Abstract
The schema of the previously described Escherischia coli database coliBASE has been applied to a number of other bacterial taxa, under the collective name xBASE. The new databases include CampyDB for Campylobacter, Helicobacter and Wolinella; PseudoDB for pseudomonads; ClostriDB for clostridia; RhizoDB for Rhizobium and Sinorhizobium; and MycoDB, for Mycobacterium, Streptomyces and related organisms. The databases provide user friendly access to annotation and genome comparisons through a web-based graphical interface. Newly developed features include whole genome displays, ‘painting’ of genes according to properties such as GC content, a pattern search system to identify conserved motifs and batch BLAST searching of every protein encoded by a region. Examples of how the databases have been, and continue to be, used to generate hypotheses for subsequent laboratory investigation are presented. xBASE is available online at http://xbase.bham.ac.uk.
INTRODUCTION
coliBASE, a database for comparative genomics of Escherichia coli and related genera, has previously been described (1). Here we present a number of recent developments to the database schema and user interface that expand the potential applications of the system. Additionally, the database schema has been applied to a number of other taxa to produce a family of databases, providing the powerful tools that have been developed to other bacterial research communities. The databases are collected together under the generic name xBASE at http://xbase.bham.ac.uk.
THE xBASE DATABASES
The xBASE collection includes six databases, the previously presented coliBASE, CampyDB and ClostriDB together with RhizoDB, MycoDB and PseudoDB. The genera included in each database are summarized in Table 1. The schema was originally designed to include complete annotated genomes and unannotated data from incomplete genome projects. The unfinished genomes are often in multiple contigs and are subjected to automated gene prediction using Glimmer (2). The schema has been relaxed to a more generalized model to allow the inclusion of data from complete but unannotated genomes, such as E.coli 042, and incomplete but annotated genomes such as Campylobacter coli RM2228 (3). The databases store annotation derived from GenBank and Uniprot entries, together with codon usage data obtained using CodonW (http://codonw.sourceforge.net) (4). Comparative data include whole genome alignments, as determined using MUMmer and PROmer (4), and putative orthologues, as determined using a mutual best BLAST hits approach. The databases are accessed through a web-based interface that has been designed to be accessible to laboratory biologists who may have limited bioinformatics skills.
Table 1.
Component databases of xBASE, and the genera for which genome sequence data are currently included
| Database | Included genera | URL |
|---|---|---|
| coliBASE | Escherichia/Shigella | http://colibase.bham.ac.uk |
| Salmonella | ||
| Citrobacter | ||
| Photorhabdus | ||
| Proteus | ||
| Serratia | ||
| Erwinia | ||
| Yersinia | ||
| Blochmannia | ||
| Buchnera | ||
| Wigglesworthia | ||
| CampyDB | Campylobacter | http://campy.bham.ac.uk |
| Helicobacter | ||
| Wolinella | ||
| ClostriDB | Clostridium | http://clostri.bham.ac.uk |
| PseudoDB | Pseudomonas | http://pseudo.bham.ac.uk |
| MycoDB | Mycobacterium | http://myco.bham.ac.uk |
| Streptomyces | ||
| Corynebacterium | ||
| Tropheryma | ||
| RhizoDB | Rhizobium | http://rhizo.bham.ac.uk |
| Sinorrhizobium | ||
| Mesorhizobium | ||
| Bradyrhizobium | ||
| Bartonella | ||
| Brucella | ||
| Rhodopseudomonas |
RECENT DEVELOPMENTS
The previously described search methods for locating genes by annotation, homology or coordinate have been supplemented with an additional ‘Pattern Search’. This is intended for the identification of conserved DNA motifs such as transcription factor binding sites, based on a (possibly degenerate) consensus pattern. The search can be restricted to intragenic or intergenic regions, or within a set region upstream of a gene. The pattern matching is performed using fuzznuc, part of the EMBOSS suite, and can be relaxed to allow a specified number of mismatches. The coordinates of any matches are reported, along with their genetic context. This tool has recently been applied to identify potential targets for the Campylobacter jejuni regulator NssR (5).
