Abstract
The mosaic structure and molecular evolution of the leukotoxin operon (lktCABD) was investigated by nucleotide sequence comparison of the lktC, lktB, and lktD genes in 23 Mannheimia (Pasteurella) haemolytica, 6 Mannheimia glucosida, and 4 Pasteurella trehalosi strains. Sequence variation in the lktA gene has been described previously (R. L. Davies et al., J. Bacteriol. 183:1394–1404, 2001). The leukotoxin operon of M. haemolytica has a complex mosaic structure and has been derived by extensive inter- and intraspecies horizontal DNA transfer and intragenic recombination events. However, the pattern of recombination varies throughout the operon and among the different evolutionary lineages of M. haemolytica. The lktA and lktB genes have the most complex mosaic structures with segments derived from up to four different sources, including M. glucosida and P. trehalosi. In contrast, the lktD gene is highly conserved in M. haemolytica. The lktC, lktA, and lktB genes of strains representing the major ovine lineages contain recombinant segments derived from bovine or bovine-like serotype A2 strains. These findings support the previous conclusion that host switching of bovine A2 strains from cattle to sheep has played a major role in the evolution of the leukotoxin operon in ovine strains of M. haemolytica. Homologous segments of donor and recipient alleles are identical, or nearly identical, indicating that the recombinational exchanges occurred relatively recent in evolutionary terms. The 5′ and 3′ ends of the operon are highly conserved in M. haemolytica, which suggests that multiple horizontal exchanges of the complete operon have occurred by a common mechanism such as transduction. Although the lktA and lktB genes both have complex mosaic structures and high nucleotide substitution rates, the amino acid diversity of LktB is significantly lower than that of LktA due to a higher degree of evolutionary constraint against amino acid replacement. The recombinational exchanges within the leukotoxin operon have had greatest effect on LktA and probably provide an adaptive advantage against the host antibody response by generating novel antigenic variation at surface-exposed sites.
Mannheimia (Pasteurella) haemolytica is a gram-negative bacterium that is responsible for economically important respiratory tract infections of cattle and sheep known as pneumonic pasteurellosis (22, 24). Although the overall pathologies of bovine and ovine pneumonic pasteurellosis are very similar (22, 24), M. haemolytica consists of genetically distinct subpopulations (15, 17) that are differentially adapted to, and elicit disease in, either cattle or sheep. Strains that were previously classified as serotype A11 of M. haemolytica represent a divergent lineage (15, 18) and are now recognized as a separate species, namely, Mannheimia glucosida (2). M. glucosida comprises a heterogeneous group of organisms that have low virulence and are mainly opportunistic pathogens of sheep (2, 15). Strains that were once recognized as the T biotype of M. haemolytica are also now classified as a separate species, namely, Pasteurella trehalosi (52). However, unlike M. haemolytica, which occurs in cattle and sheep, P. trehalosi infects only sheep causing a systemic disease that is pathogically distinct from pneumonic pasteurellosis (24).
M. haemolytica produces a leukotoxin that is considered to be an important virulence factor in the pathogenesis of both bovine and ovine pneumonic pasteurellosis (3, 8, 36, 43, 56, 57). Leukotoxin is also produced by strains of M. glucosida and P. trehalosi (7, 23, 38, 47), but its role in infection in these two species is less well documented. The toxin is a member of the RTX family of gram-negative bacterial pore-forming cytotoxins that includes the alpha-hemolysin of Escherichia coli (35, 54, 58). Most RTX toxins interact with different cell types from a variety of species, but leukotoxin is specific for ruminant lymphoid cells (4, 6, 10, 28, 48). It has recently been shown that β2 integrins are the putative leukotoxin receptor on bovine leukocytes (1, 27, 33). At high concentrations leukotoxin forms pores in the cell membrane that rapidly lead to cell swelling and lysis (10, 13). However, at low or sublytic concentrations, leukotoxin causes activation of neutrophils (14), production of inflammatory cytokines (60), degranulation and generation of oxygen-derived free radicals (37), and morphologic changes consistent with apoptosis (53).
