Abstract
Shewanella putrefaciens strain 200 respires anaerobically on a wide range of compounds as the sole terminal electron acceptor, including ferric iron [Fe(III)] and manganese oxide [Mn(IV)]. Previous studies demonstrated that a 23.3-kb S. putrefaciens wild-type DNA fragment conferred metal reduction capability to a set of respiratory mutants with impaired Fe(III) and Mn(IV) reduction activities (T. DiChristina and E. DeLong, J. Bacteriol. 176:1468–1474, 1994). In the present study, the smallest complementing fragment was found to contain one open reading frame (ORF) (ferE) whose translated product displayed 87% sequence similarity to Aeromonas hydrophila ExeE, a member of the PulE (GspE) family of proteins found in type II protein secretion systems. Insertional mutants E726 and E912, constructed by targeted replacement of wild-type ferE with an insertionally inactivated ferE construct, were unable to respire anaerobically on Fe(III) or Mn(IV) yet retained the ability to grow on all other terminal electron acceptors. Nucleotide sequence analysis of regions flanking ferE revealed the presence of one partial and two complete ORFs whose translated products displayed 55 to 70% sequence similarity to the PulD, -F, and -G homologs of type II secretion systems. A contiguous cluster of 12 type II secretion genes (pulC to -N homologs) was found in the unannotated genome sequence of Shewanella oneidensis (formerly S. putrefaciens) MR-1. A 91-kDa heme-containing protein involved in Fe(III) reduction was present in the peripheral proteins loosely attached to the outside face of the outer membrane of the wild-type and complemented (Fer+) B31 transconjugates yet was missing from this location in Fer mutants E912 and B31 and in uncomplemented (Fer−) B31 transconjugates. Membrane fractionation studies with the wild-type strain supported this finding: the 91-kDa heme-containing protein was detected with the outer membrane fraction and not with the inner membrane or soluble fraction. These findings provide the first genetic evidence linking dissimilatory metal reduction to type II protein secretion and provide additional biochemical evidence supporting outer membrane localization of S. putrefaciens proteins involved in anaerobic respiration on Fe(III) and Mn(IV).
Dissimilatory Fe(III) and Mn(IV) reduction are relatively recent additions (3, 26, 35) to the suite of known anaerobic respiratory processes carried out by microorganisms. Compared to the wealth of knowledge existing on the molecular details of aerobic respiration, denitrification, sulfate reduction, and methanogenesis, little is known about dissimilatory Fe(III) and Mn(IV) reduction (28). Although Fe(III)- and Mn(IV)-reducing proteins have been isolated from several metal-reducing microorganisms (6, 13, 16, 29, 34, 48), an Fe(III) or Mn(IV) terminal reductase gene or enzyme has yet to be definitively identified. Ironically, recent microbiological (55) and geological (5) evidence indicates that dissimilatory Fe(III) reduction may have been one of the first respiratory processes to have evolved on early Earth. In the modern biosphere, dissimilatory metal reduction is central to a variety of globally significant processes, including the biogeochemical cycling of Fe, Mn, trace elements, and phosphate, degradation of natural and contaminant organic matter, weathering of Fe(III)-containing clays and minerals, and biomineralization of Fe(II)-bearing minerals such as magnetite (12, 23, 25, 37).
Respiration-linked terminal reductases are widely considered (15, 28) to be an integral part of or associated with the cytoplasmic membranes of gram-negative microorganisms. This view is based on respiratory electron transfer to soluble terminal electron acceptors such as dissolved oxygen, nitrate, sulfate, or carbon dioxide which diffuse passively into the cell or are taken up by transmembrane transport systems (15, 28). At neutral pH, Fe(III) and Mn(IV) oxides are largely found in crystalline form or as amorphous (oxy)hydroxide particles (52). Neutrophilic Fe(III)- and Mn(IV)-reducing microorganisms are therefore required to generate energy via electron transfer to highly insoluble terminal electron acceptors which are presumably unable to contact the inner membrane. To overcome this problem, Fe(III)- and Mn(IV)-reducing microorganisms have been postulated to contact the mineral surface directly (4, 26) or to transfer electrons to Fe(III) via extracellular electron shuttles such as humic acids, quinones, or c-type cytochromes (24, 38, 39, 48). Fe(III)-reducing c-type cytochromes have been found in the soluble fraction, inner and outer membrane fractions, and spent anaerobic growth medium of Fe(III)-reducing microorganisms (13, 33, 48). These findings have led to the hypothesis that c-type cytochromes are targeted to either the outer membrane or cell exterior, where they may contact Fe(III) and catalyze the terminal electron transfer reaction (6, 13, 33, 48). Genetic evidence linking outer membrane-targeted or extracellular protein secretion and dissimilatory Fe(III) or Mn(IV) reduction has yet to be reported.
The Fe(III)- and Mn(IV)-reducing gamma-proteobacterium Shewanella putrefaciens strain 200 is a nonfermenting heterotroph that respires on a remarkable number of terminal electron acceptors, including oxygen, nitrate (NO3−), nitrite (NO2−), trimethylamine-N-oxide (TMAO), sulfite (SO32−), thiosulfate (S2O32−), elemental sulfur [S(0)], fumarate, uranyl carbonate [U(VI)], selenite [Se(IV)], Fe(III), Mn(IV), and potentially several others (8, 11, 53, 57). A set of rapid screening techniques was previously used to isolate S. putrefaciens respiratory mutants which had impaired Fe(III) and Mn(IV) reduction capability (8, 10, 11) yet which retained the ability to grow anaerobically on all other terminal electron acceptors (8, 11, 53, 57). A genetic complementation strategy was subsequently used to isolate a 23.3-kb complementing DNA fragment (designated B4) that conferred the Fe(III) reduction (Fer+) phenotype to several of the respiratory mutants, including strain B31 (11). In the present study, we report on subcloning of the 23.3-kb complementing fragment B4, sequence analysis of a 4.2-kb DNA fragment containing the complementing gene, and a comparison of the Fe(III)-reducing protein profiles of wild-type and mutant subcellular fractions. We demonstrate that a homolog of the pulE (gspE) type II protein secretion gene (designated ferE) is required for anaerobic growth on Fe(III) and Mn(IV) and provide preliminary evidence that disruption of ferE causes a 91-kDa heme-containing protein involved in Fe(III) reduction to accumulate at the S. putrefaciens inner membrane rather than be secreted to the outer membrane, which appears to be its physiological location in the wild-type strain.
MATERIALS AND METHODS
Growth media and cultivation conditions.
