Abstract
DBTSS was first constructed in 2002 based on precise, experimentally determined 5′ end clones. Several major updates and additions have been made since the last report. First, the number of human clones has drastically increased, going from 190 964 to 1 359 000. Second, information about potential alternative promoters is presented because the number of 5′ end clones is now sufficient to determine several promoters for one gene. Namely, we defined putative promoter groups by clustering transcription start sites (TSSs) separated by <500 bases. A total of 8308 human genes and 4276 mouse genes were found to have putative multiple promoters. Third, DBTSS provides detailed sequence comparisons of user-specified TSSs. Finally, we have added TSS information for zebrafish, malaria and schyzon (a red algae model organism). DBTSS is accessible at http://dbtss.hgc.jp.
INTRODUCTION
Recently, a huge amount of comprehensive expression profile data obtained by various experiments, such as microarrays, has been made available. It is a challenging problem to uncover the regulatory networks among the expressed genes from these data. Information about promoters, which contain most of the binding sites of transcription factors, is indispensable for solving this question. To define promoter regions, precise information about transcription start sites (TSSs) is also required. Such data, however, are not easily obtained because the cDNA sequence data in repository sequence databases provide no guarantees regarding the 5′ end of the sequences and because the computational prediction of promoters and TSSs still remains problematic (1). To overcome these difficulties several databases (2), including DBTSS (DataBase of Transcription Start Sites) have been constructed. DBTSS contains TSS information of genes based on specific experiments (3,4). Clones constructed by full-length cDNA methods such as oligo-capping (5,6) or CAP-trapper (7,8) are mapped on to genome sequences to determine TSSs. Each TSS is determined based on the 5′ end of the corresponding clone. DBTSS was first constructed in 2002, and has been improved by several major and minor updates. The original version (version 1) contained only human data (3). Two years later, we reported the addition of mouse TSS information (9) in version 3 (4). Here we introduce the new updates and additions since version 3, the most important one being the addition of putative alternative promoter information.
NEW FEATURES
The current version of DBTSS, version 5, includes some notable improvements since the previous report, in addition to minor updates such as modifications of the interface and the result views.
One major improvement is that the amount of data for human TSSs has been significantly increased: in our report in 2002, we described 190 964 human clones which corresponded to 11 234 NCBI reference sequence cDNAs (RefSeq) (4). Because we added data from a new full-length cDNA project (10), DBTSS now contains 1 359 000 clones corresponding to 19 753 RefSeq cDNAs (Table 1). Since RefSeq cDNAs contain splicing variants as separate entries, we performed clustering of clones' information depending on their coordinate in the genome sequence; if their sequences overlapped, we regard them as the same locus. After clustering, our data correspond to 15 262 genes (Table 1). This is one of the largest collections of human 5′ end cDNA sequences.
Table 1.
Statistics of DBTSS
| No. of genes/no. of RefSeq | No. of promoters | No. of TSSs | No. of clones | |
|---|---|---|---|---|
| Human | 15 262/19 753 | 30 964 | 452 117 | 1 359 000 |
| Mouse | 14 162/14 746 | 19 023 | 149 876 | 364 487 |
| Zebrafish | 3061/3075 | 3382 | 15 198 | 32 263 |
| Malaria | 1527/NA | NA | 6908 | 10 236 |
| Schyzon | 3635/NA | NA | 14 029 | 22 923 |
To check the quality of our TSS data, we compared DBTSS with the Eukaryote Promoter Database (EPD) (2). In EPD Release 82, there are 1871 promoters collected from the literature. Among them, we could map 1767 promoter sequences to the human genome; 1639 of them mapping within 100 bases of the DBTSS TSSs, indicating that the data in DBTSS are consistent with the data obtained from ordinary methods.
In the next two sections, we will discuss two other major updates: alternative promoters (APs) and promoter comparison.
