Abstract
As more archaeal genomes are sequenced, effective research and analysis tools are needed to integrate the diverse information available for any given locus. The feature-rich UCSC Genome Browser, created originally to annotate the human genome, can be applied to any sequenced organism. We have created a UCSC Archaeal Genome Browser, available at http://archaea.ucsc.edu/, currently with 26 archaeal genomes. It displays G/C content, gene and operon annotation from multiple sources, sequence motifs (promoters and Shine-Dalgarno), microarray data, multi-genome alignments and protein conservation across phylogenetic and habitat categories. We encourage submission of new experimental and bioinformatic analysis from contributors. The purpose of this tool is to aid biological discovery and facilitate greater collaboration within the archaeal research community.
INTRODUCTION
As more genomic sequences become available, analysis and interpretation of existing annotation becomes increasingly difficult without useful tools (1). This large quantity of data provides a challenge regarding what information should be provided at the top level so that the data can inform but not overwhelm. The extendibility of the UCSC Genome Browser allows easy integration of new organisms into a system that has a well-developed database and user interface, in the form of a graphical web page (2). BLAT, an alignment tool designed to quickly find sequences of 95% and greater similarity (3), allows quick look-up of sequences of interest. The UCSC Genome Browser was first implemented for the relatively large human genome, containing 24 linear chromosomes and dozens of annotation ‘tracks’ (2,4). Because the genome and annotation datasets are comparatively small for all prokaryotic species, loading new archaeal genomes into the browser requires only modest computational resources. To date, the Archaeal Genome Browser contains all 24 complete archaeal genomes, plus two draft assemblies (Table 1). Four bacterial genomes are also provided for comparative purposes. The genomes of these species, along with a variety of annotation tracks from our own group and other public datasets, provide a simple, interactive genome information resource for which the UCSC Genome Browser is renowned.
Table 1.
Species available in the Archaeal Genome Browser at time of publication
| Archaeal species | Hyperthermophile | Halophile | Acidophile | Methanogen | Other extreme |
|---|---|---|---|---|---|
| Pyrobaculum aerophilum | X | ||||
| Aeropyrum pernix | X | ||||
| Sulfolobus acidocaldarius | X | ||||
| Sulfolobus solfataricus | X | ||||
| Sulfolobus tokodaii | X | ||||
| Nanoarchaeum equitans | X | ||||
| Thermococcus kodakaraensis | X | ||||
| Pyrococcus abyssi | X | ||||
| Pyrococcus furiosus | X | ||||
| Pyrococcus horikoshii | X | ||||
| Methanopyrus kandleri | X | ||||
| Methanothermobacter thermautotrophicus | X | ||||
| Methanocaldococcus jannaschii | X | ||||
| Methanococcus maripaludis | X | ||||
| Ferroplasma acidarmanus | X | ||||
| Picrophilus torridus | X | X | |||
| Thermoplasma acidophilum | X | ||||
| Thermoplasma volcanium | X | ||||
| Archaeoglobus fulgidus | X | ||||
| Halobacterium sp. | X | ||||
| Haloarcula marismortui | X | ||||
| Natronomonas pharaonis | X | Alkaliphile | |||
| Methanococcoides burtonii | X | Psychrotroph | |||
| Methanosarcina barkeri | X | ||||
| Methanosarcina mazei | X | ||||
| Methanosarcina acetivorans | X | ||||
| Bacterial species | |||||
| Aquifex aeolicus | X | ||||
| Thermotoga maritima | X | ||||
| Escherichia coli K12 | |||||
| Vibrio cholerae | |||||
All 24 fully sequenced archaeal genomes are included. In addition, two draft archaeal genomes (M.burtonii, F.acidarmanus) and four bacterial genomes are also displayed. Each species is characterized by features of its optimal growth environment.
MATERIALS AND METHODS
Archaeal browser tracks
The data displayed in the Archaeal Genome Browser give the user the ability to integrate information to make new observations. Each track presents a different biological perspective on the organism. A detailed overview of the tracks can be found on corresponding description pages in the browser. We briefly describe the 11 currently available tracks here. The Pyrobaculum aerophilum and Pyrococcus furious browsers have the most complete sets of tracks.
Basic tracks provide the framework for understanding a genome. These include G/C (guanine/cytosine) content, annotated protein gene predictions (ORFs) from Genbank (5) and the Institute for Genome Research (TIGR, www.tigr.org), all possible start/stop codons in all reading frames and current known or predicted non-coding RNA (ncRNA) genes. Clicking on a Genbank ORF opens a page detailing the available RefSeq annotation for that gene, InterPro (6) and Pfam (7) domains, COG group (8), GO categories (9), KEGG pathway information (10) and ModBase (11) structural predictions when available.
