Abstract
The SNP500Cancer database provides sequence and genotype assay information for candidate SNPs useful in mapping complex diseases, such as cancer. The database is an integral component of the NCI Cancer Genome Anatomy Project (http://cgap.nci.nih.gov). SNP500Cancer reports sequence analysis of anonymized control DNA samples (n = 102 Coriell samples representing four self-described ethnic groups: African/African-American, Caucasian, Hispanic and Pacific Rim). The website is searchable by gene, chromosome, gene ontology pathway, dbSNP ID and SNP500Cancer SNP ID. As of October 2005, the database contains >13 400 SNPs, 9124 of which have been sequenced in the SNP500Cancer population. For each analysed SNP, gene location and >200 bp of surrounding annotated sequence (including nearby SNPs) are provided, with frequency information in total and per subpopulation as well as calculation of Hardy–Weinberg equilibrium for each subpopulation. The website provides the conditions for validated sequencing and genotyping assays, as well as genotype results for the 102 samples, in both viewable and downloadable formats. A subset of sequence validated SNPs with minor allele frequency >5% are entered into a high-throughput pipeline for genotyping analysis to determine concordance for the same 102 samples. In addition, the results of genotype analysis for select validated SNP assays (defined as 100% concordance between sequence analysis and genotype results) are posted for an additional 280 samples drawn from the Human Diversity Panel (HDP). SNP500Cancer provides an invaluable resource for investigators to select SNPs for analysis, design genotyping assays using validated sequence data, choose selected assays already validated on one or more genotyping platforms, and select reference standards for genotyping assays. The SNP500Cancer database is freely accessible via the web page at http://snp500cancer.nci.nih.gov.
INTRODUCTION
SNP500Cancer is a component of the Cancer Genome Anatomy Project (CGAP) of the National Cancer Institute (NCI) and is specifically designed to generate resources for the identification and characterization of genetic variation in genes important in cancer (1). The database reports the validation of SNPs by sequence analysis and optimizes genotyping assays for SNPs of interest to molecular epidemiology studies in cancer.
CGAP is dedicated to the development of technology, including both assays and utilization of technical platforms, and to determining the gene expression profiles of normal, precancer and cancer cells (2). Accordingly, data pertaining to genes and their variation are available on the public website http://cgap.nci.nih.gov. SNP500Cancer represents one of several initiatives designed to characterize sequence variation and is a resource for studying common germ-line genetic variation in the etiology of different cancers as well as related phenotypes. A validated SNP in the database has 100% concordance between sequence analysis and genotyping results on one or more platforms. Assays are developed and optimized in the Core Genotyping Facility (CGF) of the NCI for associations studies conducted in the Division of Cancer Epidemiology and Genetics (DCEG) and the Center for Cancer Research within the Intramural Program of the NCI. The primary focus of the DCEG's intramural studies is to conduct population-based research on environmental and genetic determinants of cancer.
DNA SAMPLES
Sequence and genotype analysis are conducted in a set of 102 unique anonymized individuals of diverse geographic origin with self-described ethnic group affiliation information, chosen to represent four major ethnic groups in the USA; it includes 24 African/African-American, 31 Caucasian, 23 Hispanic and 24 Pacific Rim individuals. The 102 anonymized samples can be obtained from the Coriell Cell Repositories (Coriell Institute for Medical Research, Camden, NJ). The sets of individuals are not a random sampling of one or more human populations and thus the predictive value of the sequence and genotype data provided can vary for different population samples.
A subset of SNPs has been genotyped in 280 anonymized individuals drawn from the Human Diversity Panel (HDP) (3), using DNA that has been amplified by whole genome amplification (4). The ethnic distribution of the 280 individuals approximates the 102 of the SNP500Cancer set, namely, the four self-described ethnic groups listed here. It is notable that there are 49 Native Americans, 25 of Mayan background and 24 of Piman background.
