Abstract
Online Mendelian Inheritance in Animals (OMIA) is a comprehensive, annotated catalogue of inherited disorders and other familial traits in animals other than humans and mice. Structured as a comparative biology resource, OMIA is a comprehensive resource of phenotypic information on heritable animal traits and genes in a strongly comparative context, relating traits to genes where possible. OMIA is modelled on and is complementary to Online Mendelian Inheritance in Man (OMIM). OMIA has been moved to a MySQL database at the Australian National Genomic Information Service (ANGIS) and can be accessed at http://omia.angis.org.au/. It has also been integrated into the Entrez search interface at the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=omia). Curation of OMIA data by researchers working on particular species and disorders has also been enabled.
INTRODUCTION
Information on the occurrence and inheritance of disorders and other familial traits in non-laboratory animal species was collected and catalogued by Nicholas from 1978. Designed to be complementary to McKusick's catalogue of human genes and genetic disorders Online Mendelian Inheritance in Man (OMIM) (1), this animal catalogue emphasizes comparative biology by highlighting homologous traits across the animal kingdom. Well-established as a valuable resource for comparative genetics, clinical research and tertiary education the electronic version of this catalogue, Online Mendelian Inheritance in Animals (OMIA), has been available online at the Australian National Genomic Information Service (ANGIS) since 1995 (2).
The earlier version of OMIA (2) was maintained by the curator as a desktop database, from which static web pages were exported periodically. The considerable limitations imposed by this configuration have now been overcome by the transfer of the master database to a MySQL platform on an ANGIS server, creation of new web pages interacting dynamically with the new database and the integration of OMIA within the Entrez system at the NCBI.
This report describes these enhancements to this important resource for inherited traits in animals.
STRUCTURE
Primarily, OMIA is a knowledgebase of inherited disorders and other familial traits, collectively called ‘phenes’ (V. A. McKusick, personal communication). It covers all animals, other than humans and mice, with an emphasis on domesticated species. Each phene has been assigned a unique six-digit OMIA ID. The most common type of record in OMIA is a phene-species, containing information on a phene in a particular species. Whenever a phene is determined to have a human homolog, the OMIM descriptor has been adopted, and a reciprocal hyperlink is provided to the relevant OMIM entry. OMIA has been structured to incorporate the growing knowledgebase of animal genes. Following the convention of the domestic animal gene-mapping community, when there is convincing evidence that a particular gene in one or more animal species is orthologous to a human gene, the human gene symbol is used, and a reciprocal hyperlink to OMIM is provided. For genes not yet recognized as having a human orthologue the gene symbol(s) found in the literature are used, with the official symbol indicated, if appropriate. Our goal is to generate a comprehensive resource documenting gene–phene relationships across all animal species.
NEW FEATURES
Significant enhancements have been made to both the appearance and functionality of OMIA. For administrators, the move to a MySQL database allows the content and structure of OMIA to be visualized or modified quickly and easily, using freely available software. Curators are now able to edit and update entries using a dynamic interface created using the Python programming language. For end-users, our new website dynamically queries the parent database, allowing immediate access to the information as soon as it is entered in the database. The integration of OMIA into NCBI's Entrez interface offers more advanced search facilities enabling complex queries and provides comprehensive direct links to other NCBI (3) databases and external bioinformatics resources. NCBI OMIA provided links between OMIA phenes and the Mammalian Phenotype Ontology (4). This mapping allows OMIA records to be associated with data from other resources linked to the same controlled term. In addition to the curated links between phenes and genes culled from the literature, NCBI OMIA contains links computed between particular phene-species records and associated records in Entrez Gene, Protein, OMIM, PubMed, Taxonomy, UniGene, HomoloGene and UniSTS. The Links menu, found at the right-hand margin of each NCBI OMIA record, provides convenient access to these associated records.
Statistics
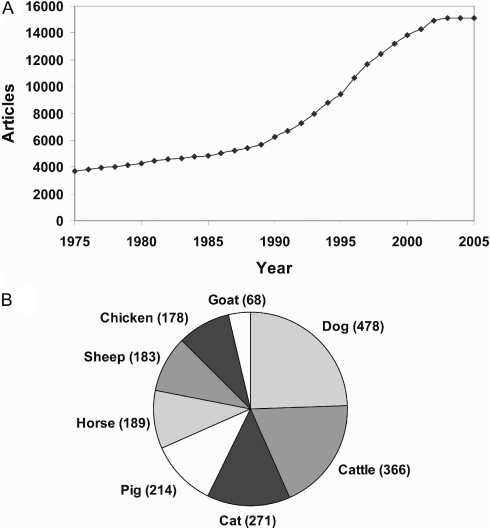
Currently, the database comprises over 15 000 publications dating back to the turn of the century (data from 1975 are shown in Figure 1A), describing 2508 phene-species across 135 species, with a third of phenes presenting in more than one species. The distribution of phenes by species (Figure 1B) reflects general research emphases, with common livestock and companion species comprising the majority of entries in OMIA.
