Abstract
The EMBL Nucleotide Sequence Database (www.ebi.ac.uk/embl) at the EMBL European Bioinformatics Institute, UK, offers a comprehensive set of publicly available nucleotide sequence and annotation, freely accessible to all. Maintained in collaboration with partners DDBJ and GenBank, coverage includes whole genome sequencing project data, directly submitted sequence, sequence recorded in support of patent applications and much more. The database continues to offer submission tools, data retrieval facilities and user support. In 2005, the volume of data offered has continued to grow exponentially. In addition to the newly presented data, the database encompasses a range of new data types generated by novel technologies, offers enhanced presentation and searchability of the data and has greater integration with other data resources offered at the EBI and elsewhere. In stride with these developing data types, the database has continued to develop submission and retrieval tools to maximise the information content of submitted data and to offer the simplest possible submission routes for data producers. New developments, the submission process, data retrieval and access to support are presented in this paper, along with links to sources of further information.
INTRODUCTION
The EMBL Nucleotide Sequence Database is the European node of the three way International Nucleotide Sequence Database Collaboration (Box 1), whose aim is to collect and present nucleotide sequence and annotation with comprehensive global coverage.
INSDC
The International Nucleotide Sequence Database Collaboration (INSDC) comprises the EMBL Nucleotide Sequence Database at EMBL-EBI, The DNA Databank of Japan (1) and GenBank (2) in the USA. The collaboration aims to gather and present nucleotide sequence and annotation with comprehensive global coverage. Principles of the INSDC include the provision of free and unrestricted access to data for all users and the maintenance of permanently accessible records (3). Mandatory submission policies put in place by publishers of the leading life sciences journals continue to allow INSDC to offer global and timely coverage of public domain sequence and annotation. Amongst its functions is the maintenance of the INSDC Feature Table Definition (available from www.insdc.org)
In its twenty-seventh year, the EMBL Nucleotide Sequence Database continues to serve its users in the provision of submission and data management tools and services for submitters, as well as an increasing variety of means to search and download data of interest. A key goal of the EMBL Nucleotide Sequence Database is to integrate nucleotide sequence and annotation into the wealth of bioinformatics resources also offered at the EBI (4,5) and elsewhere. Through database cross-referencing and the extensive integrative capacity of the Sequence Retrieval System (SRS), (6), the data can be viewed in the context of over 200 locally held data libraries, ranging from protein resources such as UniProt (7) and InterPro (8), interaction databases such as IntAct (9), through to ontology collections such as GOA (10) and literature resources.
In the last year, the EMBL Nucleotide Sequence Database has continued to grow exponentially, both in terms of the numbers of entries and the numbers of bases stored. The database celebrated the 100 Gigabase milestone in August 2005. Standing at over 58 million entries at the time of going to press (September 2005), sequence from over 200 000 organisms is represented, from studies with methodological approaches as diverse as conventional sequencing, shotgun sequencing, high-throughput sequencing of concatenated tag molecules and environmental sampling techniques.
Organized into methodological and taxonomic divisions, the database is presented as individual entries, each carrying sequence, submission information (submission and update dates, version numbers and submitter details), literature citations and annotation in the form of a feature table, where biological features and their qualifiers are mapped to specific nucleotide locations within the sequence. Full details of database flatfile format are available in the user manual (see Table 1 for all hyperlinks). Details of feature table format are available in the INSDC Feature Table Definition.
Table 1.
