of the flaB promoter region. Bent arrow, transcription start point. The putative transcription signals are boxed, and halobacterial consensus sequences are shown under them. The putative ribosome binding site is underlined.
Search for flaB1 to flaB4 gene products among proteins isolated from flagella.
N. magadii flagella consist of four different proteins, FM1 to FM4, with molecular masses of 105, 60, 59, and 45 kDa, respectively, as estimated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (11). In this study we identified four transcribed flagellin genes, flaB1 to flaB4, which encode proteins FlaB1 to FlaB4, with molecular masses of 21.4, 26.5, 41.7, and 27.0 kDa, respectively. The poor agreement between the molecular masses estimated by SDS-gel electrophoresis and those expected from the gene sequences is common for flagellins due to (i) the high content of acidic amino acids, which can decrease SDS binding and result in slower migration of the proteins in gels (26), and (ii) posttranslation modifications of flagellins (36). To identify the genes encoding flagellum proteins FM1 and FM4, we performed N-terminal sequencing of tryptic fragments derived from these flagellins. Sequences AEVVFELDGXPGSYD and VPSTFIEDE from FM4 match the amino acid sequences of the FlaB1 protein at positions 144 to 158 and 188 to 196, respectively. Thus, flaB1 encodes flagellin FM4. In turn, FM1-derived sequence AIELP coincides with the unique amino acid sequence of FlaB3 (positions 117 to 121); therefore, FM1 is encoded by the flaB3 gene. Since our attempts to obtain proteins FM2 and FM3 without cross-contamination failed due to the similar molecular masses of these flagellins, we performed tryptic digestion and sequencing of their mixture. Sequence IDTTIAVGTVST, determined in the course of this analysis, is identical to the unique sequence of the FlaB2 protein (positions 61 to 72), encoded by flaB2. We strongly suggest that FlaB2 is the FM2 protein based on the observation that only FM2 and FM4 are glycosylated (11). A search for potential N-glycosylation sites (Asn-X-Thr/Ser) (4, 14, 16, 21) revealed the presence of these signals in FlaB1 (positions 81, 98, and 114), identified here as FM4, and FlaB2 (positions 105 and 158) proteins. Since the sequencing result indicates that FlaB2 is present in the FM2-FM3 mixture, only the product of the flaB2 gene, the FlaB2 protein, can be considered to be glycosylated protein FM2. It was shown that FM2 and FM3 give similar proteolysis patterns (11); therefore, they should be encoded by the same gene or similar genes. FlaB4 has a high degree of homology to FlaB2; thus, FM3 might be the flaB4 gene product. The slightly faster SDS-gel migration of this flagellin compared to that of FM2 (11) may be attributed to the absence of glycosylation. In theory, the FM3 protein can also contain the nonglycosylated product of the flaB2 gene. These results and transcription analyses suggest that all four flagellin genes of N. magadii are expressed and that at least three of them participate in the assembly of flagella. It is unclear why archaeal flagella are composed of different flagellins, whereas bacterial flagella contain only a single flagellin. The work from our laboratory has recently shown that in H. halobium only flagellins FlaA1 and FlaA2 are needed for the formation of the flagellar spiral filament. The function of the other three flagellins is auxiliary (37). Similar conclusions were also made for M. voltae flagellins (19).
Possible mode of N. magadii flagellin gene evolution.