ALTERNATIVE PROMOTERS
Several genes are known to have multiple promoters which could be regulated in a different manner. These promoters, labeled as APs, could be useful to maximally exploit the relatively limited number of genes in the genome (11). However, no estimation of how many genes might have alternative promotes is available to date. Since DBTSS now has enough 5′ end clones from human and mouse, we performed this estimate. This is the most important addition in version 5. Although the details of our analysis will be reported elsewhere (12), the procedure is summarized below.
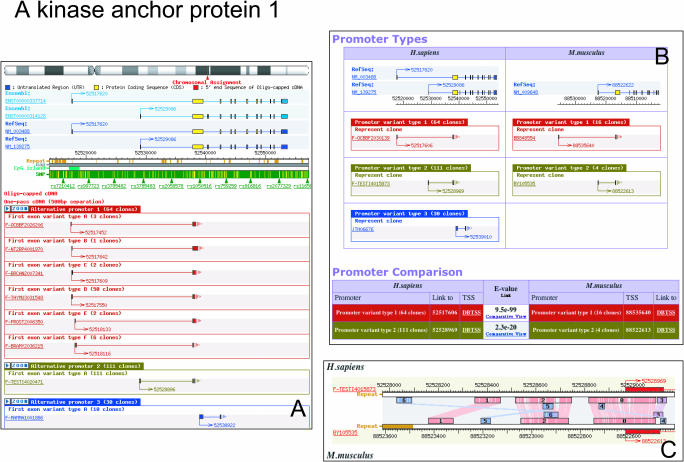
To determine APs, we first collected all the TSSs from the same locus. TSSs located inside a RefSeq gene exon, with the exception of the first one, were removed in order to avoid artifacts caused by truncated 5′ ends. We used several intervals to define AP clusters. The distribution of the number of putative alternative promoter containing genes shows a plateau before the interval size reaches 500 bp (12). We, therefore, clustered the clones using a 500 base interval, and defined each cluster as an promoter. We obtained 30 964 promoters, and 26 784 (86.5%) of them are within 500 bp. According to this procedure, 6954 human loci and 9886 mouse loci have only one promoter while 8308 human loci and 4276 mouse loci have two promoters or more. Figure 1A shows the three alternative promoters found in the gene encoding human A kinase anchor protein 1 (AKAP1). It is notable that DBTSS also provides comparative information between human and mouse promoters. Figure 1B shows an example of comparative promoter analysis between orthologous genes. Two promoters were identified for the mouse gene for AKAP1. From this view, the representative APs are also available for alignment. By clicking ‘Comparative View’ in ‘Promoter Comparison’ in Figure 1B, the LALIGN-based alignment view, shown in Figure 1C is obtained.
Figure 1.
An example of alternative promoter view. Here we show AKAP1 (NM_003488) as an example. (A) The putative promoter clusters are given using different colors; therefore, there are three putative promoters in human AKAP1. We observed several patterns of first exon in promoter type 1, so we clustered them and show them as ‘First exon variant type A–F’. (B) Comparative analysis between human and mouse alternative promoters. There are two putative promoters in mouse. The best match between two promoters is available in ‘Promoter Comparison’. (C) Clicking the ‘Comparative View’, the user can obtain the alignment between these promoters.