Additional tracks provide data from computational analyses and experimental genomics studies. The Shine-Dalgarno track indicates positions with base pairing potential to the 3′ end of 16S rRNA, which is required for translation initiation. A companion promoter track indicates similarity to the consensus transcription factor IIB (BRE)-TATA box, which highlights possible sites of transcription initiation. The promoter track can be useful to detect incorrect start codons of annotated genes or novel genes. There are two operon tracks: published predictions by Ermolaeva and colleagues (12) at TIGR and a less stringent set of predictions using a simple distance-rule method integrated with TIGR-predicted operons. Microarray tracks display results from experiments within our laboratory. The MULTIZ conservation track provides visual display of a full-genome multiple sequence alignment to closely related species (13). The browser also graphs a conservation score based on a phylogenetic hidden Markov model (14), useful for finding candidate ncRNAs, repetitive elements or features that are not annotated in the genome. A track displaying BLASTZ (15) hits to other loci within the same genome is useful for identifying repetitive elements and paralogous genes.
Finally, a protein conservation track displays each ORF's alignment score to other species by phylogenetic category or native environment (i.e. hyperthermophile, halophile, acidophile). This similarity database was created locally using BLASTP (16) against NCBI's non-redundant protein database. The information page for each ORF contains a ranked table of hits—this includes the gene name and annotated function, a link to the full Genbank entry and a link to the corresponding genome locus in the browser of the other organism (if available). The latter provides a level of ‘genome-interoperability’ not available in single-species genome browser tools. An additional link allows one to re-run the BLASTP search against the most current protein database at NCBI. Together, these data help improve or update functional prediction of poorly characterized genes.
Archaeal browser features
Every organism's browser includes a BLAT server (3) for performing sequence similarity searches. Given a DNA or protein sequence, BLAT returns a list of links to all genome positions that share 95% or greater identity with the input sequence. A browser position search utility takes one or more terms (but not sequence) as input and returns all matching genome positions. Together, BLAT and the browser position search utility allow one to quickly search a genome by sequence, keyword or locus.
The table browser is a powerful tool for retrieving raw data and performing intersections and unions between data in different tracks (4). For example, with a few mouse clicks, one can obtain a list of all GenBank coding genes that overlap known non-coding RNAs or all places where GenBank and TIGR ORF annotations disagree. A correlation feature computes and plots the correlation coefficient between any two tracks at a user-specified genome location and base resolution.
The browser also provides a ‘Custom Track’ tool, which allows one to easily and privately upload datasets for display along side the publicly available tracks described earlier. Detailed help pages explain how to use the browser and format data for custom tracks.
RESULTS
Archaeal browser data mining
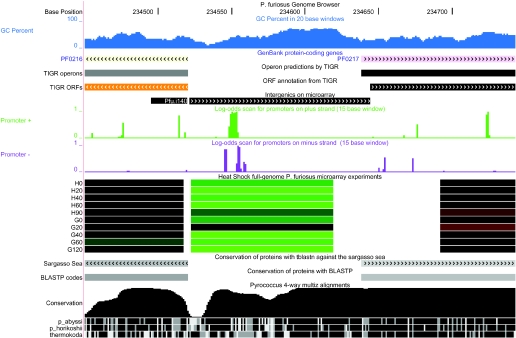
The Archaeal Genome Browser is a powerful research tool that has facilitated new discoveries in our laboratory. For example, it has allowed us to integrate sequence annotations with whole genome microarray data for two species to date, P.aerophilum and P.furiosus. Visualizing expression patterns along with gene conservation and sequence motifs has been particularly useful for these species where 60–80% of protein coding genes do not have a well-annotated function. By comparison, the Escherichia coli K12 genome has <24% genes with no function (17) and Saccharomyces cerevisiae has <37% [http://www.yeastgenome.org/ (18)]. Furthermore, in these and other archaea, the start codons of many ORFs are likely to be incorrect because of frequent use of UUG and GUG as alternatives to AUG. We find many ‘intergenic’ regions of the genome to be transcribed, but with no annotation clues in GenBank entries. Figure 1 illustrates an intergenic region for which the browser track data strongly suggests the presence of a novel gene or untranslated region (UTR). While a single piece of information (e.g. microarray data) alone would be only suggestive, the combination of evidence across multiple tracks helps to build confidence in new observations. Promising hypotheses such as these can then be followed up by further computational analysis or traditional experimental testing.
Figure 1.