S.putrefaciens cultures were grown at 30°C in the presence of 50 μg of rifamycin per ml. Growth medium (pH 7.5 to 8.0) consisted of either nutrient agar (Difco) or a defined salts medium (SM) (35) supplemented with lactate (30 mM) and rifamycin. For SM agar medium, Bacto agar (Difco) was added at 1.5% (wt/vol). Anaerobic growth experiments were carried out in 13-ml Hungate tubes (Bellco Glass, Inc.) filled with 10 of ml SM and sealed with black butyl rubber stoppers under an N2 atmosphere. Filter-sterilized terminal electron acceptor stocks were added at the following final concentration (terminal electron acceptor synthesis is described in reference 11, except where indicated): NO3−, 15 mM; NO2−, 3 mM; Fe(III) citrate, 50 mM; amorphous Fe(III) oxide, 100 mM; Mn(IV), 10 mM as colloidal MnO2 (27, 41), TMAO, 25 mM; SO32−, 10 mM; S2O32−, 10 mM; fumarate, 5 mM; Se(IV), 2 mM (53); and U(VI), 2 mM as uranyl carbonate (57). When required, other antibiotics were added at the following final concentrations: chloramphenicol, 25 μg/ml; gentamicin, 25 μg/ml; ampicillin, 50 μg/ml; and streptomycin, 50 μg/ml.
Analytical techniques.
Cell growth was monitored bymeasuring cell counts and terminal electron acceptor depletion or end product production. Acridine orange-stained cells were counted directly via epifluorescence microscopy (Nikon Diaphot 300 microscope) according to previously described procedures (26). Cell numbers at each time point were calculated as the average of 10 counts from two parallel yet independent anaerobic incubations. U(VI) was measured spectrophotometrically at 380 nm with benzohydroxamic acid in 1-hexanol (30, 57). NO2− was measured spectrophotometrically with sulfanilic acid-N-1-naphthylethylene-diamine dihydrochloride solution (31). Fe(III) reduction was monitored by measuring Fe(II) production with the ferrozine technique (51). Mn(IV) reduction was monitored by measuring Mn(IV) depletion with the benzidine blue colorimetric technique (8). Sulfide concentration was measured colorimetrically by the method of Cline (9). Anaerobic growth on NO3−, TMAO, and fumarate was monitored by cell growth only. Control experiments consisted of incubating strain 200R in SM with a terminal electron acceptor omitted.
Subcloning, nucleotide sequencing, and sequence analysis.
Subcloning of complementing fragment B4 was carried out in broad-host-range cloning vector pBBR1MCS (20) by standard cloning (45) and previously described biparental mating (11) procedures. DNA fragment B4-2S-C (an internal region of complementing fragment B4-2S) was PCR amplified using primers C-forward (TTTAATGCCGGATACTCAAG) and C-reverse (TGACACGACCAATCACAGG) synthesized by Integrated DNA Technologies, Inc. (Coralville, Iowa). PCR mixtures (50-μl total volume) consisted of 20 ng of recombinant pBBR1MCS (containing B4-2S) as template DNA, 250 μM deoxynucleoside triphosphates, 1.5 mM MgCl2, and 50 ng of each oligonucleotide primer with the supplied reaction buffer. Cycling included 95°C for 5 min and 72°C for 5 min, followed by addition of Taq DNA polymerase (2.5 U; Promega) and 35 cycles of 94°C for 1 min 30 s, 47°C for 2 min, and 72°C for 3 min, with a final cycle of 72°C for 10 min. PCR products were extracted from agarose gels using a Qiaex II agarose gel extraction kit (Qiagen) and cloned with a TOPO TA cloning kit (Invitrogen) prior to insertion in pBBR1MCS. Fragment B4-2S was sequenced by the University of Georgia Molecular Genetics Facility using an Applied Biosystems, Inc., model 373A automated sequencer. Open reading frames (ORFs) were identified with GeneMark software (7), and sequence analysis was performed with Gapped BLAST search programs (1). Preliminary Shewanella oneidensis (formerly S. putrefaciens) strain MR-1 genome sequence data were obtained from The Institute for Genomic Research website at http://www.tigr.org.
Gene replacement mutagenesis.
ferE was disrupted in the S. putrefaciens chromosome by insertion of a 1.6-kb gentamicin resistance cassette derived from plasmid pGMΩ1 (47). The disruption strategy entailed a two-step process: (i) construction of an insertionally inactivated ferE construct (designated ferE′) and (ii) replacement of wild-type ferE with ferE′ in the S. putrefaciens chromosome via homologous recombination. ferE-containing DNA fragment B4-2S-C was cloned into suicide vector pRTP1 (encoding ampicillin resistance and streptomycin sensitivity) (50), and the gentamicin resistance cassette was inserted at a unique BamHI restriction site in ferE (see Fig. 4) using standard cloning procedures (45). Recombinant pRTP1 (containing ferE′) was electroporated into Escherichia coli donor strain S17-1 (49) and mobilized into a spontaneous rifamycin- and streptomycin-resistant S. putrefaciens recipient (designated strain 200RS) as described above (11). Recipient strains in which recombinant pRTP1 was integrated into the chromosome by a single crossover event were identified by the acquired resistance to gentamicin and ampicillin. To identify those recipients in which a second crossover event resulted in the loss of pRTP1 (yet retention of ferE′), 1,300 gentamicin-resistant recipients were screened on nutrient agar for acquired sensitivity to ampicillin and resistance to streptomycin, and two gentamicin- and streptomycin-resistant exconjugants (designated strains E726 and E912) were identified. Replacement of ferE with ferE′ in strains E726 and E912 was verified via both PCR amplification of ferE′ and Southern hybridization analysis according to standard procedures (45). The C-forward and C-reverse primers were used to amplify ferE′ from chromosomal DNAs isolated from strains E726 and E912. PCRs were essentially identical to those described above, with E726 and E912 chromosomal DNAs replacing cloned fragment B4-2S.
FIG. 4.
Nucleotide sequence and deduced amino acid sequence of the 2,655-bp PCR product B4-2S-C, which contains the 1,535-bp orf1 (ferE). The deduced amino acid sequence is shown below each gene. Predicted ferE (pulE homolog) signature regions are noted (nucleotide positions): primer C-forward (1 to 20), ferD stop codon (158 to 160), ferE start codon (315 to 317), Walker box A nucleotide binding site (1131 to 1154), two putative CXXC metal binding sites (1530 to 1541 and 1629 to 1640), Walker box B nucleotide binding site (1659 to 1697), BamHI restriction site (1669 to 1674) used as the insertion site for gentamicin resistance cassette disruption, ferE stop codon (1851 to 1853), ferF start codon (1857 to 1859). and primer C-reverse (2637 to 2655). Stop codons are indicated by asterisks.
Isolation of peripherally attached proteins and cell fractionation.