Points of entry to the EMBL Nucleotide Sequence Database: submissions, retrieval and support
| Tool | Point of entry | Comment |
|---|---|---|
| Submission | ||
| Webin: submission of new data | http://www.ebi.ac.uk/embl/Submission/webin.html | For direct submissions of small scale sequencing projects, bulk data (e.g. rRNA and EST), large genomes, TPA, etc. |
| Update existing data | http://www.ebi.ac.uk/webin/update.html | For updates to directly submitted entries |
| Account and WGS submissions | datasubs@ebi.ac.uk | Contact us to discuss establishing project accounts and pipelines for WGS projects. |
| Retrieval | ||
| SRS | http://srs.ebi.ac.uk | Data retrieval by term search and through links to/from other databases |
| Homology search | http://www.ebi.ac.uk/Tools/similarity.html | Data retrieval by sequence similarity and homology |
| SVA | http://www.ebi.ac.uk/cgi-bin/sva/sva.pl | Access to current and historic data by accession number |
| FTP | http://www.ebi.ac.uk/embl/Access/index.html#ftp | Access to complete datasets in flatfile format, for both release and updated data |
| DVD | datasubs@ebi.ac.uk | Release DVD delivered by post |
| Genomes | http://www.ebi.ac.uk/genomes/ | Completed genomes and proteomes |
| Dbfetch | http://www.ebi.ac.uk/cgi-bin/emblfetch | Retrieval by accession number through web browser |
| Wsdbfetch | http://www.ebi.ac.uk/Tools/webservices/WSDbfetch.html | Retrieval by accession number through Webservice |
| Netserv | netserv@ebi.ac.uk | Email retrieval of entries by accession number; send e-mail with ‘HELP’ in message body for details |
| Custom datasets | datasubs@ebi.ac.uk | For data requirements not supported by existing tools |
| Specific help | datasubs@ebi.ac.uk | For help with all EMBL Nucleotide Sequence Database services; with direct submissions, account and WGS projects, data access tools, etc. |
| Support | ||
| General information | http://www.ebi.ac.uk/embl/ | Website, including user manual and INSDC Feature Table Definition |
| News | http://www.ebi.ac.uk/embl/News/news.html | EMBL Nucleotide Sequence Database news |
| Forthcoming changes | http://www.ebi.ac.uk/embl/Documentation/forthcomingchanges.html | Forthcoming data and format changes |
| XML documentation | http://www.ebi.ac.uk/embl/Documentation/xml/ | Details of INSDC XML and EMBL XML |
| Educational information | http://www.ebi.ac.uk/2can/ | Background bioinformatics educational resources |
NEW DEVELOPMENTS
New data
In the last year, we have seen an increase in the diversity of methodological approaches by our submitters to sequence generation. In terms of numbers of nucleotides, the rapid growth of the Whole Genome Shotgun (WGS) section of the database is notable. At the time of going to press, there are 16 finished WGS projects, 14 having been completed in the last year.
The recently developed Mass Genome Annotation (MGA) data type is used for high-throughput sequencing studies that generate large numbers of short tag sequences that provide valuable information when mapped to sequence that already exists in the database. Libraries within this dataset will soon include HTPSELEX sets of transcription factor binding sites (11,12). Specific keywords in MGA entries indicate the methodology used for tag generation.
The Genomes webserver showed 2494 completed genomes (including organellar and plasmid) at the time of going to press. This year's additions include recently completed assemblies of Leishmania major chromosomes (13). A related EBI resource, the Genome Reviews project, where added value genome annotation is drawn from UniProt and a variety of other sources, currently shows 231 completed genomes (14).
In June, 2005, we partitioned off the environmental sampling division (ENV) of the database. This division contains anonymously sequenced environmental samples, where source organism identification is based only on the sequence generated, rather than conventional taxonomic techniques. A driving force for creation of the ENV division was the need of users to access datasets offering reliable taxonomic source information; users now searching against the prokaryotic (PRO) division, for example, will resolve only those entries that have not been generated by anonymous environmental sample sequencing techniques and that have reliable taxonomic annotation, beyond that produced by sequence comparison methods.