SELECTION OF GENES AND SNPS
This database is biased towards SNPs that lie within or are situated close to ‘candidate’ genes. The selection of genes and SNPs for analysis has been drawn from the following sources: (i) genes that fit a plausible model for cancer studies (e.g. by pathway), (ii) review of the published literature on SNPs and cancer, (iii) SNPs reported in public databases with some associated non-in silico determined frequency and (iv) SNPs verified during re-sequence analysis of candidate genes (5). In addition, the database contains SNPs analysed in the Breast and Prostate Cohort Consortium (http://epi.grants.cancer.gov/BPC3/ and http://cgf.nci.nih.gov/cohort.cfm), which targets the genes of the sex steroid metabolism and insulin growth factor pathways (n = 55 in total).
As of October 2005, the database contains SNPs in 812 known genes. The average number of SNPs per gene is 14.1 and the median is 8 SNPs per gene; the range is between 1 and 237 SNPs per gene.
SEQUENCING PROTOCOL
A PCR amplicon of ∼600 bp is generated for each SNP, which is localized to the center, creating flanking regions of ∼300 bp in each direction. Additional putative SNPs (determined from dbSNP) are annotated on the sequence of the amplicon. Oligonucleotide primers are designed for bi-directional sequence analysis using Primer3 software (6) and ProbeITy primer design software (Celadon Laboratories, Hyattsville MD). Each oligonucleotide primer is extended with a universal sequencing primer, M13 forward (TGTAAAACGACGGCCAGT) or M13 reverse (CAGGAAACAGCTATGACC). The sequencing assay protocols and conditions are displayed on the SNP500Cancer website. Sequence tracings are analysed in Seqscape v2.5 (Applied Biosystems, Foster City, CA). A sequencing call is deemed acceptable if the Seqscape quality score is >20 for at least 98 of the 102 individuals. Genotype calls are determined for each of the 102 individuals. The genotype and allele frequencies are maintained in an Oracle database and displayed on the SNP500Cancer website.
GENOTYPING PROTOCOLS AND VALIDATION
For SNPs that are determined to have >5% minor allele frequency (MAF) in at least one of the SNP500Cancer subpopulations, ∼200 bp of DNA sequence surrounding each SNP is submitted for design on one of the CGF's genotyping platforms: (i) Applied Biosystems' TaqMan™ ‘Assay by Design’ service and (ii) EPOCH Biosciences' MGB Eclipse™ probes. The genotyping assay procedures and conditions are displayed on the SNP500Cancer website. Genotyping assays are validated if there is complete concordance between the genotype results and the primary sequence analysis for the 102 samples. In addition, the 280 HDP samples are genotyped for SNPs with >5% MAF. The frequencies found in the four HDP subpopulations are displayed on the SNP500Cancer website.
ANALYSIS OF ALLELE FREQUENCIES
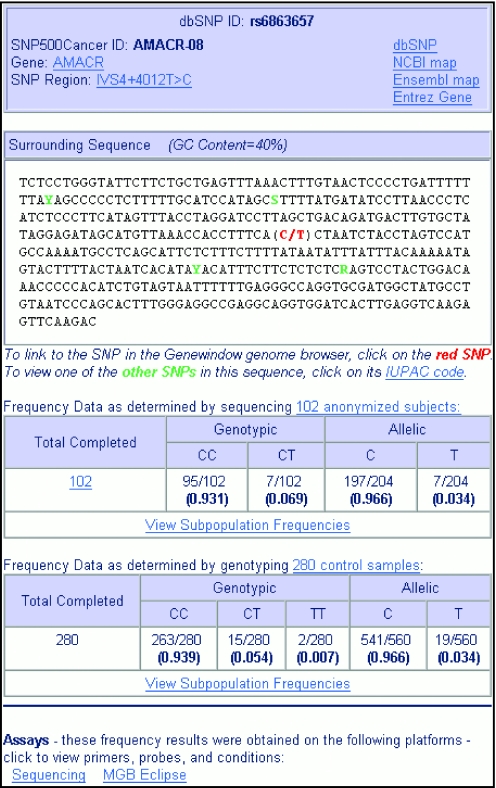
For each validated SNP, allele and genotype frequencies are displayed for the 102 individuals overall (Figure 1) and for each SNP500Cancer subpopulation. The result of a test for Hardy–Weinberg equilibrium, χ2 with one degree of freedom for two alleles (7) is posted for each subpopulation.