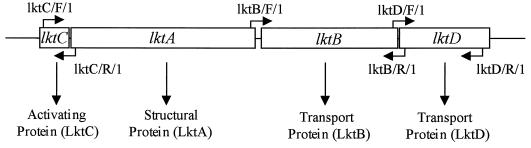
The genetic organization of the leukotoxin operon is similar to that of the E. coli hemolysin and consists of four contiguous genes designated lktCABD (25, 26, 35, 55). The lktA gene encodes the inactive protoxin, the lktC gene is required for posttranslational activation of the protoxin prior to secretion, and the lktB and lktD genes are required for secretion of the toxin from the organism (Fig. 1).
FIG. 1.
Structure and function of the lktCABD operon of M. haemolytica, M. glucosida, and P. trehalosi. The location of the amplification primers for the lktC, lktB, and lktD genes are indicated (lktC/F/1, etc.).
Horizontal transfer and recombination of DNA segments (intragenic) or entire genes (assortative) are now recognized as important evolutionary mechanisms, complementing mutation, in the diversification of bacteria (30, 31, 34, 42, 51). It has been suggested that the effective recombination rate varies among genes in relation to functional type, being highest for genes encoding cell surface and other proteins for which there is an adaptive advantage in structural diversity (40). Indeed, there is evidence that horizontal DNA transfer and recombination are responsible for the diversification of various virulence factors, including capsular polysaccharide (11), lipopolysaccharide (44), outer membrane proteins (21), and flagellar antigens (34, 51).
The objective of the present study was to investigate the amount of DNA polymorphism and molecular divergence of the leukotoxin operon (lktCABD) among pathogenic strains of M. haemolytica, M. glucosida, and P. trehalosi. This study was inspired by previous work that has shown that the leukotoxin structural gene (lktA) of M. haemolytica is highly polymorphic with multiple alleles. Phylogenetic analysis indicates that novel lktA alleles have been derived by recombination between lktA genes of M. glucosida and P. trehalosi (20). To what extent does the diversifying selection on lktA affect levels of variation in the other genes of the lkt operon? How does the variability of lktA influence the molecular evolution of lktC whose product directly interacts with and activates leukotoxin? Is there evidence that horizontal gene transfer and recombination has generated mosaic operons and, if so, how do these recombinant leukotoxin operons function in pathogenesis? To address these questions, we determined the nucleotide sequences in the lktC, lktB, and lktD genes of 33 strains representing pathogenic pasteurellae isolated from diseased cows and sheep and used statistical tools to analyze these data from an evolutionary perspective.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The lktC, lktB, and lktD genes were sequenced in 23 M. haemolytica, 6 M. glucosida, and 4 P. trehalosi isolates. The isolates have been well characterized in previous studies (15,16,17,18,19,20) and were selected to represent specific multilocus enzyme electrophoretic types (ETs), capsular serotypes, hosts of origin, and lktA alleles. The properties of these isolates are presented in Table 1.
TABLE 1.