The collaboration has broadened the scope of the Third Party Annotation (TPA) project in recognition of the value of annotation from submitters whose approach does not draw on direct experimental evidence; until recently, the TPA project, where submitters present novel peer-reviewed annotation of existing sequence, of which they are not the generators, was open only to experimentalists. In order to retain the quality, we continue to require that the study be specifically discussed in a peer-reviewed publication before making available to our users. The new two-tiered TPA dataset will be rolled out in the coming months. In addition to this, the entire TPA dataset is included in the EMBL Nucleotide Sequence Database release from December 2005 onwards.
The value of annotation is increased when users are able to trace evidence for a particular annotation and make a balanced judgement about its validity. Evidence tagging in EMBL annotation is highly complex because of the wide breadth of features that are supported. Over the coming year, we will extend the functionality of our current evidence labelling system from /evidence = EXPERIMENTAL and /evidence = NOT_EXPERIMENTAL to /experiment and /inference, where we will encourage the use of concise free text to provide the experimental techniques used and a structured inference text that details whether the inference has been made on the basis of sequence similarity or profile, details of the nature of the sequence to which it is similar or the profile matching algorithm that was used and an additional reference to the sequence or profile that has been matched.
The ever-increasing diversity and volume of data have required a comprehensive review of the technology behind the EMBL Nucleotide Sequence Database. So far, schema and data distribution changes have been largely transparent to users, in order to preserve compatibility between our data and users' further processing tools. An important flatfile format change over the coming months is an improvement of the ID line information content and parsability. All forthcoming changes to data format and content are announced on a regular basis (Table 1).
Data integration and presentation
The integration of EMBL Nucleotide Sequence Database data into other EBI resources traditionally operates through database cross-references, at entry-level (DR line) and feature level (/db_xref qualifier). The EMBL Nucleotide Sequence Database offers cross-references to 25 resources. 46.8 million entry-level and 8.5 million feature-level references are available at the time of going to press. Within a year, we have seen a growth in cross-references of 63 and 185% at entry and feature level, respectively.
Although cross-references are presented in flatfiles, additional inferred relationships both within the EMBL Nucleotide Sequence Database and between database entries and objects in external resources are presented when viewing data at the EBI. Using the EBI SRS and homology search tools, the EMBL Nucleotide Sequence Database now projects many of these further inferred relationships to create enhanced views. This information appears under ‘EMBLextras’ in the SRS ‘Emblentry’ view (set as default). Currently, inferred relationships such as citation of an entry in TPA or in EMBL Align are presented (15). In addition, SRS offers integration of EMBL Nucleotide Sequence Database data with a further collection of in excess of 200 library resources through cross-references offered by the database and through inferred links routed via cross-referenced resources.
From April 2005, we have increased the cross-referencing frequency for UniProt, InterPro and GOA references from 3 months to 6 weeks. This provides users with more up-to-date cross-referencing information for some of the more dynamic resources to which we cross-refer.
While the term and homology search tools on offer at the EBI provide flexible access to EMBL Nucleotide Sequence Database data, in some cases, the databases are able to provide custom datasets and views (see Data Access section and Table 1).
The EMBLCDS dataset arose from user requests for whole database dumps of coding sequence. Because of user demand, EMBLCDS is now offered as a regularly maintained dataset, available by anonymous FTP (under the CDS directory, Table 1). This year, we have launched the non-redundant coding sequence dataset, EMBLCDSnr, where each sequence is represented once. EMBLCDSnr can also be downloaded from the FTP site. There are currently 4.1 million EMBLCDS entries and 3.6 million EMBLCDSnr items.
Work continues on development of XML formats for EMBL Nucleotide Sequence Database data. In the last year, the collaborative INSDXML DTD 1.3 has been made available and EMBLXML offers more stability and finer-grained modelling of nucleotide sequence information.