The G+C content of the sequenced DNA locus is 54%. This is less than the average G+C content in randomly sequenced DNA fragments from N. magadii (63%) (data not presented), in known genes encoding proteins from other haloalkaliphilic archaea (63 and 61%), and in known genes from halophiles (64%, excluding gas vesicle genes [18]). Such a value is due to a low G+C frequency (49 to 60%) in the third codon position (GC3), which is considerably less than that found in halophilic and haloalkaliphilic species (77 to 85%) and even in the sequence of the downstream ORF5 (74%). To prove unusual codon bias in N. magadii flagellin genes, we estimate the differences in codon usage in distinct organisms by a simple pairwise comparison using the following espression:
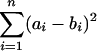 |
where ai and bi are the frequencies (in percent) of the same codon within a codon family of two organisms under comparison and n is the number of codon pairs analyzed. C, H, K, R, W, Y, and P codons are poorly represented or not represented at all in the N. magadii flagellins and were therefore excluded from the analyses. As expected, our calculations revealed the least difference in codon usage, with a value of 2,350 to 4,050, among the genes from halophiles and haloalkalophiles H. halobium, Natrialba asiatica, and Natromonas (Natronobacterium) pharaonis. The maximum differences found, 33,050 and 27,100, were between the most divergent pairs of organisms analyzed, Escherichia coli and H. halobium, and E. coli and N. asiatica, respectively. Pairs with N. magadii flagellins demonstrate intermediate values (11,335 to 20,056), but, strikingly, the smallest value found (8,374) was that for the average E. coli codon usage. Thus, the codon usage of N. magadii flagellin genes is rather different from that for the genes of its relatives and surprisingly resembles that for typical mesophilic bacterium E. coli. Two other observations, low G+C average and GC3 content, also raise questions as to the origins of the N. magadii flagellin genes and strongly support an idea that the locus has a mixed, probably unique, evolutionary history. Remarkably, the conserved 5′ regions of each N. magadii flagellin gene (encoding aa 13 to 60) are enriched by G and C (58 to 60%) compared to the rest of the sequence (47 to 57%) and demonstrate a similar distribution of G+C in codon positions. The values for the remaining parts, especially the GC3 content, vary significantly (data not shown). The least conserved central region (aa 71 to 280) of the largest flagellin FlaB3 shows the lowest GC3 percentage, 15 to 20% less than those for other fragments. Since this region does not show any similarity to the other N. magadii flagellin genes and known archaeal genes, it has most likely been transferred to the locus, probably from another organism. Calculation of the codon usage differences in distinct parts of the flagellin genes, although less relevant because of poorer statistics, also emphasizes the strong divergence of the 71-to-280 region. Indeed, the sum of the differences between the 71-to-280 region and other flagellin sequences, namely, the N-terminal (aa 11 to 60) and the middle-to-C-terminal parts (aa 281 to 395 for FlaB3 and from aa 60 to the end for the other proteins) is 124,334. This is higher than the corresponding values, which range from 69,382 to 104,231, calculated for the other regions. Such diverse codon usage in the central region of the flaB3 gene may result in lower expression levels of the gene and may explain the relatively low content of the FlaB3 protein in the flagella (11). Remarkably, BLAST (2) and FASTA (29) searches revealed homology between this region (extended to aa 57 and 353) and central variable parts of four E. coli flagellins of the EPEC 1 group (31) (Fig. 2B). These homologies (about 25% identity and 41% similarity) are plausible, since their scores and E values are comparable to the values for some archaeal flagellins. These include the flagellins from the close relative H. halobium, found in the same search, even if the comparison was between the genes encoding conserved N termini. These results suggest that different parts of the N. magadii flagellin genes might have a different evolutionary histories. Horizontal transfer may be one of the causes of such diversity. A mosaic gene structure resulting from horizontal transmission of gene parts for some extracellular systems has been already described: in the pilin gene cluster from gram-negative bacteria (5, 12) and in bacterial flagellin genes from E. coli (31) and Salmonella (34). Growing evidence for horizontal gene transfer between Archaea and Bacteria (13), including pathogenic bacteria (10), provides support for our model of flagellin gene exchange between E. coli and N. magadii. Although the mechanism of such transfers and the possible involvement of mediators remain to be investigated, our hypothesis is indirectly supported by the fact that one of the EPEC 1 group representatives, E. coli strain DEC 2a, and N. magadii have been found in geographical areas close to each other, e.g., in Congo and Kenya, respectively (31, 38). Moreover, the borders between constant and variable parts of the E. coli flagellin coincide with the borders found in the archaeal flagellin region. Therefore, this region of the E. coli flagellin genes may have been moved to the archaeal genome and may have been integrated into the analogous locations between the most homologous regions of the E. coli flagellin gene and the N. magadii flaB3 gene (positions 55 to 70 and 280 to 353). It is necessary to mention that the sequence similarity between archaeal and bacterial flagellins was not described earlier, although it was intensively searched (3, 8, 15).
Nucleotide sequence accession number.
The nucleotide sequence determined in this study was assigned GenBank accession no. AJ277988.
Acknowledgments
We are indebted to N. Matvienko, T. Ivashina, and J. Coyle for helpful discussions and carefully reading the manuscript.
This work was supported by a grant from the Russian Foundation for Basic Research (no. 98-04-48318).