Properties of 23 M. haemolytica, 6 M. glucosida, and 4 P. trehalosi isolates
| Isolate | ETa | Capsular serotype | Host species | Allelesb
|
GenBank accession no. | |||
|---|---|---|---|---|---|---|---|---|
| lktC | lktAc | lktB | lktD | |||||
| M. haemolytica | ||||||||
| PH2 | 1 | A1 | Bovine | lktC1.1 | lktA1.1 | lktB1.1 | lktD1.1 | AF314503 |
| PH376 | 1 | A6 | Bovine | lktC1.1 | lktA1.1 | lktB1.1 | lktD1.1 | |
| PH346 | 1 | A12 | Ovine | lktC1.1 | lktA1.2 | lktB1.1 | lktD1.1 | |
| PH540 | 2 | A1 | Bovine | lktC1.1 | lktA1.1 | lktB1.1 | lktD1.1 | |
| PH388 | 4 | A7 | Ovine | lktC1.1 | lktA1.3 | lktB1.1 | lktD1.1 | |
| PH238 | 5 | A9 | Ovine | lktC1.1 | lktA1.4 | lktB1.1 | lktD1.1 | |
| PH8 | 6 | A1 | Ovine | lktC1.1 | lktA1.5 | lktB2.1 | lktD4.1 | AF314506 |
| PH398 | 7 | A1 | Ovine | lktC1.1 | lktA1.5 | lktB2.1 | lktD4.1 | |
| PH284 | 8 | A6 | Ovine | lktC1.1 | lktA1.2 | lktB1.1 | lktD1.1 | |
| PH66 | 10 | A14 | Ovine | lktC1.3 | lktA9 | lktB5.1 | lktD1.1 | AF314508 |
| PH706 | 11 | A16 | Ovine | lktC1.3 | lktA7 | lktB5.2 | lktD1.1 | AF314509 |
| PH296 | 12 | A7 | Ovine | lktC1.3 | lktA8.1 | lktB5.1 | lktD1.1 | AF414141 |
| PH484 | 14 | A7 | Ovine | lktC1.3 | lktA8.1 | lktB5.1 | lktD1.1 | |
| PH588 | 15 | A13 | Ovine | lktC2.3 | lktA6 | lktB3.1 | lktD1.1 | AF314510 |
| PH494 | 16 | A2 | Ovine | lktC2.4 | lktA2.1 | lktB6.1 | lktD1.1 | AF314511 |
| PH550 | 17 | A2 | Bovine | lktC2.4 | lktA2.1 | lktB6.1 | lktD1.1 | |
| PH196 | 18 | A2 | Bovine | lktC1.2 | lktA3 | lktB7.1 | lktD2.1 | AF314512 |
| PH202 | 21 | A2 | Bovine | lktC2.1 | lktA2.2 | lktB6.2 | lktD1.1 | AF314513 |
| PH470 | 21 | A2 | Bovine | lktC2.1 | lktA2.2 | lktB6.2 | lktD1.1 | |
| PH278 | 21 | A2 | Ovine | lktC2.1 | lktA10.1 | lktB1.2 | lktD1.1 | AF314514 |
| PH372 | 21 | A2 | Ovine | lktC2.1 | lktA10.1 | lktB1.2 | lktD1.1 | |
| PH292 | 22 | A2 | Ovine | lktC2.2 | lktA8.1 | lktB5.3 | lktD1.1 | AF314515 |
| PH392 | 22 | A2 | Ovine | lktC2.2 | lktA8.2 | lktB5.3 | lktD1.1 | |
| M. glucosida | ||||||||
| PH344 | 1 | A11 | Ovine | lktC3.1 | lktA4.1 | lktB8.1 | lktD3.1 | AF314517 |
| PH498 | 3 | A11 | Ovine | lktC3.1 | lktA4.2 | lktB8.2 | lktD3.1 | AF314518 |
| PH240 | 5 | A11 | Ovine | lktC3.1 | lktA4.3 | lktB8.6 | lktD3.2 | AF314519 |
| PH496 | 7 | UG3 | Ovine | lktC1.4 | lktA4.4 | lktB8.4 | lktD3.4 | AF314520 |
| PH574 | 10 | UG3 | Ovine | lktA4.5 | lktB8.3 | lktD3.3 | AF314521 | |
| PH290 | 16 | UG3 | Ovine | lktC1.4 | lktA4.6 | lktB8.5 | lktD3.4 | AF314522 |
| P. trehalosi | ||||||||
| PH246 | 2 | T4 | Ovine | lktC4.1 | lktA5.1 | lktB4.1 | lktD5.1 | AF314523 |
| PH252 | 4 | T10 | Ovine | lktC4.3 | lktA5.2 | lktB4.2 | lktD5.2 | AF314524 |
| PH254 | 15 | T15 | Ovine | lktC4.1 | lktA5.3 | lktB4.3 | lktD5.4 | AF314525 |
| PH68 | 19 | T3 | Ovine | lktC4.2 | lktA5.4 | lktB4.1 | lktD5.3 | AF314526 |
Bacteria from −85°C stock cultures in 50% (vol/vol) glycerol in brain heart infusion broth (BHIB) were subcultured on blood agar (brain heart infusion agar containing 5% [vol/vol] sheep’s blood) and incubated aerobically overnight at 37°C. For preparation of DNA a few colonies were inoculated into 10-ml volumes of BHIB and grown overnight at 37°C at 120 rpm.