THE CONTINUED NEED TO SUBMIT AND UPDATE DATA
A publicly available record of the body of biological knowledge built up over the years has proved to be an indispensable tool in addressing new areas of research. As part of this body of knowledge, the EMBL Nucleotide Sequence Database and its INSDC partners provide a vital service that allows archiving of novel sequence to be allied to the publication process. The major scientific journals have a mandatory database submission policy for novel sequence and many have been keen to promote TPA submission as part of the publication process. As a result, the goal of the database to provide comprehensive coverage of publicly discussed sequence is feasible and will continue to form the backbone of our policy.
Data submitted to the EMBL Nucleotide Sequence Database, or either of the other INSDC partners are made available to the user community very rapidly; data are exchanged on a nightly basis to maximise synchrony. Because each of the three nodes attracts large numbers of users, submitters can expect maximal exposure of their data to target users in the shortest possible time.
Nucleotide sequence and annotation remain within the ownership, and hence editorial control, of the original submitters. The EMBL Nucleotide Sequence Database maintains database syntax and applies some biological consistency controls to existing data, but essentially, responsibility for biological content remains with the submitters. Updates to existing entries are encouraged as soon as new sequence, biological and citation information becomes available. Third party users who discover errors in existing sequence and annotation are encouraged to relay their findings to the original submitters of the entry or entries. Where such users generate novel sequence and annotation relating to source nucleic acid molecules already recorded in the database, the novel data should be submitted. At the level of a given source molecule, then, some EMBL Nucleotide Sequence Database data are redundant, but novel sequence relating to a particular source nucleotide generated by the original submitters is treated as an update to the existing entry or entries.
USER SERVICES
For details of URLs, email addresses and other contact information, please refer to Table 1, where details of submission, retrieval and support access points to the EMBL Nucleotide Sequence Database are presented in full.
Submission and update
As the variety and volume of nucleotide data grow, the EMBL Nucleotide Sequence Database has successfully provided technical solutions to assist in sequence versioning, annotation and presentation to the community. Data are submitted to EMBL through project accounts or, increasingly, the direct submissions department. Project account submissions require extensive bioinformatics capabilities of the submitting team, while direct submissions call on an expert in house curation staff to assist in annotation and preparation of data for the database.
The core direct submissions tool, the web-based Webin, comprises a variety of submission procedures, offering a smooth submission process for single, manually annotated sequences through to fasta format submission of large numbers of similarly annotated sequences (e.g. cytochrome oxidase genes in a barcoding study) and extensively annotated complete genome submission.
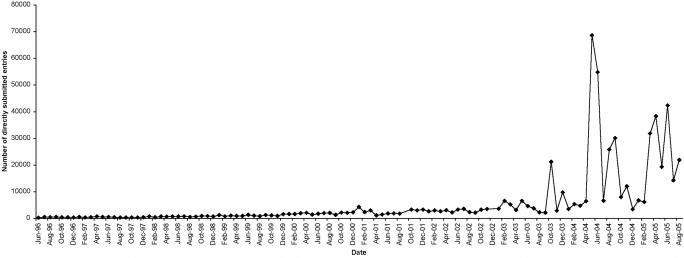
Over the year, while the number of data submitters has grown gently, as new functionality has been rolled out into Webin, the database has seen a significant increase in entries submitted through direct submissions (Figure 1).
Figure 1.
Increase in scale of direct submissions. Monthly newly submitted entry counts are shown.
For large-scale direct submissions, starting with the Webin tool, users submit a representative sample entry from their dataset. They also detail which fields will vary between entries. Using the submission information and, in some cases, further communication with the submitter, a database curator offers the most appropriate (for submitter and curator alike) means of submission. This can involve the creation of web-based templates for completion or invitation to submit a single file in fasta, or some other format.
For large single entries that are likely to have extensive annotation, such as complete bacterial genomes or eukaryotic chromosomes, Webin users submit a ‘blank’ entry, including details of the source organism, submission and publication details and sequence. Following a curation step, submitters are invited to submit their annotation in a suitable format, such an output file from Artemis (16).