Batch cultures (500 ml) were grown to a target optical density (A600 = 1.0) in a shaker flask without aeration (30°C, pH 7.5) in a defined liquid medium (35) amended with antibiotics as described above. Cells were harvested by centrifugation (4°C) at 10,000 × g and washed once with phosphate-buffered saline solution (11). Growth was halted by chloramphenicol addition prior to harvesting. Cell suspensions were subsequently washed in 0.5 M KCl by previously described procedures (13, 40). Spent wash buffer was filtered through 0.22-μm-pore-size filters, and proteins contained in filtrates were captured on Millipore YM1 disk filters (1-kDa-molecular-mass cutoff). Spent culture medium (collected immediately after centrifugation) was also processed in an identical manner, and little if any protein was detected. For subcellular fractionation, unwashed cell suspensions were lysed by three passes (12,000 lb/in2) through a French pressure cell. Membrane and soluble fractions were separated via centrifugation at 200,000 × g for 1 h. Outer and inner membrane fractions were separated by extraction of total membrane fractions with Triton X-100 (46). To measure contamination between fractions during separation, NADH oxidase activity (2) and 2-keto-3-deoxyoctonate concentration (18) were determined for each cell fraction.
Detection of heme-containing and Fe(III)-reducing proteins.
Proteins containing covalently attached heme or expressing Fe(III) reduction activity were identified by heme and ferrozine staining after separation via denaturing (sodium dodecyl sulfate [SDS]) or nondenaturing (native) polyacrylamide gel electrophoresis (PAGE) (21). Heme (54) and ferrozine (32) staining procedures were adapted from previously described protocols. Heme staining solution consisted of 2.5 mM diaminobenzidine and 0.12% (vol/vol) hydrogen peroxide. Horse heart cytochrome c was used as a positive control in heme staining. Ferrozine staining solution consisted of 15 mM duroquinol (prepared by reducing duroquinone as previously described) (59), 2.5 mM Fe(III) citrate, and 1.5 mM ferrozine. All gel images were captured with a Stratagene Eagle Eye II gel documentation system and were processed with National Institutes of Health Image analysis software.
Nucleotide sequence accession number.
The nucleotide sequence of S. putrefaciens strain 200 genome fragment B4-2S has been deposited in GenBank under accession no. AF188713.
RESULTS
A pulE homolog is required for Fe(III) and Mn(IV) reduction by S. putrefaciens.
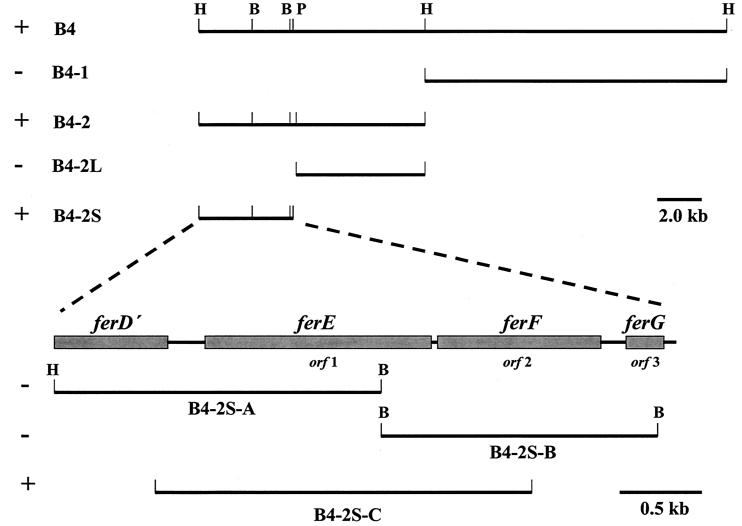
The 23.3-kb complementing fragment B4 was subcloned, and a 4.2-kb BamHI-PstI internal region (B4-2S) was the smallest subclone that conferred the Fe(III) reduction phenotype to mutant B31 (Fig. 1 and 2). Subclone B4-2S was sequenced and found to contain three complete ORFs (Fig. 1), including orf1 (designated ferE, located on subclone B4-2S-C), which was subsequently isolated by PCR amplification and found to confer Fe(III) and Mn(IV) reduction phenotypes (Fer+ and Mnr+, respectively) to B31 (Fig. 2 and 3). The abilities of the wild-type, B31, B31 transconjugate, E726, and E912 strains to reduce amorphous Fe(III) oxide (data not shown) were identical to that reported with Fe(III) citrate. The translated product of orf1 (FerE) (Fig. 4 Table 1) displayed 87% amino acid sequence similarity to the PulE family of proteins found in type II protein secretion systems (42, 44), including ExeE (17) of Fe(III)-reducing (25) A. hydrophila. FerE also displayed moderate to high sequence similarity to putative PulE homologs found in recently sequenced genomes of the Fe(III)-reducing (55) archaeon Archaeoglobus fulgidus (19) (22% identity and 47% similarity) and the Fe(III)- and Mn(IV)-reducing (26, 35) proteobacterium S. oneidensis (formerly S. putrefaciens) (56) strain MR-1 (Table 2). FerE also contained predicted PulE homolog signature regions, including Walker box A and B nucleotide binding sites and two putative CXXC metal binding sites (Fig. 4).
FIG. 1.
Schematic representation of complementing fragment B4 subcloning strategy. +, restores Fe(III) and Mn(IV) reduction capability to B31; −, does not restore Fe(III) and Mn(IV) reduction capability to B31. H, HindIII; B, BamHI; P, PstI. Note the different scale bar with B4-2S subcloning.
FIG. 2.
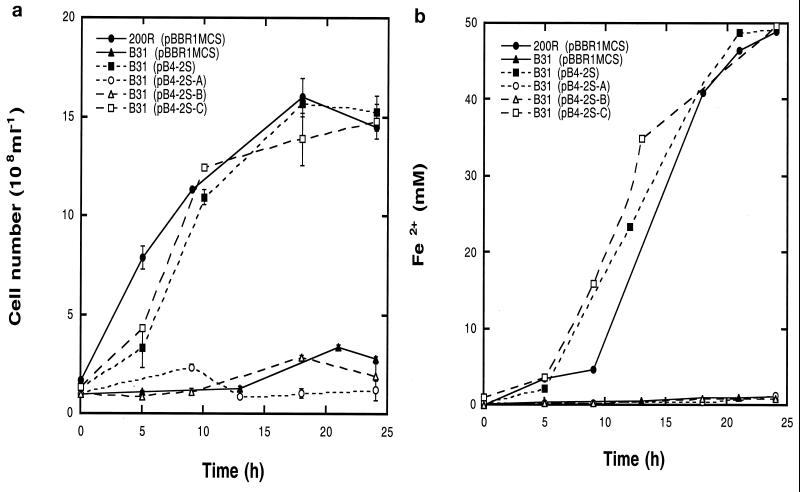
Anaerobic growth on Fe(III) (a) and corresponding Fe2+ production (b) by S. putrefaciens strains 200R(pBBR1MCS) and B31(pBBR1MCS) and a series of B31 transconjugate strains. Error bars indicate standard errors.
FIG. 3.