The intergenic region denoted Pfu.i140, located between P.furious ORFs PF0216 and PF0217, shows high conservation among closely related species (‘Conservation’ track), high ‘G/C Percent’ values (indicative of RNA structure in hyperthermophiles), and a different expression pattern from the neighboring ORFs (green: suppressed relative to baseline, red: induced relative to baseline in a heat shock time course). The ‘Promoter +’ track with green peaks indicates a TATA box that is ∼88 bp upstream of PF0217, not the −38 position that is typical for this species. Together, these pieces of evidence suggest that transcription starts 50 bp upstream of the predicted coding region, which implies one of three possibilities: (i) this is a non-coding RNA gene that is co-transcribed with PF0217 but later processed away from the transcript; (ii) this is a regulatory UTR of PF0217 with secondary structure; or (iii) the start codon is incorrectly annotated forPF0217.
DISCUSSION
Future developments
The Archaeal Genome Browser is run off the same source code and mySQL database as the UCSC Genome Browser, which allows for easy code, data and feature updating. When new computational or functional analyses become available for an organism with an existing genome browser, the database can be quickly be updated to create a new track of genome annotation. Development of browsers for additional sequenced archaeal species is planned. We strongly encourage members of the archaeal research community to contribute genome-wide datasets to the browser project.
CONCLUSIONS
The Archaeal Genome Browser is a simple, graphic navigation tool and annotation database designed to help researchers perform effective research and analysis, while being relatively easy to improve in content and function.
Acknowledgments
We thank Jim Kent and the Genome Bioinformatics Group of UC Santa Cruz, in particular Hiram Clawson, for providing the UCSC Genome Browser and assisting us in developing the Archaeal Genome Browser. Lowe laboratory members Patricia Chan, Michael Pearson and Matt Weirauch helped develop annotation tracks. Thanks also to Pernille Nielsen of the Krogh laboratory for contributing the EasyGene gene prediction track. Funding to pay the Open Access publication charges for this article was provided by an Alfred P. Sloan Research Fellowship (T.M.L.).
Conflict of interest statement. Andy Pohl receives royalties for the sale of the original UCSC genome browser (on which the Archaeal Genome Browser is based) to commercial entities. No authors receive royalties for sale of the Archaeal genome browser.
REFERENCES
- 1.Karolchik D., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kent W.J. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karolchik D., Hinrichs A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L. GenBank: update. Nucleic Acids Res. 2005;33:D34–D38. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mulder N.J., Apweiler R., Attwood T.K., Bairoch A., Bateman A., Binns D., Bradley P., Bork P., Bucher P., Cerutti L., et al. InterPro, progress and status in 2005. Nucleic Acids Res. 2005;33:D201–D205. doi: 10.1093/nar/gki106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L.L., et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tatusov R.L., Koonin E.V., Lipman D.J. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 9.The Gene Ontology Consortium. Gene Ontology: tool for the unification of biology. Nat. Genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pieper U., Eswar N., Braberg H., Madhusudhan M.S., Davis F., Stuart A.C., Mirkovic N., Rossi A., Marti-Renom M.A., Fiser A., et al. MODBASE, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res. 2004;32:D217–D222. doi: 10.1093/nar/gkh095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ermolaeva M.D., White O., Salzberg S.L. Prediction of operons in microbial genomes. Nucleic Acids Res. 2001;29:1216–1221. doi: 10.1093/nar/29.5.1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siepel A., Haussler D. Combining phylogenetic and hidden Markov models in biosequence analysis. J. Comput. Biol. 2004;11:413–428. doi: 10.1089/1066527041410472. [DOI] [PubMed] [Google Scholar]
- 15.Schwartz S., Kent W.J., Smit A., Zhang Z., Baertsch R., Hardison R., Haussler D., Miller W. Human-mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altschul S., Madden T., Schaffer A., Zhang J., Zhang Z., Miller W., Lipman D. Gapped BLAST and PSI-BLAST: a new generation of protein database search algorithms. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keseler I.M., Collado-Vides J., Gama-Castro S., Ingraham J., Paley S., Paulsen I.T., Peralta-Gil M., Karp. P.D. EcoCyc: A comprehensive database resource for Escherichia coli. Nucleic Acids Res. 2005;33:D334–D337. doi: 10.1093/nar/gki108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christie K.R., Weng S., Balakrishnan R., Costanzo M.C., Dolinski K., Dwight S.S., Engel S.R., Feierbach B., Fisk D.G., Hirschman J.E., et al. Saccharomyces Genome Database (SGD) provides tools to identify and analyze sequences from Saccharomyces cerevisiae and related sequences from other organisms. Nucleic Acids Res. 2004;32:D311–D314. doi: 10.1093/nar/gkh033. [DOI] [PMC free article] [PubMed] [Google Scholar]