The EMBL Nucleotide Sequence Database has continued to operate a two day turnaround for small direct submissions (<25 entries) and a 5 day working rule for large-scale submissions (>25 entries); we aim to provide accession numbers within these time frames, provided we have been supplied with all of the information we require to enter the data into the database.
Data access
Access points to EMBL Nucleotide Sequence Database data include SRS, homology search tools, the Sequence Version Archive (SVA), (17), the FTP site, the Genomes webserver (for completed genomes) and sequence retrieval by accession number (Dbfetch, Wsdbfetch and netserv), (6). These facilities offer a wide range of opportunities to search and download data. Assistance is available while using the tools from the EBI toolbox help documentation, linked from toolbox pages, by clicking on support at the foot of many EBI web pages, or by contacting datasubs@ebi.ac.uk directly. The EBI also offers bioinformatics educational resources at the 2can site (see Table 1 for all URLs).
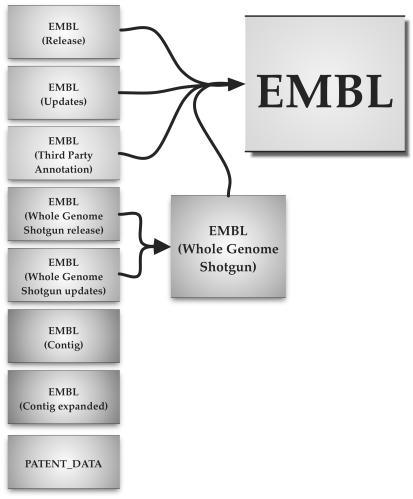
SRS presents EMBL Nucleotide Sequence Database data in the form of a number of component libraries. For many uses, the virtual library, EMBL, is sufficient and is set by default for quick searches. EMBL libraries represented in SRS are shown in Figure 2. Assistance with SRS is available at the SRS help centre and by email (see Table 1). Specific help with EMBL data are also available from datasubs@ebi.ac.uk.
Figure 2.
SRS Library organization.
While the database access tools available are suitable for the majority of uses of the data, there are some users who have unusual queries to run on the data, or require presentations of the data that are unavailable through the tools offered. Typically, these users lack the bioinformatics resources required to download whole datasets from the FTP server and implement their own database for analysis, but have extensive data manipulation to perform. In many of these cases, the database is able to provide custom datasets to assist the user. Please send custom dataset requests to datasubs@ebi.ac.uk and we will be able to advise.
Helpdesk facilities
As a key service provider, we operate extensive helpdesk facilities, where users, both submitters and data readers, are able to resolve issues that relate to the EMBL Nucleotide Sequence Database, its data and tools. Moreover, helpdesk functions are integrated with the broader EBI support operations, where holistic solutions are provided by those at the working end of the resources concerned. The helpdesk team can be contacted at datasubs@ebi.ac.uk.
Staff members at the EMBL Nucleotide Sequence Database are able to assist users of the database with problems relating to data format for submission, the submission process, through to searching, downloading and making sense of the data. All queries are welcome and we aim to respond rapidly where possible, please provide any accession numbers or submission identifiers that we have supplied to help us deal with the issue quickly.
Announcements are posted as appropriate on the EMBL news page and changes to database format and contents are announced on the Forthcoming Changes page (see Table 1 for all URLs).
Acknowledgments
Funding to pay the Open Access publication charges for this article was provided by EMBL.
Conflict of interest statement. None declared.