The inclusion of unfinished genomes allows such data, which can otherwise be left languishing on FTP sites for prolonged periods during the assembly and annotation process, to prompt laboratory-based studies (see e.g. Ref. 6). Genome alignments can be used to identify the Glimmer predicted ORFs, but this approach is not useful for novel gene clusters. To remedy this situation we have developed a tool that will BLAST all the proteins encoded in a region against the NCBI non-redundant protein database. When the results are ready the top hits can be quickly inspected by the same ‘mouseover’ technique used to display annotation in xBASE. This approach can also be applied to annotated genomes, to facilitate reannotation in cases where the published annotation is inaccurate or outdated (as in Ref. 7).
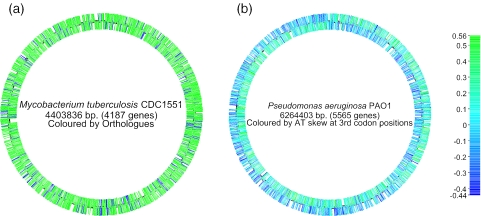
The xBASE databases now include a facility to visualize a complete chromosome or plasmid using the Genome Browser tool (see Figure 1). This image can be ‘painted’ using a variety of criteria, including GC content, GC skew, codon adaptation index (CAI), or according to the presence or absence of orthologues of a gene within selected other genomes from the database. This functionality is similar to that provided by the CBS Genome Atlas database (11); however integration with the rest of the database means that our system is dynamic. The genome map is clickable, and it is possible to zoom in on any region of interest that has been highlighted. The gene visualization tools have been updated to also support alternate colour schemes, so that the ‘painting’ is maintained when moving between different views.
Figure 1.
Whole genome displays generated by xBASE. (a) The genome of Mycobacterium tuberculosis CDC1551 (8), coloured according to the presence (red) or absence (blue) of orthologues in Mycobacterium bovis AF2122/97 (9). This display illustrates the regions of the M.tuberculosis backbone that have been deleted during the evolution of M.bovis. (b) The genome of Pseudomonas aeruginosa PAO1 (10), coloured by AT skew (A−T/A+T) at synonymous third codon positions. This colour scheme reflects an asymmetric mutation bias during chromosomal replication. The ‘switch’ from leading to lagging strand at the origin and terminus of replication can be seen. They are not directly opposite due to a large 2.2 Mb inversion that spans the replication origin of the P.aeruginosa genome.
xBASE IN ACTION
The versatility of the xBASE gene painting and tools means that it is easy to gather and visualize evidence from several lines of investigation. One cogent example is the detection of horizontally transferred genes in E.coli, in particular genes that encode targets of type-III secretion systems. These systems translocate so-called ‘effector proteins’ into the cytoplasm of eukaryotic cells, subverting cellular processes to the bacterium's advantage. Although there is no obvious signature in the sequence of the effector proteins that makes them instantly recognizable, effector genes show several characteristics that help distinguish them from E.coli house-keeping genes: they show evidence of horizontal genes transfer (including a lower than average GC content), they tend to cluster with other effector genes and their protein products show homology to proven translocated effectors from other species or systems.
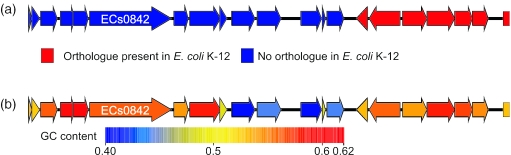
Figure 2 shows two views of a 20 kb section of the genome of a strain of enterohaemorrhagic E.coli (EHEC) O157:H7, centred on the gene ECs0847. In the first view (Figure 2a), genes are painted according to whether they have orthologues in the laboratory strain K-12. The run of blue to the left highlights genes present in EHEC and absent from K-12, while the run of red to the right highlights genes common to both strains (a similar conclusion can be reached by visualizing the MUMmer comparison between the two genomes). Mousing over the blue genes quickly, to call up their descriptions, identifies them as one end of an EHEC-specific prophage. However, in this view it is impossible to distinguish genes involved in phage replication from ‘passenger genes’ that could have hitched a lift on the phage but play no role in phage replication. However, when genes are instead painted by GC content (Figure 2b), it immediately becomes obvious that four substantial genes stand out in green as separate from both the phage replication genes to their left and the house-keeping genes to their right. BLAST searches launched against the NCBI database using the protein sequences encoded by these genes identifies homology to known type-III-secreted proteins from other pathogens, while coliBLAST searches quickly identify homologues in several as yet unpublished genomes of related organisms. With several lines of evidence identifying them as likely effectors, they thus become good candidates for experimental investigation.
Figure 2.