COMPARATIVE PROMOTER ANALYSIS
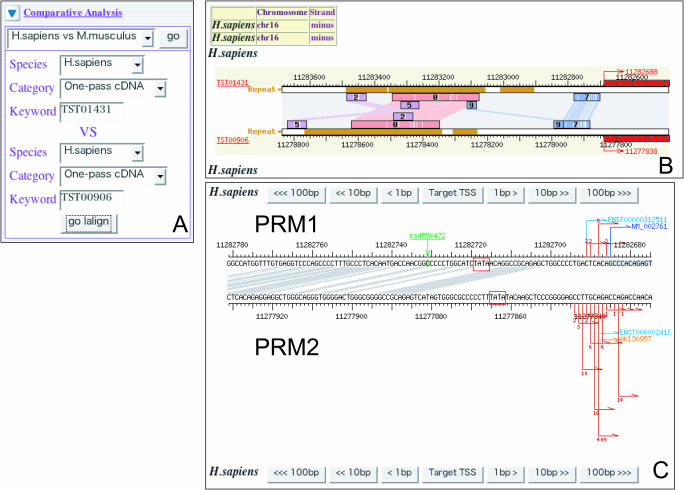
In the previous section, we showed an example of alternative promoter comparison between human and mouse. Before version 5 of DBTSS, these were precomputed, and the user could only obtain alignments between orthologous human and mouse genes. Despite being a useful idea, this sometimes failed to answer the user's need for alignments of arbitrary promoter pairs, for instance, promoters of paralogous genes. We therefore implement a dynamic viewer allowing the alignment of any two TSSs present in DBTSS. Such analyses are necessary to understand how transcriptional regulatory elements were conserved or diverged during gene and exon duplication. For example, in Figure 2A, the clones TST01431 of protamine 1 (PRM1: NM_002761) and TST00906 of protamine 2 (PRM2: NM_002762) are selected for alignment. Both genes are expressed in testis and are paralogous to each other. PRM1 is found in nearly all mammals while PRM2 is observed in relatively few mammals including human and mouse (13). In human, both genes are on chromosome 16, separated by ∼5 kb (14). The obtained alignment and the determined conserved regions are shown in Figure 2B. In this case, the blocks ‘0’ and ‘7’ are highly conserved. The details of the alignment of both TSS regions are also available, as shown in Figure 2C. Especially, it is noteworthy that the putative TATA-box is inside block ‘7’ for the PRM1 promoter and outside of it for the PRM2 promoters (15).
Figure 2.
An example of comparative analysis with any pair of TSSs. We show paralogous genes, protamine 1 (PRM1: NM_002761) and protamine 2 (PRM2: NM_002762), as an example. (A) By inputting the IDs of clones of PRM1 (TST01431) and PRM2 (TST00906) representative TSSs, users can obtain the results (B and C). (B) LALIGN analysis between two sequences. Note: smaller numbers indicate more highly conserved blocks. In this figure, the most conserved region between a pair is block 0; however, it includes Alu repeats. (C) The detail of the alignment of block 7. The putative TATA-boxes are marked with boxes.
FUTURE PERSPECTIVE
As shown in Table 1, we have added data from 32 263 zebrafish (Danio rerio) (16), 10 236 malaria (Plasmodium falciparum) (17) and 22 923 schyzon (Cyanidioscyzon merolae) (18) 5′ end clones. These correspond to 3061 zebrafish, 1527 malaria and 3635 schyzon genes. We will continue to expand DBTSS by adding TSS information for other species, such as macaque, when the relevant data become publicly available. Such data will give us a deeper insight into how the transcriptional regulatory networks have been shaped into their current form in humans, in terms of the molecular evolution of the promoters.
Acknowledgments
We thank all the members of the HGC-IMSUT sequencing team for their excellent sequencing work. We are also thankful to Nicolas Sierro for critically reading the manuscript. This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas and by special coordination funds for promoting science and technology (SCF), both from the Ministry of Education, Culture, Sports, Science and Technology in Japan. Computation time was provided by the Super Computer System, Human Genome Center, Institute of Medical Science, University of Tokyo. Funding to pay the Open Access publication charges for this article was provided by MEXT.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bajic V.B., Tan S.L., Suzuki Y., Sugano S. Promoter prediction analysis on the whole human genome. Nat. Biotechnol. 2004;22:1467–1473. doi: 10.1038/nbt1032. [DOI] [PubMed] [Google Scholar]