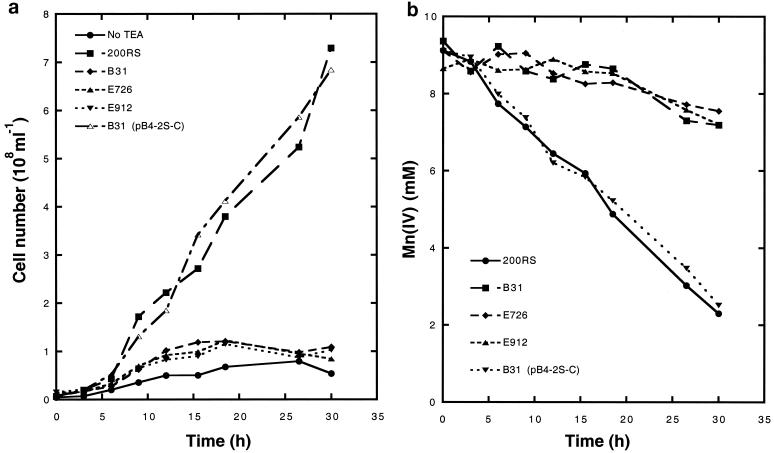
Anaerobic growth on Mn(IV) (a) and corresponding Mn(IV) depletion (b) of S. putrefaciens strains 200RS and Fer− B31, insertional mutants E726 and E912 carrying a disrupted ferE construct, and complemented transconjugate B31(pB4-2S-C). No TEA, no terminal electron acceptor [200RS anaerobic control incubation with Fe(III) omitted].
TABLE 1.
Results of Gapped BLAST protein database searches with S. putrefaciens strain 200 chromosome fragment B4-2S
| Gene | %
|
Best hit | |
|---|---|---|---|
| Identity | Similarity | ||
| ferD | 57 | 70 | Vibrio cholerae EpsD |
| ferE | 75 | 87 | Aeromonas hydrophila ExeE |
| ferF | 56 | 69 | A.hydrophila ExeF |
| ferG | 41 | 55 | Erwinia carotovora OutG |
TABLE 2.
Comparison of pul homologs of S. putrefaciens strain 200 and S. oneidensis strain MR-1
| Gene | %
|
|
|---|---|---|
| Identity | Positive | |
| ferD | 85 | 90 |
| ferE | 84 | 90 |
| ferF | 81 | 89 |
| ferG | 86 | 90 |
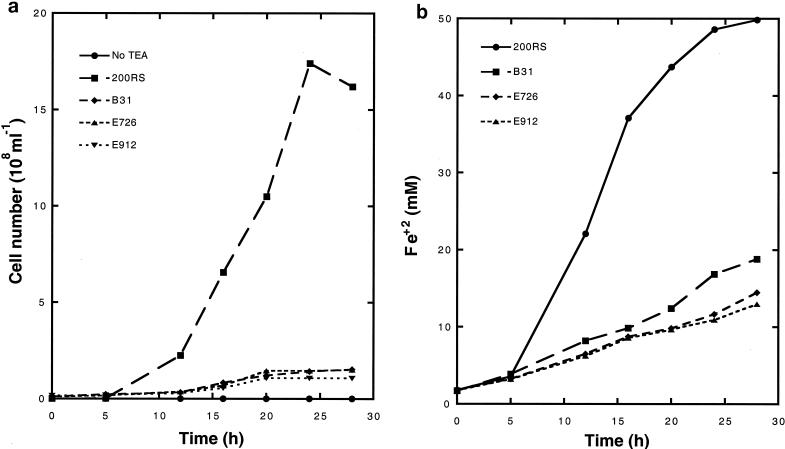
To confirm that ferE was required for anaerobic growth on Fe(III) and Mn(IV), insertional mutants E726 and E912 were constructed by replacing wild-type ferE with an insertionally inactivated ferE construct, ferE′. E726 and E912 were unable to grow anaerobically on Fe(III) (Fig. 5) or Mn(IV) (Fig. 3) yet retained the ability to reduce NO3−, NO2−, TMAO, SO32−, S2O32−, fumarate, Se(IV), and U(VI) (data not shown). Insertion of the 1.6-kb gentamicin resistance cassette in E726 and E912 was verified by Southern hybridization and by noting a 1.6-kb increase in the PCR product amplified by the C-forward and C-reverse primers (data not shown).
FIG. 5.
Anaerobic growth on Fe(III) (a) and corresponding Fe2+ production (b) of S. putrefaciens strains 200RS and Fer− B31, insertional mutants E726 and E912 carrying a disrupted ferE construct, and complemented transconjugate B31(pB4-2S-C). No TEA, no terminal electron acceptor [200RS anaerobic control incubation with Fe(III) omitted].
Other type II protein secretion genes are located proximal to ferE in the S. putrefaciens genome.
To determine if other type II secretion genes were located proximal to ferE on the S. putrefaciens 200 chromosome, upstream and downstream regions of B4-2S were analyzed for predicted protein similarities. The translated product of the 823-nucleotide region immediately upstream of ferE (designated ferD′) displayed strong similarity to the highly conserved carboxy terminus of the PulD superfamily of proteins (14), while downstream, the translated products of orf2 and orf3 (designated ferF and ferG, respectively) displayed strong similarity to PulF and PulG homologs (42, 44), respectively (Table 1). Nucleotide sequence analysis of the S. oneidensis MR-1 genome indicated that the translated products of pulD to -G homologs of strain MR-1 displayed 90% sequence similarity to ferD to -G, respectively, of strain 200 (Table 2). In addition, a contiguous cluster of 12 putative type II protein secretion genes (homologs of pulC to -N), including the ferE homolog, was found in the unannotated MR-1 genome (data not shown). Sequence analysis upstream of the pulC homolog in the MR-1 genome revealed the absence of the pullulanase structural gene (pulA) homolog that normally occupies this position in pullulanase-secreting Klebsiella oxytoca (42, 44, 58).
Identification of peripherally attached proteins displaying Fe(III) reduction activity.