Figure 1.
The SNP page for a typical SNP from the SNP500Cancer website, showing links to external resources, the SNP within flanking sequence, frequency data and links to assay information.
The observed allele frequencies in the SNP500Cancer population of n = 102 can provide a useful estimate overall, as well as for the four subgroups. In an analysis of 1164 SNPs genotyped in the additional 280 individuals compared with the data for the 102 which were both genotyped and sequenced, the coefficient of correlation (r2) for each group was as follows: Caucasian 0.948, African-American 0.922, Pacific Rim 0.919 and Hispanic 0.742. The latter is notable because of the differences in the degree of admixture between the Hispanic subgroup of the 102 and the two Native American populations, of Maya and Pima heritage. Analysis of the SNP500Cancer dataset can be useful in estimating gene diversity. For instance, consistently reduced genetic diversity has been observed for SNP loci causing amino acid changes, especially radical shifts in protein structure (8); a similar trend was observed for 5′-untranslated regions (5′-UTRs). Moreover, the reduction of genetic diversity of nonsynonymous SNPs and those within the 5′-UTR are evidence that purifying selection could have acted at these sites, leading to reduced interpopulation divergence (9).
dbSNP SUBMISSION
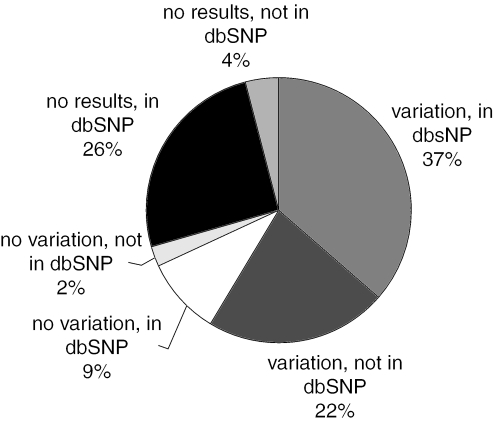
All analysed SNPs from the SNP500Cancer project are submitted to dbSNP (10)—http://www.ncbi.nlm.nih.gov/SNP. This information includes flanking sequence, observed variation, assay primers, probes, and conditions, and frequency of the sequence variation among the SNP500Cancer total population and subpopulations. Figure 2 depicts the distribution of SNPs already in dbSNP and those newly observed in close proximity to target SNPs. It is also notable that ∼10% of targeted SNPs are monoallelic in the 102 samples when compared with data in dbSNP build 124.
Figure 2.
The distribution of 13.437 SNPs in SNP500Cancer—those already in dbSNP, those not yet in dbSNP (submitted for next build). Almost 10% of SNP500Cancer SNPs were found to have no variation in the set of 102 individuals of the SNP500Cancer population overall, even though they are in dbSNP.
DATABASE AND WEB SERVER SPECIFICATIONS
The SNP500Cancer database is implemented using Oracle 9i (Oracle Corporation, Redwood Shores, CA). The web interface is written in ColdFusion (Macromedia, San Francisco, CA). The web server is a Compaq (Cupertino, CA) DL585 running Linux version 2.6.5. The database server is a Sun Microsystems (Santa Clara, CA) Sunfire 4800 running Oracle version 9.2.0.6.0. Both servers are supported by NCI Computer Services (Rockville, MD).
SNP500Cancer data are available through the caGRID, part of the Cancer Biomedical Informatics Grid (caBIG), a common interface providing data sharing among cancer researchers worldwide (http://cabig.nci.nih.gov) (11).
USING THE SNP500CANCER WEBSITE
Searching for genes
The SNP500Cancer website provides capabilities for searching for genes in several ways: (i) gene name or alias (including wild card searches), (ii) chromosome location and (iii) Gene Ontology (GO) pathway (12)—numeric or text. The gene is displayed with a list of SNPs that have been validated, and those that were not found to occur in the SNP500Cancer population.