Use of the ‘gene painting’ facility of xBASE to highlight potential type-III secretion effectors that are passenger genes within an E.coli O157:H7 specific prophage. (a) Illustrates the boundary of the phage, as determined by comparison with the E.coli K12 MG1655 genome. (b) Highlights four large genes within the prophage that have a low GC content (blue) and represent putative passenger genes. Homology searches indicate that these genes are potential type-III effectors.
FUTURE DIRECTIONS
The facilities described here and in our previous paper (1) represent the completion of xBASE version 1. We are currently working on the second version, intended for release in early 2006. We intend to move the database to a newly developed schema, using the OBDA BioSQL system (http://obda.open-bio.org) for archiving sequence data and annotation and providing additional tables for accessory data such as genome alignments and predicted orthology groups. It is planned to remove the distinction between the component databases, so that it will be possible to compare regions of homology that reside in distantly related genomes due to horizontal transfer. The ‘gene painting’ facility will be expanded to allow the display of generic data, derived from sources such as microarray experiments and whole genome PCR scanning. Additional methods of data visualization including an XY plot are also in development. We also intend to provide a facility for community-based reannotation, perhaps using a Wiki-style interface, to allow the dissemination of experimental observations that may not warrant formal publication.
Acknowledgments
We would like to thank Charles Penn and the other members of the Universiy of Birmingham E.coli group (UBEC). This work has been funded by the BBSRC (grant reference number EGA16107). Funding to pay the Open Access publication charges for this article was provided by JISC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Chaudhuri R.R., Khan A.M., Pallen M.J. coliBASE: an online database for Escherichia coli, Shigella and Salmonella comparative genomics. Nucleic Acids Res. 2004;32:D296–D299. doi: 10.1093/nar/gkh031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Delcher A.L., Harmon D., Kasif S., White O., Salzberg S.L. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fouts D.E., Mongodin E.F., Mandrell R.E., Miller W.G., Rasko D.A., Ravel J., Brinkac L.M., DeBoy R.T., Parker C.T., Daugherty S.C., et al. Major structural differences and novel potential virulence mechanisms from the genomes of multiple campylobacter species. PLoS Biol. 2005;3:e15. doi: 10.1371/journal.pbio.0030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delcher A.L., Phillippy A., Carlton J., Salzberg S.L. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 2002;30:2478–2483. doi: 10.1093/nar/30.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elvers K.T., Turner S.M., Wainwright L.M., Marsden G., Hinds J., Cole J.A., Poole R.K., Penn C.W., Park S.F. NssR, a member of the Crp-Fnr superfamily from Campylobacter jejuni, regulates a nitrosative stress-responsive regulon that includes both a single-domain and a truncated haemoglobin. Mol. Microbiol. 2005;57:735–750. doi: 10.1111/j.1365-2958.2005.04723.x. [DOI] [PubMed] [Google Scholar]
- 6.Ren C.P., Beatson S.A., Parkhill J., Pallen M.J. The Flag-2 locus, an ancestral gene cluster, is potentially associated with a novel flagellar system from Escherichia coli. J. Bacteriol. 2005;187:1430–1440. doi: 10.1128/JB.187.4.1430-1440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Betts H.J., Chaudhuri R.R., Pallen M.J. An analysis of type-III secretion gene clusters in Chromobacterium violaceum. Trends Microbiol. 2004;12:476–482. doi: 10.1016/j.tim.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 8.Fleischmann R.D., Alland D., Eisen J.A., Carpenter L., White O., Peterson J., DeBoy R., Dodson R., Gwinn M., Haft D., et al. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 2002;184:5479–5490. doi: 10.1128/JB.184.19.5479-5490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garnier T., Eiglmeier K., Camus J.C., Medina N., Mansoor H., Pryor M., Duthoy S., Grondin S., Lacroix C., Monsempe C., et al. The complete genome sequence of Mycobacterium bovis. Proc. Natl Acad. Sci. USA. 2003;100:7877–7882. doi: 10.1073/pnas.1130426100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stover C.K., Pham X.Q., Erwin A.L., Mizoguchi S.D., Warrener P., Hickey M.J., Brinkman F.S., Hufnagle W.O., Kowalik D.J., Lagrou M., et al. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 11.Hallin P.F., Ussery D.W. CBS Genome Atlas Database: a dynamic storage for bioinformatic results and sequence data. Bioinformatics. 2004;20:3682–3686. doi: 10.1093/bioinformatics/bth423. [DOI] [PubMed] [Google Scholar]