REFERENCES
- 1.Alam, M., and D. Oesterhelt. 1984. Morphology, function and isolation of halobacterial flagella. J. Mol. Biol. 176:459–475. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- 3.Bayley, D. P., and K. F. Jarrell. 1998. Further evidence to suggest that archaeal flagella are related to bacterial type IV pili. J. Mol. Evol. 46:370–373. [PubMed] [Google Scholar]
- 4.Bayley, D. P., V. Florian, A. Klein, and K. F. Jarrell. 1998. Flagellin genes of Methanococcus vannielii: amplification by the polymerase chain reaction, demonstration of signal peptides and identification of major components of the flagellar filament. Mol. Gen. Genet. 258:639–645. [DOI] [PubMed] [Google Scholar]
- 5.Boyd, E. F., and D. L. Hartl. 1999. Analysis of the type 1 pilin gene cluster fim in Salmonella: its distinct evolutionary histories in the 5′ and 3′ regions. J. Bacteriol. 181:1301–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bult, C., O. White, G. J. Olsen, L. Zhou, R. D. Fleischmann, et al. 1996. Complete genome sequence of the methanogenic archaeon Methanococcus jannaschii. Science 273:1058–1072. [DOI] [PubMed] [Google Scholar]
- 7.Danner, S., and J. Soppa. 1996. Characterization of the distal promoter element of halobacteria in vivo using saturation mutagenesis and selection. Mol. Microbiol. 19:1265–1276. [DOI] [PubMed] [Google Scholar]
- 8.Faguy, D. M., K. F. Jarrell, J. Kuzio, and M. L. Kalmokoff. 1994. Molecular analysis of archaeal flagellins: similarity to the type IV pilin-transport superfamily widespread in bacteria. Can. J. Microbiol. 40:67–71. [DOI] [PubMed] [Google Scholar]
- 9.Faguy, D. M., and K. F. Jarrell. 1999. A twisted tale: the origin and evolution of motility and chemotaxis in prokaryotes. Microbiology 145:279–281. [DOI] [PubMed] [Google Scholar]
- 10.Faguy, D. M., and W. F. Doolittle. 2000. Horizontal transfer of catalase-peroxidase genes between Archaea and pathogenic bacteria. Trends Genet. 16:196–197. [DOI] [PubMed] [Google Scholar]
- 11.Fedorov, O. V., M. G. Pyatibratov, A. S. Kostyukova, N. K. Osina, and V. Y. Tarasov. 1994. Protofilament as a structural element of flagella of haloalkalophilic archaebacteria. Can. J. Microbiol. 40:45–53. [Google Scholar]
- 12.Fussenegger, M., T. Rudel, R. Barten, R. Ryll, and T. F. Meyer. 1997. Transformation competence and type-4 pilus biogenesis in Neisseria gonorrhoeae. Gene 192:125–134. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Vallve, S., A. Romeu, and J. Palau. 2000. Horizontal gene transfer in Bacterial and archaeal complete genomes. Genome Res. 10:1719–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gavel, Y., and G. von Heijne. 1990. Sequence differences between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites: implications for protein engineering. Protein Eng. 3:433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerl, L., and M. Sumper. 1988. Halobacterial flagellins are encoded by a multigene family. Characterization of the flagellin genes. J. Biol. Chem. 263:13246–13251. [PubMed] [Google Scholar]
- 16.Gerl, L., R. Deutzmann, and M. Sumper. 1989. Halobacterial flagellins are encoded by a multigene family. Identification of all five gene products. FEBS Lett. 244:137–140. [DOI] [PubMed] [Google Scholar]
- 17.Guffanti, A. A., and H. C. Eisenstein. 1983. Purification and characterisation of flagella from the alkalophile Bacillus firmus RAB. J. Gen. Microbiol. 129:3239–3242. [Google Scholar]
- 18.Horne, M., C. Englert, and F. Pfeifer. 1988. Two genes encoding gas vacuole proteins in Halobacterium halobium. Mol. Gen. Genet. 213:459–464. [DOI] [PubMed] [Google Scholar]
- 19.Jarrell, K., D. P. Bayley, V. Florian, and A. Klein. 1996. Isolation and characterization of insertional mutants in flagellin genes in the archaeon Methanococcus voltae. Mol. Microbiol. 20:657–666. [DOI] [PubMed] [Google Scholar]