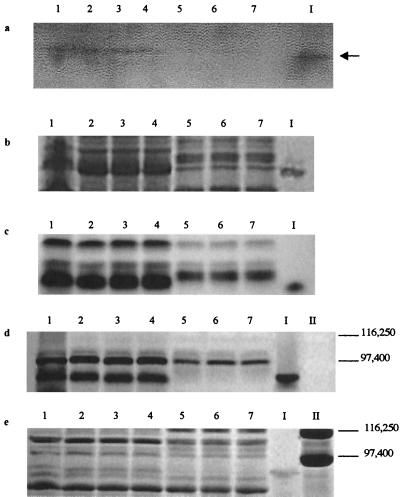
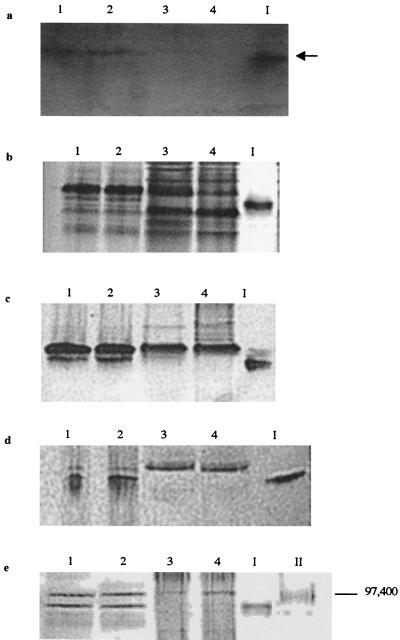
Weakly bound peripheral proteins were detached from the surface of the wild-type, B31, and B31 transconjugate strains and analyzed for Fe(III) reduction activity in native gels. A single band displaying strong Fe(III) reduction activity was detected in the peripheral proteins of the wild-type and Fer+ B31 transconjugate cells but was missing from the B31 and the Fer− B31 transconjugate cells (Fig. 6a). The Fe(III)-reducing wild-type band was excised from an unstained native gel and included as a marker in all gels. Coomassie blue (Fig. 6b) and heme (Fig. 6c) staining of otherwise identical native gels revealed that a heme-positive band similar in size to the Fe(III)-reducing band was missing from the peripheral proteins of all Fer− strains yet was detected with all Fer+ strains. A similar set of native gel-based analyses were done with peripheral proteins detached from insertionally inactivated ferE (and Fer−) mutant E912, and an identical set of results were obtained: a single band displaying strong Fe(III) reduction activity was detected in the peripheral proteins of the wild type and all Fer+ cells yet was missing from Fer− E912 (Fig. 7a). Coomassie blue (Fig. 7b) and heme (Fig. 7c) staining of otherwise identical native gels revealed that a heme-positive band nearly identical in size to the Fe(III)-reducing band was missing from the peripheral proteins of Fer− E912 yet was detected in all Fer+ strains. Analogous native gel-based Mn(IV) reduction activity stains were not possible, because all artificial electron donors tested (methyl viologen, benzyl viologen, dithionite, ascorbate, and duroquinol) rapidly reduced Mn(IV) chemically (thereby masking any enzymatic activity) (data not shown).
FIG. 6.
Separation of peripheral proteins via native PAGE with ferrozine staining (5 min) (a), Coomassie blue staining (b), and heme staining (c) and via SDS-PAGE with heme staining (d) and Coomassie blue staining (e). Lanes contain peripheral proteins washed from whole cells of Fe(III)-grown 200R(pBBR1MCS) (lanes 1), aerobically grown 200R(pBBR1MCS) (lanes 2), Fer+ transconjugates B31(pB4-2S) (lanes 3) and B31 (pB4-2S-C) (lanes 4), Fer− B31(pBBR1MCS) (lanes 5), and Fer− transconjugates B31(pB4-2S-A) (lanes 6) and B31(pB4-2S-B) (lanes 7). The 91-kDa heme-containing band displaying Fe(III) reduction activity was excised from an unstained native gel and is included as a marker in lane I of all gels. Lanes II contain SDS-PAGE molecular mass standards (Bio-Rad) E. coli β-galactosidase (116,250 kDa) and rabbit muscle phosphorylase b (97,400 Da). The arrow indicates a band displaying Fe(III) reduction activity (the black color on the image corresponds to magenta-colored Fe2+-ferrozine complex on the actual gel).
FIG. 7.
Separation of peripheral proteins of insertional mutant E912 via native PAGE with ferrozine staining (5 min) (a), Coomassie blue staining (b), and heme staining (c) and via SDS-PAGE with heme staining (d) and Coomassie blue staining (e). Lanes contain peripheral proteins washed from whole cells of 200R(pBBR1MCS) (lanes 1), complemented transconjugate B31(pB4-2S-C) (lanes 2), Fer− transconjugate B31(pBBR1MCS) (lanes 3), and insertional mutant E912 (lanes 4). The 91-kDa heme-containing band displaying Fe(III) reduction activity was excised from an unstained native gel and is included as a marker in lane I of all gels. Lanes II contain SDS-PAGE molecular mass standards Bio-Rad) E. coli β-galactosidase (116,250 Da) and rabbit muscle phosphorylase b (97,400 Da). The arrow indicates a band displaying Fe(III) reduction activity (the black color on the image corresponds to magenta-colored Fe2+-ferrozine complex on the actual gel).
Polypeptide profile and heme content of the peripherally attached Fe(III)-reducing band.
SDS-PAGE analyses indicated that the excised Fe(III)-reducing band contained a single polypeptide with an apparent molecular mass of 91 kDa (Fig. 6e, lane I), migrated to the corresponding location of a polypeptide missing from the B31 and Fer− B31 transconjugate strains (Fig. 6d and e), and stained heme positive (Fig. 6d, lane I). A similar set of SDS-PAGE-based analyses were done with peripheral proteins detached from insertional mutant E912, and an identical set of results were obtained: a single heme-positive polypeptide was detected in peripheral proteins from wild-type and Fer+ cells but was not detected with E912 (Fig. 7d). Coomassie blue staining of an otherwise identical SDS gel revealed that a polypeptide nearly identical in size to the 91-kDa heme-containing polypeptide was missing from the proteins peripherally attached to E912 yet was detected with the Fer+ strains (Fig. 7e). The 91-kDa heme-containing protein appeared to be present in both aerobically and anaerobically [Fe(III) citrate] grown S. putrefaciens cells: All native PAGE and SDS-PAGE analyses indicated that Fe(III)-grown wild-type cells (Fig. 6, lanes 1) contained a peripheral, heme-containing protein that displayed Fe(III) reduction activity and comigrated with the peripheral, 91-kDa heme-containing protein of aerobically grown cells (Fig. 6, lanes 2).
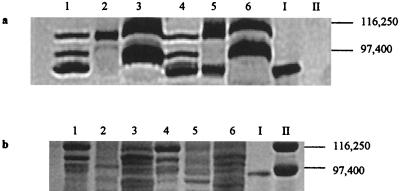
The 91-kDa heme-containing protein appears to be localized to the outer membrane fraction of wild-type S. putrefaciens.
Heme-containing proteins were found in all wild-type fractions analyzed via SDS-PAGE with heme staining (Fig. 8a, lanes 1 to 3). However, the previously excised 91-kDa heme-containing protein (Fig. 8a, lane I) comigrated with a heme-containing polypeptide found largely in the outer membrane fraction (Fig. 8a, lane 1) and not the inner membrane (Fig. 8a, lane 2) or soluble (Fig. 8a, lane 3) fraction of the wild-type strain. Identical results were obtained with Coomassie blue-stained SDS gels (Fig. 8b, lanes 1 to 3, I). NADH oxidase activity and 2-keto-3-deoxyoctonate concentration measurements for each fraction indicated that contamination between the soluble, inner membrane, and outer membrane fractions was 5 to 10% for both strains 200R and B31 (data not shown).
FIG. 8.