Figure 1.
(A) Growth of the database in terms of number of publications referenced annually in OMIA as of September 1, 2005. (B) Distribution of phene entries in OMIA for the top seven species.
The NCBI Entrez system links 989 OMIA phenes to one (53%) or more (up to 37 for OMIA ID 000197) OMIM records, 198 to Gene and HomoloGene, 188 to Protein, 1528 to PubMed, 128 to UniGene clusters and 127 to UniSTS sequence tagged sites. Correspondingly, 86% of OMIM records with OMIA links map to a single OMIA entry.
SEARCHING OMIA
OMIA@ANGIS
Entering terms for phenes in the search window (http://omia.angis.org.au) allows users to locate relevant entries. Entering a species name generates a complete list of phenes for that species. Searching for author name or gene symbol is best done by searching within the relevant fields, using the drop-down menus on the Advanced Search page. More complex queries can be constructed from various combinations of words and field restrictions in this same facility. A special feature of the OMIA home page is a searchable table summarizing the numbers of phenes, single-locus phenes and phenes with a known molecular basis, for each of the major species in OMIA. The Advanced Search page facilitates searches across all species (or for any particular species) for single-locus phenes or for phenes with a molecular basis. It is also possible to use the Advanced Search page to generate lists of other categories of phenes such as bleeding disorders and lysosomal storage disorders.
OMIA@NCBI
NCBI OMIA can be accessed at http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=omia. The database has been indexed such that queries can be constructed based on Entrez Gene ID, gene name, OMIM ID, Taxonomy ID, organism's scientific or common name, phenotype term, Mammalian Phenotype Ontology ID, PubMed ID, OMIA ID or species-specific OMIA Phene ID. In addition, filters for linking to other NCBI resources can be found by clicking on the Preview/Index tab, selecting Filter and clicking on Index. For example, a query to find OMIA phenes from dog, containing genes associated with a cholesterol phenotype can be phrased as: ‘omia gene’[Filter] AND ‘Canis familiaris’[Organism] AND cholesterol[Phenotype]. OMIA phenes from any organism linked to a particular OMIM ID for osteogenesis imperfecta can simply be obtained using the OMIM ID itself for searching: 259410[OMIM ID]. This is a simple and powerful interface for retrieving subsets of OMIA data. Links to associated genes, references, taxa and phenotype ontology terms are made available in the text for each record.
UPDATING OMIA
An important new feature is the introduction of remote administration capabilities, permitting experts on particular species and/or disorders to add to the database or edit existing entries. We intend to expand the curation process by forming an international team of curators and editors. Members of the scientific community who register will be able to submit corrections and updates, although these will require vetting by a curator before being made publicly available. All curation will take place within the master ANGIS database; with weekly updates ported to the NCBI database.
A link from the top left corner of the ANGIS site provides access for curation. Authorized experts are guided through a series of web forms allowing them to search for and edit existing entries. The addition of new information is facilitated by the automated suggestion of names and keywords from existing data in order to minimize typographical errors and redundancy.
After a new submission is made, the OMIA curator will be automatically notified. Only after the changes have been approved will the submission be made public. In opening OMIA to global collaboration, we aim to increase the quality of available data through expert curation while simultaneously enabling OMIA to cope with the increasing publication rate of research concerning heritable disorders and traits.
CONCLUSION
Rebuilt on a flexible, modular foundation of MySQL and Python, OMIA is now easily extensible and better positioned to cope with the ever-increasing influx of new genomic and phenotypic information. Future directions include annotating gene mutations found to be associated with particular phenes by linking dbSNP (5) records (which encompass a number of species included in OMIA) with phenes, and the incorporation of an alias file to cover any synonyms ever used for a particular gene. Integration into the NCBI's Entrez system will allow OMIA to play a similar role in the support of animal biomedical and genomic research as OMIM has played for the human biomedical and genomic science. The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hamosh A., Scott A.F., Amberger J.S., Bocchini C.A., McKusick V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nicholas F.W. Online Mendelian Inheritance in Animals (OMIA): a comparative knowledgebase of genetic disorders and other familial traits in non-laboratory animals. Nucleic Acids Res. 2003;31:275–277. doi: 10.1093/nar/gkg074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wheeler D.L., Barrett T., Benson D.A., Bryant S.H., Canese K., Church D.M., DiCuccio M., Edgar R., Federhen S., Helmberg W., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2005;33:D39–D45. doi: 10.1093/nar/gki062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith C.L., Goldsmith C.A., Eppig J.T. The Mammalian Phenotype Ontology as a tool for annotating, analyzing and comparing phenotypic information. Genome Biol. 2005;6:R7. doi: 10.1186/gb-2004-6-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]