- 2.Praz V., Perier R., Bonnard C., Bucher P. The Eukaryotic Promoter Database, EPD: new entry types and links to gene expression data. Nucleic Acids Res. 2002;30:322–324. doi: 10.1093/nar/30.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzuki Y., Yamashita R., Nakai K., Sugano S. DBTSS: DataBase of human Transcriptional Start Sites and full-length cDNAs. Nucleic Acids Res. 2002;30:328–331. doi: 10.1093/nar/30.1.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suzuki Y., Yamashita R., Sugano S., Nakai K. DBTSS, DataBase of Transcriptional Start Sites: progress report 2004. Nucleic Acids Res. 2004;32:D78–D81. doi: 10.1093/nar/gkh076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maruyama K., Sugano S. Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. Gene. 1994;138:171–174. doi: 10.1016/0378-1119(94)90802-8. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki Y., Yoshitomo-Nakagawa K., Maruyama K., Suyama A., Sugano S. Construction and characterization of a full length-enriched and a 5′-end-enriched cDNA library. Gene. 1997;200:149–156. doi: 10.1016/s0378-1119(97)00411-3. [DOI] [PubMed] [Google Scholar]
- 7.Carninci P., Kvam C., Kitamura A., Ohsumi T., Okazaki Y., Itoh M., Kamiya M., Shibata K., Sasaki N., Izawa M., et al. High-efficiency full-length cDNA cloning by biotinylated CAP trapper. Genomics. 1996;37:327–336. doi: 10.1006/geno.1996.0567. [DOI] [PubMed] [Google Scholar]
- 8.Carninci P., Westover A., Nishiyama Y., Ohsumi T., Itoh M., Nagaoka S., Sasaki N., Okazaki Y., Muramatsu M., Schneider C., et al. High efficiency selection of full-length cDNA by improved biotinylated cap trapper. DNA Res. 1997;4:61–66. doi: 10.1093/dnares/4.1.61. [DOI] [PubMed] [Google Scholar]
- 9.Okazaki Y., Furuno M., Kasukawa T., Adachi J., Bono H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H., et al. Analysis of the mouse transcriptome based on functional annotation of 60 770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 10.Ota T., Suzuki Y., Nishikawa T., Otsuki T., Sugiyama T., Irie R., Wakamatsu A., Hayashi K., Sato H., Nagai K., et al. Complete sequencing and characterization of 21 243 full-length human cDNAs. Nature Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- 11.Landry J.R., Mager D.L., Wilhelm B.T. Complex controls: the role of alternative promoters in mammalian genomes. Trends Genet. 2003;19:640–648. doi: 10.1016/j.tig.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 12.Kimura K., Watanabe A., Suzuki Y., Ota T., Nishikawa T., Yamashita R., Yamamoto J., Sekine M., Tsuritani K., Ishii S., et al. Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 2005 doi: 10.1101/gr.4039406. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang X., Miller W., Schwartz S., Hardison R.C. Parallelization of a local similarity algorithm. Comput. Appl. Biosci. 1992;8:155–165. doi: 10.1093/bioinformatics/8.2.155. [DOI] [PubMed] [Google Scholar]
- 14.Domenjoud L., Nussbaum G., Adham I.M., Greeske G., Engel W. Genomic sequences of human protamines whose genes, PRM1 and PRM2, are clustered. Genomics. 1990;8:127–133. doi: 10.1016/0888-7543(90)90234-l. [DOI] [PubMed] [Google Scholar]
- 15.Wykes S.M., Krawetz S.A. Conservation of the PRM1 –> PRM2 –> TNP2 domain. DNA Seq. 2003;14:359–367. doi: 10.1080/10425170310001599453. [DOI] [PubMed] [Google Scholar]
- 16.Gerhard D.S., Wagner L., Feingold E.A., Shenmen C.M., Grouse L.H., Schuler G., Klein S.L., Old S., Rasooly R., Good P., et al. The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC) Genome Res. 2004;14:2121–2127. doi: 10.1101/gr.2596504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watanabe J., Suzuki Y., Sasaki M., Sugano S. Full-malaria 2004: an enlarged database for comparative studies of full-length cDNAs of malaria parasites, Plasmodium species. Nucleic Acids Res. 2004;32:D334–D338. doi: 10.1093/nar/gkh115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuzaki M., Misumi O., Shin -I.T., Maruyama S., Takahara M., Miyagishima S.Y., Mori T., Nishida K., Yagisawa F., Yoshida Y., et al. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature. 2004;428:653–657. doi: 10.1038/nature02398. [DOI] [PubMed] [Google Scholar]