Separation of subcellular fractions of wild-type S. putrefaciens 200R and Fer− B31 via SDS-PAGE with heme (a) and Coomassie blue (b) staining. Lanes correspond to 200R outer membrane (lanes 1), 200R inner membrane (lanes 2), 200R soluble fraction (lanes 3), B31 outer membrane (lanes 4), B31 inner membrane (lanes 5), and B31 soluble fraction (lanes 6). The 91-kDa heme-containing band displaying Fe(III) reduction activity was excised from an unstained native gel and is included as a marker in lane I of both gels. Lanes II contain SDS-PAGE molecular mass standards (Bio-Rad) E. coli β-galactosidase (116,250 Da) and rabbit muscle phosphorylase b (97,400 Da).
Identical analyses of B31 subcellular fractions were carried out to determine the subcellular location of the 91-kDa heme-containing protein in a ferE mutant background. As with the wild-type strain, heme-containing proteins were detected in all B31 subcellular fractions (Fig. 8a, lanes 4 to 6). Interestingly, a heme-containing protein that comigrated with the excised 91-kDa heme-containing protein (Fig. 8a, lane I) was detected in both the inner (Fig. 8a, lane 4) and outer (Fig. 8a, lane 5) membrane fractions but not in the soluble fraction (Fig. 8a, lane 6) of B31. Identical results were obtained with Coomassie blue-stained SDS gels (Fig. 8b, lanes 4 to 6, I).
DISCUSSION
Neutrophilic Fe(III)- and Mn(IV)-reducing microorganisms are presented with a unique physiological challenge: they are required to generate energy via electron transfer to highly insoluble terminal electron acceptors. To overcome this problem, Fe(III)- and Mn(IV)-reducing microorganisms have been postulated to excrete electron shuttles which reduce Fe(III) extracellularly in a secondary chemical reaction (39, 48) or localize Fe(III) and Mn(IV) terminal reductases to the outer membrane and directly transfer electrons to insoluble metals in contact with the cell surface (4, 13, 33, 34). Type II protein secretion systems are used by a variety of gram-negative bacteria to translocate exoproteins into and across the outer membrane (42, 44, 58). The present study demonstrates that ferE, a homolog of the pulE type II protein secretion gene, is required for dissimilatory Fe(III) and Mn(IV) reduction by S. putrefaciens: a ferE-containing recombinant plasmid confers wild-type Fe(III) and Mn(IV) reduction capability to mutant B31, and insertional mutants (E726 and E912) carrying a disrupted ferE gene are unable to grow anaerobically on Fe(III) or Mn(IV). ferE appears to be part of a complete type II secretion system, since other putative type II secretion genes are found upstream and downstream of ferE in the S. putrefaciens strain 200 chromosome and a contiguous cluster of 12 putative type II secretion genes is found clustered on the S. oneidensis MR-1 genome. Type II secretion systems are part of the main terminal branch of the sec-dependent general secretory pathway (42, 44, 58) and are generally comprised of 12 to 14 proteins encoded by a contiguous cluster of moderately to highly conserved pul (or gsp) genes, almost invariably in the same order.
Pullulanase secretion by the plant cell wall-degrading microorganism K. oxytoca is one of the best characterized type II secretion systems, and a working model for pullulanase secretion has been proposed (44). Nascent pullulanase is first directed into and across the cytoplasmic membrane, where it folds and is transiently anchored to the periplasmic aspect of the cytoplasmic membrane. After processing (signal peptide cleavage, disulfide bond formation, and fatty acylation), the mature pullulanase is guided across the periplasmic space by the type II secretion apparatus and interacts with the outer membrane-associated, multimeric PulD channel. Pullulanase is subsequently attached to the outside face of the outer membrane via a fatty acid tail. The ferE homolog pulE is postulated to encode a secretion ATPase that drives the entire secretion process. Peripherally attached pullulanase cleaves alpha-1,6 linkages in branched maltodextrin polymers such as glycogen or amylopectin and releases linear dextrins for cellular uptake and metabolism. Based on the K. oxytoca type II pullulanase secretion model and the previously reported involvement of S. putrefaciens outer membrane proteins in dissimilatory Fe(III) and Mn(IV) reduction (6, 13, 16, 29, 34, 36, 48), we postulate that the Fer− and Mnr− phenotypes of B31, E726, and E912 are due to their inability to secrete Fe(III) and Mn(IV) terminal reductases to the outside face of the S. putrefaciens outer membrane.
To investigate this possibility, the Fe(III)-reducing protein profiles of subcellular fractions of the S. putrefaciens wild-type and respiratory mutant strains were compared. A 91-kDa heme-containing protein with Fe(III) reduction activity is found peripherally attached to the wild-type and Fer+ B31 transconjugate strains yet is missing from this location in Fer− mutants E912 and B31 and Fer− B31 transconjugate strains. Interestingly, a 91-kDa heme-containing protein appears to be associated largely with the outer membrane fraction of the wild-type strain yet is detected with both the inner and outer membrane fractions of mutant B31. Confirmation of the physiological location of the 91-kDa heme-containing protein in the wild type and ferE mutants will require Western blot analysis of S. putrefaciens membrane and soluble fractions using antibodies directed against the purified 91-kDa protein. If our findings are confirmed, then a defect in ferE results in accumulation of the 91-kDa protein at the inner membrane with partial transport to the outer membrane. This phenotype is also observed in K. oxytoca, where the type II secretion system transports the entire pool of pullulanase to the wild-type cell surface, while defects in one or more type II proteins result in accumulation of pullulanase at the inner membrane with partial (up to 20%) transport to the cell surface (43, 44). If the pool of 91-kDa protein is partially transported to the outer membrane of ferE mutant B31 and E912, it may not be properly exposed at the cell surface, since a protein of similar size is not detected (by heme or ferrozine staining) in the set of peripheral proteins weakly bound to the mutant cell surfaces.
Our preliminary results suggest that a type II secretion system in S. putrefaciens may be required to efficiently target the 91-kDa heme-containing protein to the outside face of the outer membrane, where it is involved in dissimilatory Fe(III) reduction. The strategy of linking Fe(III) reduction and type II secretion is logical, since type II secretion systems are able to translocate exoproteins in their fully mature form to the outside face of outer membranes, where they may contact extracellular substrates and be fully active without further maturation (42, 44, 58). This strategy also circumvents problems associated with extracellular secretion of electron shuttles, a mechanism that is predicted to be energetically wasteful (22).
The 91-kDa heme-containing protein may also be involved in Mn(IV) reduction, since ferE confers Mn(IV) reduction capability to B31 and insertional mutants carrying disrupted ferE constructs are unable to respire on Mn(IV). However, a native gel-based Mn(IV) reduction activity stain (analogous to the ferrozine stain used in the present study) is not available to test the Mn(IV) reduction activity of the 91-kDa heme-containing protein (see above). In addition, it is possible that defects in ferE block transport of not only the 91-kDa heme-containing protein but also other outer membrane proteins (e.g., MtrB, OmcA, and OmcB) (6, 36) essential for anaerobic growth on Fe(III) and Mn(IV). Further work to examine these possibilities will include (i) purification of the 91-kDa heme-containing protein and tests of the purified enzyme for Fe(III) and Mn(IV) reduction activities, (ii) targeted disruption of the gene encoding the 91-kDa heme-containing protein and tests of the resulting insertional mutant for anaerobic growth on Fe(III) and Mn(IV), and (iii) subcellular localization of MtrB, OmcA, and OmcB in the ferE mutants B31 and E912.