REFERENCES
- 1.Okubo K., Sugawara H., Gojobori T., Tateno Y. DDBJ in preparation for overview of research activities behind data submissions. Nucleic Acids Res. 2006;34:D6–D9. doi: 10.1093/nar/gkj111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L. GenBank. Nucleic Acids Res. 2006;34:D16–D20. doi: 10.1093/nar/gkj157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunak S., Danchin A., Hattori M., Nakamura H., Shinozaki K., Matise T., Preuss D. Nucleotide sequence database policies. Science. 2002;298:1333. doi: 10.1126/science.298.5597.1333b. [DOI] [PubMed] [Google Scholar]
- 4.Brooksbank C., Camon E., Harris M.A., Magrane M., Martin M.J., Mulder N., O'Donovan C., Parkinson H., Tuli M.A., Apweiler R., et al. The European Bioinformatics Institute's data resources. Nucleic Acids Res. 2003;31:43–50. doi: 10.1093/nar/gkg066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brooksbank C., Cameron G., Thornton J. The European Bioinformatics Institute's data resources: towards systems biology. Nucleic Acids Res. 2005;33:D46–D53. doi: 10.1093/nar/gki026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harte N., Silventoinen V., Quevillon E., Robinson S., Kallio K., Fustero X., Patel P., Jokinen P., Lopez R. Public web-based services from the European Bioinformatics Institute. Nucleic Acids Res. 2004;32:W3–W9. doi: 10.1093/nar/gkh405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu C.H., Apweiler R., Bairoch A., Natale D.A., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., Martin M.J., Mazumder R., O'Donovan C., Redaschi N., Suzek B. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mulder N.J., Apweiler R., Attwood T.K., Bairoch A., Bateman A., Binns D., Bradley P., Bork P., Bucher P., Cerutti L., et al. InterPro, progress and status in 2005. Nucleic Acids Res. 2005;33:D201–D205. doi: 10.1093/nar/gki106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hermjakob H., Montecchi-Palazzi L., Lewington C., Mudali S., Kerrien S., Orchard S., Vingron M., Roechert B., Roepstorff P., Valencia A., et al. IntAct-an open source molecular interaction database. Nucleic Acids Res. 2004;32:D452–D455. doi: 10.1093/nar/gkh052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Camon E., Magrane M., Barrell D., Lee V., Dimmer E., Maslen J., Binns D., Harte N., Lopez R., Apweiler R. The Gene Ontology Annotation (GOA) Database: sharing knowledge in Uniprot with Gene Ontology. Nucleic Acids Res. 2004;32:D262–D266. doi: 10.1093/nar/gkh021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roulet E., Busso S., Camargo A.A., Simpson A.J., Mermod N., Bucher P. High-throughput SELEX SAGE method for quantitative modeling of transcription-factor binding sites. Nat. Biotechnol. 2002;20:831–835. doi: 10.1038/nbt718. [DOI] [PubMed] [Google Scholar]
- 12.Jagannathan V., Roulet E., Delorenzi M., Bucher P. HTPSELEX-a database of high-throughput SELEX libraries for transcription factor binding sites. Nucleic Acids Res. 2006;34:D90–D94. doi: 10.1093/nar/gkj049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ivens A.C., Peacock C.S., Worthey E.A., Murphy L., Aggarwal G., Berriman M., Sisk E., Rajandream M.A., Adlem E., Aert R., et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kersey P., Bower L., Morris L., Horne A., Petryszak R., Kanz C., Kanapin A., Das U., Michoud K., Phan I., et al. Integr8 and Genome Reviews: integrated views of complete genomes and proteomes. Nucleic Acids Res. 2005;33:D297–D302. doi: 10.1093/nar/gki039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lombard V., Camon E.B., Parkinson H.E., Hingamp P., Stoesser G., Redaschi N. EMBL-Align: a new public nucleotide and amino acid multiple sequence alignment database. Bioinformatics. 2002;18:763–764. doi: 10.1093/bioinformatics/18.5.763. [DOI] [PubMed] [Google Scholar]
- 16.Rutherford K., Parkhill J., Crook J., Horsnell T., Rice P., Rajandream M.A., Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 17.Leinonen R., Nardone F., Oyewole O., Redaschi N., Stoehr P. The EMBL SVA. Bioinformatics. 2003;19:1861–1862. doi: 10.1093/bioinformatics/btg248. [DOI] [PubMed] [Google Scholar]