Searching for SNPs
SNPs can be searched for using the dbSNP ID (rs cluster number, e.g. rs799917), or the internal SNP500Cancer polymorphism ID (gene symbol followed by a sequence number, e.g. BRCA1-02). The SNP is displayed in the center of surrounding sequence, and other SNPs in the sequence are annotated with IUPAC codes (Figure 1). Clicking on the SNP variation links to Genewindow (13), a publicly available genome browser developed at the CGF.
Viewing genotypic and allelic frequencies
For the SNP of interest, genotypic and allelic frequencies for the entire SNP500Cancer population of 102 individuals are displayed. The ‘view subpopulation frequencies’ link displays a page with genotypic and allelic frequencies for each subpopulation—African/African-American, Caucasian, Hispanic and Pacific Rim. Each subpopulation link leads to a list of individual genotypes for the samples within that subpopulation.
Viewing haplotype and htSNP data
For the gene of interest, haplotypes and haplotype-tagging SNPs (htSNP) data can be displayed for the entire control population or by subpopulation. Two different programs are available for determination of htSNPs: TagSNPs (14) and Tagger (http://www.broad.mit.edu/mpg/tagger/). The haplotype block structure can be visualized using Haploview (15).
Displaying assay conditions
For the SNP of interest, links are displayed for all validated assays (sequencing, MGB Eclipse™, TaqMan™). The link displays a page with detailed information on primers, probes, temperature and procedural steps.
Downloading information via FTP
FTP files are posted for SNPs by gene (including identifiers, genomic location and amino acid change), assays (identifier, primers, probes and conditions), frequencies and genotypes for each SNP with variation, as well as genotypes for all SNPs with >5% allelic frequency. These files can be found at ftp://ftp-snp500cancer.nci.nih.gov.
Connecting to other information sources
Each gene and SNP page on the SNP500Cancer website includes links to external resources; for genes: Entrez (16) and GO database; for SNPs: dbSNP, NCBI MapViewer (17) and Ensembl (18).
FUTURE DIRECTIONS
SNP500Cancer is a genetic resource designed to provide data on SNPs with neighboring sequence, for design and optimization of genotyping assays to be used in molecular epidemiology studies of cancer. A major goal of SNP500Cancer is to increase the breadth of genes and specifically develop sets of genes within a common pathway (e.g. DNA repair or telomere stability) (19). To this end, the project will utilize public resources to identify and validate SNPs across each gene at a greater density (e.g. one SNP every 1–3 kb) (20). The additional SNPs needed to densely cover the genes of interest will be selected based on their location, as well as suitability for inclusion as an htSNP in one or more population. The project will also add SNPs of great importance to cancer based on high quality citations in the published literature (21). There will be a special emphasis on SNPs drawn from coding and regulatory regions (including microRNAs) as well as those SNPs that have been shown to be associated with cancer susceptibility or outcome, particularly in pharmacogenomics.
SNPs with validated sequencing performed in other public projects will be imported, and assays optimized based on the concordance between sequence and genotype analysis. For instance, current plans are underway to optimize and validate assays for SNPs identified in the NIEHS Environmental Genome Project (NIEHS SNPs. NIEHS Environmental Genome Project, University of Washington, Seattle, WA, http://egp.gs.washington.edu/).
Though the primary goal of the SNP500Cancer database has been to focus on SNPs in and around known genes, future plans include the results of sequence analysis around small RNA sequences and their possible targets in cancer. Lastly, SNP500Cancer will make available results of dense, whole genome SNP scans using the 102 samples.