It will be interesting to compare the nucleotide and amino acid sequence homologies of the 91-kDa protein involved in metal reduction in S. putrefaciens with analogous proteins found in other Fe(III)- and Mn(IV)-reducing microorganisms, including other Shewanella species, members of the strict anaerobic family Geobacteracea, and hyperthermophilic Bacteria and Archaea most closely related to the last common ancestor (55). Functional analysis of genes associated with dissimilatory Fe(III) and Mn(IV) reduction may provide model systems with which to study the evolutionary history of anaerobic respiratory processes in all microorganisms.
Acknowledgments
We thank Mark Barrett, Jessica Pritchard, and Gladys Alexandre for laboratory assistance.
This work was supported by grants from the Office of Naval Research (Biosystems Science and Technology Program), the National Science Foundation (Environmental Geochemistry/Biogeochemistry and Graduate Research Traineeship Programs), and the Nelson and Bennie Abell Professorship in Biology at Georgia Tech. Sequencing of the S. oneidensis MR-1 genome was accomplished with support from the Department of Energy.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Zhang, Z., W. Miller, and S. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anwar, H., M. R. Brown, R. M. Cozens, and P. A. Lambert. 1983. Isolation and characterization of the outer and cytoplasmic membranes of Pseudomonas cepacia. J. Gen. Microbiol. 129:499–507 [DOI] [PubMed] [Google Scholar]
- 3.Arnold, R. G., T. J. DiChristina, and M. R. Hoffmann. 1986. Inhibitor studies of dissimilatory Fe(III) reduction by Pseudomonas putrefaciens sp. strain 200 (“Pseudomonas ferrireducens”). Appl. Environ. Microbiol. 52:281–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnold, R. G., T. J. DiChristina, and M. R. Hoffmann. 1988. Reductive dissolution of Fe(III) oxides by Pseudomonas species 200. Biotechnol. Bioeng. 32:1081–1096 [DOI] [PubMed] [Google Scholar]
- 5.Beard, B. L., C. M. Johnson, L. Cox, H. Sun, K. H. Nealson, and C. Aquilar. 1999. Iron isotope biosignatures. Science 285:1889–1892 [DOI] [PubMed] [Google Scholar]
- 6.Beliaev, A. S., and D. A. Saffarini. 1998. Shewanella putrefaciens MtrB encodes an outer membrane protein required for Fe(III) and Mn(IV) reduction. J. Bacteriol. 180:6292–6297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borodovsky, M., and J. McIninch. 1993. GeneMark: parallel gene recognition for both DNA strands. Comp. Chem. 17:123–133 [Google Scholar]
- 8.Burnes, B. S., M. J. Mulberry, and T. J. DiChristina. 1998. Design and application of two rapid screening techniques for isolation of Mn(IV) reduction-deficient mutants of Shewanella putrefaciens. Appl. Environ. Microbiol. 64:2716–2720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cline, J. D. 1969. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol. Oceanogr. 14:454–458 [Google Scholar]
- 10.DiChristina, T. J., R. G. Arnold, M. E. Lidstrom, and M. R. Hoffmann. 1988. Dissimilative iron reduction by the marine eubacterium Alteromonas putrefaciens strain 200. Wat. Sci. Technol. 20:69–79 [Google Scholar]
- 11.DiChristina, T. J., and E. F. DeLong. 1994. Isolation of anaerobic respiratory mutants of Shewanella putrefaciens and genetic analysis of mutants deficient in anaerobic growth on Fe(III). J. Bacteriol. 176:1468–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehrlich, E. A. 1996. Geomicrobiology. Marcel Dekker Inc., New York, N.Y.
- 13.Gaspard, S., F. Vazquez, and C. Holliger. 1998. Localization and solubilization of the iron(III) reductase of Geobacter sulfurreducens. Appl. Environ. Microbiol. 64:3188–3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Genin, S., and C. A. Boucher. 1994. A superfamily of proteins involved in different secretion pathways in gram-negative bacteria: modular structure and specificity of the N-terminal domain. Mol. Gen. Genet. 243:112–118 [DOI] [PubMed] [Google Scholar]
- 15.Gennis, R. B., and V. Stewart. 1996. Respiration, p.217–261. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 1. ASM Press, Washington, D.C
- 16.Gordon, H. J., A. D. Pike, A. E. Hill, P. M. Cuthbertson, S. K. Chapman, and G. A. Reid. 2000. Identification and characterization of a novel cytochrome c3 from Shewanella frigidimarina that is involved in Fe(III) respiration. Biochem. J. 349:153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang, B., and S. P. Howard. 1992. The Aeromonas hydrophila exeE gene, required both for protein secretion and normal outer membrane biogenesis, is a member of a general secretion pathway. Mol. Microbiol. 6:1351–1361 [DOI] [PubMed] [Google Scholar]
- 18.Karkhanis, Y. D., J. Y. Zeltner, J. J. Jackson, and D. J. Carlo. 1978. A new and improved microassay to determine 2-keto-3-deoxyoctonate. Anal. Biochem. 85:595–601 [DOI] [PubMed] [Google Scholar]
- 19.Klenk, H.-P., et al. 1997. The complete genome sequence of Archaeoglobus fulgidus. Nature 390:364–370 [DOI] [PubMed] [Google Scholar]
- 20.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop, and K. M. Peterson. 1994. pBBR1MCS: a broad host range cloning vector. BioTechniques 16:800–802 [PubMed] [Google Scholar]
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 22.Lloyd, J. R., E. L. Blunt-Harris, and D. R. Lovley. 1999. The periplasmic 9.6-kDa c-type cytochrome is not an electron shuttle to Fe(III). J. Bacteriol. 181:7647–7649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lovley, D. R. 1991. Dissimilatory Fe(III) and Mn(IV) reduction. Microbiol. Rev. 55:259–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lovley, D. R., J. D. Coates, E. L. Blunt-Harris, E. J. P. Phillips, and J. C. Woodward. 1996. Humic substances as electron acceptors for microbial respiration. Nature 382:445–448 [Google Scholar]
- 25.Lovley, D. R., J. D. Coates, D. A. Saffarini, and D. J. Lonergan. 1997. Dissimilatory Fe(III) reduction, p.187–219. In G. Winkelman and C. J. Carrano (ed.), Transition metals in microbial metabolism. Harwood Academic Publishers, Amsteldjik, The Netherlands