- 20.Jarrell, K. F., J. D. Correia, and N. A. Thomas. 1999. Is the processing and translocation system used by flagellins also used by membrane-anchored secretory proteins in archaea? Mol. Microbiol. 34:395–398. [DOI] [PubMed] [Google Scholar]
- 21.Kalmokoff, M. L., and K. F. Jarrell. 1991. Cloning and sequencing of a multigene family encoding the flagellins of Methanococcus voltae. J. Bacteriol. 173:7113–7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawarabayasi, Y., M. Sawada, H. Horikawa, Y. Haikawa, Y. Hino, et al. 1998. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 5:55–76. [DOI] [PubMed] [Google Scholar]
- 23.Kawarabayasi, Y., Y. Hino, H. Horikawa, S. Yamazaki, and Y. Haikawa. 1999. Complete genome sequence of an aerobic hyper-thermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res. 6:83–101. [DOI] [PubMed] [Google Scholar]
- 24.Klenk, H. P., R. A. Clayton, J. Tomb, et al. 1997. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 390:364–370. [DOI] [PubMed] [Google Scholar]
- 25.Lodwick, D., H. M. Ros, J. A. Walker, J. W. Almond, and W. D. Grant. 1991. Nucleotide sequence of the 16S ribosomal RNA gene from the haloalkaliphilic archaeon (archaebacterium) Natronobacterium magadii, and the phylogeny of halobacteria. Syst. Appl. Microbiol. 14:352–357. [Google Scholar]
- 26.Matagne, A., B. Joris, and J. M. Frere. 1991. Anomalous behavior of a protein during SDS/PAGE corrected by chemical modification of carboxylic groups. Biochem. J. 280:553–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagahisa, K., E. Satoshi, S. Fujiwara, T. Imanaka, and M. Takagi. 1999. Sequence and transcriptional studies of five clustered flagellin genes from hyperthermophilic archaeon Pyrococcus kodakaraensis KOD1. FEMS Microbiol. Lett. 178:183–190. [DOI] [PubMed] [Google Scholar]
- 28.Palmer, J. R., and C. J. Daniels. 1995. In vivo definition of an archaeal promoter. J. Bacteriol. 177:1844–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pearson, W. R. 1990. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 183:63–98. [DOI] [PubMed] [Google Scholar]
- 30.Pyatibratov, M. G., A. S. Kostyukova, V. Y. Tarasov, and O. V. Fedorov. 1996. Some principles of formation of the haloalkaliphilic archaeal flagellar structure. Biochemistry (Moscow) 61:1056–1062. [Google Scholar]
- 31.Reid, S. D., R. K. Selander, and T. S. Whittam. 1999. Sequence diversity of flagellins (fliC) alleles in pathogenic Escherichia coli. J. Bacteriol. 181:153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Serganova, I. S., Y. Y. Polosina, A. S. Kostyukova, A. L. Metlina, M. G. Pyatibratov, and O. V. Fedorov. 1995. Flagella of halophilic archaea: biochemical and genetic analysis. Biochemistry (Moscow) 60:953–957. [PubMed] [Google Scholar]
- 34.Smith, N. H., P. Beltar, and R. Selander. 1990. Recombination of Salmonella phase 1 flagellin genes generates new serovars. J. Bacteriol. 172:2209–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soppa, J. 1999. Normalized nucleotide frequencies allow the definition of archaeal promoter elements for different archaeal groups and reveal base-specific TFB contacts upstream of the TATA box. Mol. Microbiol. 31:1589–1601. [DOI] [PubMed] [Google Scholar]
- 36.Southam, G., M. L. Kalmokoff, K. F. Jarrell, S. F. Koval, and T. J. Beveridge. 1990. Isolation, characterization, and cellular insertion of the flagella from two strains of the archaebacterium Methanospirillum hungatei. J. Bacteriol. 172:3221–3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tarasov, V. Y., M. G. Pyatibratov, S. Tang, M. Dyall-Smith, and O. V. Fedorov. 2000. Role of flagellins from A and B loci in flagella formation of Halobacterium salinarum. Mol. Microbiol. 35:69–78. [DOI] [PubMed] [Google Scholar]
- 38.Tindall, B. J., H. N. M. Ross, and W. D. Grant. 1984. Natronobacterium gen. nov. and Natronococcus gen. nov., two new genera of haloalkalophilic archaebacteria. Syst. Appl. Microbiol. 5:41–57. [Google Scholar]
- 39.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]