Acknowledgments
This research was supported (in part) by the Intramural Research Program of the National Cancer Institute, NIH, and Office of Cancer Genomics, NCI, NIH. Funding to pay the Open Access publication charges for this article was provided by the National Cancer Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Packer B.R., Yeager M., Staats B., Welch R., Crenshaw A., Kiley M., Eckert A., Beerman M., Miller E., Bergen A., et al. SNP500Cancer: a public resource for sequence validation and assay development for genetic variation in candidate genes. Nucleic Acids Res. 2004;32:528–532. doi: 10.1093/nar/gkh005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strausberg R.L., Simpson A.J.G., Wooster R. Sequence-based cancer genomics: progress, lessons, and opportunities. Nat. Rev. Genet. 2003;4:409–418. doi: 10.1038/nrg1085. [DOI] [PubMed] [Google Scholar]
- 3.Cann H.M., de Toma C., Cazes L., Legrand M., Morel V., Piouffre L., Bodmer J., Bodmer W., Bonne-Tamir B., Cambon-Thomsen A., et al. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- 4.Bergen A.W., Haque K.A., Qi Y., Beerman M.B., Garcia-Closas M., Rothman N., Chanock S.J. Comparison of yield and genotyping performance of multiple displacement amplification and Omniplex™ whole genome amplified DNA generated from multiple DNA sources. Hum. Mutat. 2005;26:262–270. doi: 10.1002/humu.20213. [DOI] [PubMed] [Google Scholar]
- 5.Bernig T., Taylor J.G., Foster C., Staats B., Yeager M., Chanock S. Sequence analysis of the mannose-binding lectin (MBL2) gene reveals a high degree of heterozygosity with evidence of selection. Genes Immun. 2004;5:461–476. doi: 10.1038/sj.gene.6364116. [DOI] [PubMed] [Google Scholar]
- 6.Rozen S., Skaletsky H.J. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 7.Weir B.S. Genetic Data Analysis II: Methods for Discrete Population Genetics Data. Sunderland, MA: Sinauer; 1996. [Google Scholar]
- 8.Hughes A.L., Packer B.R., Welch R., Bergen A.W., Chanock S.J., Yeager M. Effects of natural selection on inter-population divergence at polymorphic sites in human protein-coding loci. Genetics. 2005;170:1181–1187. doi: 10.1534/genetics.104.037077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes A.L., Packer B.R., Welch R., Bergen A.W., Chanock S.J., Yeager M. Widespread purifying selection at polymorphic sites in human protein-coding loci. Proc. Natl Acad. Sci. USA. 2003;100:15754–15757. doi: 10.1073/pnas.2536718100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buetow K.H. Cyberinfrastructure: empowering a “third way” in biomedical research. Science. 2005;308:821–824. doi: 10.1126/science.1112120. [DOI] [PubMed] [Google Scholar]
- 12.Gene Ontology Consortium. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Staats B.S., Qi L., Beerman M., Sicotte H., Burdett L.A., Packer B., Chanock S.J., Yeager M. Genewindow: an interactive tool for visualization of genomic variation. Nat. Genet. 2005;37:109–110. doi: 10.1038/ng0205-109. [DOI] [PubMed] [Google Scholar]
- 14.Stram D.O., Pearce C.L., Bretsky P., Freedman M., Hirschhorn J.N., Altshuler D., Kolonel L.N., Henderson B.E., Thomas D.C. Modeling and E-M estimation of haplotype-specific relative risks from genotype data for a case-control study of unrelated individuals. Hum. Hered. 2003;55:179–190. doi: 10.1159/000073202. [DOI] [PubMed] [Google Scholar]
- 15.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 16.Maglott D., Ostell J., Pruitt K.D., Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005;33:D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wheeler D.L., Barrett T., Benson D.A., Bryant S.H., Canese K., Church D.M., DiCuccio M., Edgar R., Federhen S., Helmberg W., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2005;33:D39–D45. doi: 10.1093/nar/gki062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hubbard T., Andrews D., Caccamo M., Cameron G., Chen Y., Clamp M., Clarke L., Coates G., Cox T., Cunningham F., et al. Ensembl 2005. Nucleic Acids Res. 2005;33:D447–D453. doi: 10.1093/nar/gki138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Savage S.A., Stewart B.J., Eckert A., Kiley M., Liao J.S., Chanock S.J. Genetic variation, nucleotide diversity, and linkage disequilibrium in seven telomere stability genes suggest that these genes may be under constraint. Hum. Mutat. 2005;26:343–350. doi: 10.1002/humu.20226. [DOI] [PubMed] [Google Scholar]
- 20.Carlson C.S., Eberle M.A., Rieder M.J., Yi Q., Kruglyak L., Nickerson D.A. Selecting a maximally informative set of SNPs for association analyses using linkage disequilibrium. Am. J. Hum. Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.International HapMap Consortium. The International HapMap Project. Nature. 2005;437:1299–1320. [Google Scholar]