- 26.Lovley, D. R., and E. J. Phillips. 1988. Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron and manganese Appl. Environ. Microbiol. 54:1472–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luther, G. W., D. T. Ruppel, and C. Burkhard. 1999. Reactivity of dissolved Mn(III) species and Mn(IV) species with reductants: Mn redox chemistry without a dissolution step, p.265–280. In D. Sparks and T. J. Grundl (ed.), Mineral-water interfacial reaction kinetics and mechanism, vol. 715. American Chemical Society, Washington, D.C
- 28.Madigan, M. T., J. M. Martinko, and J. Parker. 2000. Brock biology of microorganisms, 9th ed. Simon & Schuster, Upper Saddle River, N.J
- 29.Magnuson, T. S., A. L. Hodges-Myerson, and D. R. Lovley. 2000. Characterization of a membrane-bound NADH-dependent Fe(III) reductase from the dissimilatory Fe(III)-reducing bacterium Geobacter sulfurreducens. FEMS Microbiol. Lett. 185:205–211 [DOI] [PubMed] [Google Scholar]
- 30.Meloan, C. E., P. Holkeboer, and W. Brandt. 1960. Spectrophotometric determination of uranium with benzohydroxamic acid in 1-hexanol. Anal. Chem. 32:791–793 [Google Scholar]
- 31.Montgomery, H. A. C., and J. F. Dymock. 1961. The determination of nitrite in water. Analyst 86:414–416 [Google Scholar]
- 32.Moody, M. D., and H. A. Dailey. 1983. Aerobic ferrisiderophore reductase assay and activity stain for native polyacrylamide gels. Anal. Biochem. 134:235–239 [DOI] [PubMed] [Google Scholar]
- 33.Myers, C. R., and J. M. Myers. 1992. Localization of cytochromes to the outer membrane of anaerobically grown Shewanella putrefaciens MR-1. J. Bacteriol. 174:3429–3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Myers, C. R., and J. M. Myers. 1997. Outer membrane cytochromes of Shewanella putrefaciens MR-1: spectral analysis and purification of the 83-kDa c-type cytochrome. Biochim. Biophys. Acta 1326:307–318 [DOI] [PubMed] [Google Scholar]
- 35.Myers, C. R., and K. H. Nealson. 1988. Bacterial manganese reduction and growth with manganese oxide as the sole terminal electron acceptor. Science 240:1319–1321 [DOI] [PubMed] [Google Scholar]
- 36.Myers, J. M., and C. R. Myers. 2001. Role for outer membrane cytochromes OmcA and Omc B of Shewanella putrefaciens MR-1 in reduction of manganese dioxide. Appl. Environ. Microbiol. 67:260–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nealson, K. H., and D. A. Saffarini. 1994. Iron and manganese in anaerobic respiration: environmental significance, physiology and regulation. Annu. Rev. Microbiol. 48:311–343 [DOI] [PubMed] [Google Scholar]
- 38.Nevin, K. P., and D. R. Lovley. 2000. Potential for enzymatic reduction of Fe(III) via electron shuttling in subsurface sediments. Environ. Sci. Technol. 34:2472–2478 [Google Scholar]
- 39.Newman, D. K., and R. Kolter. 2000. A role of excreted quinones in extracellular electron transfer. Nature 405:94–97 [DOI] [PubMed] [Google Scholar]
- 40.Ohlendieck, K. 1996. Extraction of membrane proteins., p.293–304. In S. Doonan (ed.), Protein purification protocols. Humana Press, Inc., Totowa, N.J
- 41.Perez-Benito, J. F., and E. Brillas. 1989. Identification of a soluble form of colloidal manganese(IV). Inorg. Chem. 28:390–392 [Google Scholar]
- 42.Pugsley, A. P. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57:50–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pugsley, A. P., N. Bayan, and N. Sauvaonnet. 2001. Disulfide bond formation in secretion component PulK provides a possible explanation for the role of DsbA in pullulanase secretion. J. Bacteriol. 183:1312–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pugsley, A. P., O. Francetic, K. Hardie, O. M. Possot, N. Sauvonnet, and A. Seydel. 1997. Pullulanase: model protein substrate for the general secretory pathway of gram-negative bacteria. Fol. Microbiol. 42:184–192 [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y
- 46.Schnaitman, C. A. 1971. Solubilization of the cytoplasmic membrane of Escherichia coli by Triton X-100. J. Bacteriol. 108:545–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schweizer, H. P. 1993. Small broad-host-range gentamycin resistance cassettes for site-specific insertion and deletion. BioTechniques 15:831–833 [PubMed] [Google Scholar]
- 48.Seeliger, S., R. Cord-Ruwisch, and B. Schink. 1998. A periplasmic and extracellular c-type cytochrome of Geobacter sulfurreducens acts as a ferric iron reductase and as an electron carrier to other acceptors or to partner bacteria. J. Bacteriol. 180:3686–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784–791 [Google Scholar]
- 50.Stibitz, S., W. Black, and S. Falkow. 1986. The construction of a cloning vector designed for gene replacement in Bordetella pertussis. Gene 50:133–140 [DOI] [PubMed] [Google Scholar]
- 51.Stookey, L. L. 1970. Ferrozine: a new spectrophotometric reagent for iron. Anal. Chem. 42:779–782 [Google Scholar]
- 52.Stumm, W., and J. J. Morgan. 1996. Aquatic chemistry: chemical equilibria and rates in natural waters. John Wiley & Sons, New York, N.Y
- 53.Taratus, E. M., S. G. Eubanks, and T. J. DiChristina. 2000. Design and application of a rapid screening technique for isolation of selenite reduction-deficient mutants of Shewanella putrefaciens. Microbiol. Res. 155:79–85 [DOI] [PubMed] [Google Scholar]
- 54.Thomas, P. E., D. Ryan, and W. Levin. 1976. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 75:168–176 [DOI] [PubMed] [Google Scholar]
- 55.Vargas, M., K. Kashevi, E. L. Blunt-Harris, and D. R. Lovley. 1998. Microbial evidence for Fe(III) reduction on early Earth. Nature 395:705–724 [DOI] [PubMed] [Google Scholar]
- 56.Venkateswaran, K., D. P. Moser, M. E. Dollhopf, D. P. Lies, D. A. Saffarini, B. J. MacGregor, D. B. Ringelberg, D. C. White, M. Nishijima, H. Sano, J. Burghardt, E. Stackebrandt, and K. H. Nealson. 1999. Polyphasic taxonomy of the genus Shewanella and description of Shewanella oneidensis sp. nov. Int. J. Syst. Bacteriol. 49:705–724 [DOI] [PubMed] [Google Scholar]
- 57.Wade, R., and T. J. DiChristina. 2000. Isolation of U(VI) reduction-deficient mutants of Shewanella putrefaciens. FEMS Microbiol. Lett. 184:143–148 [DOI] [PubMed] [Google Scholar]
- 58.Wandersman, C. 1996. Secretion across the bacterial outer membrane., p.955–966. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 1. ASM Press, Washington, D.C
- 59.Zahn, J. A., and A. A. DiSpirito. 1996. Membrane-associated methane monooxygenase from Methylococcus capsulatus (Bath). J. Bacteriol. 178:1018–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]