Preparation of chromosomal DNA.
Cells from 1.0 ml of overnight cultures were harvested by centrifugation for 1 min at 13,000 × g and washed once in sterile, distilled H2O. DNA was prepared with the InstaGene Matrix (Bio-Rad) according to the manufacturer’s instructions and then stored at −20°C.
PCR amplification and DNA sequence analysis.
The lktC, lktB, and lktD genes from each of the 33 strains were amplified from the chromosomal DNA with the primers shown in Table 2. With the exception of the 5′ and 3′ ends of the lktC and lktD genes, respectively, the primer sites were located in the adjacent gene so that the noncoding flanking regions were also amplified (Fig. 1). Incomplete gene sequences were obtained for lktC (89%) and lktD (98%) because the 5′ (lktC) and 3′ (lktD) primers were located within the genes. The primers were designed from the published sequences of the leukotoxin operon of M. haemolytica serotype A1 isolates (25, 35, 55), the lktCA sequence of a M. glucosida (previously P. haemolytica serotype A11) isolate (7), and the lktA sequences of 41 M. haemolytica, M. glucosida, and P. trehalosi isolates (20). Primers were designed by using the computer program Primer Designer (version 2.0) and synthesized by Sigma-GenoSys (Cambridge, United Kingdom). The three genes were amplified with a Taq DNA polymerase kit (Boehringer Mannheim) according to the manufacturer’s instructions. PCRs were carried out in a Perkin-Elmer 480 DNA thermal cycler with the following amplification parameters: denaturation at 94°C for 45 s, annealing at 57°C for 45 s, and extension at 72°C for 2 min. Thirty cycles were performed, and a final extension step of 72°C for 10 min was used. Production of a PCR amplicon of the expected size was confirmed by agarose gel electrophoresis and the DNA purified with a QIAquick PCR purification kit (Qiagen, Chatsworth, Calif.). The DNA was finally eluted in 30 μl of sterile, distilled H2O and stored at −20°C. Sequence reactions were performed with the ABI Prism Big Dye Terminator cycle sequencing kit (Applied Biosystems) in a GeneAmp PCR System 9700 (Applied Biosystems) thermal cycler and sequence analysis carried out with an Applied Biosystems 377 DNA Sequencer (University of Glasgow Sequencing Service). Both strands of the genes were sequenced and, in the case of the lktB and lktD genes, internal pairs of primers were designed as sequence data became available.
TABLE 2.
Details of PCR primers used to amplify the lktC, lktB, and lktD genes of M. haemolytica, M. glucosida, and P. trehalosi
| Gene and strain(s) | Primer details
|
|||
|---|---|---|---|---|
| Name | Orientation | Sequence | Positiona | |
| lktC | ||||
| PH2, PH8, PH196, PH238, PH284, PH346, | lktC/F/1 | Forward | 5′-TTGGCTATGGATGAACTC-3′ | lktC/bp39 |
| PH376, PH388, PH398, PH540 | lktC/R/1 | Reverse | 5′-GGTTGCCGTTAAAGTGTT-3′ | lktA/bp57 |
| PH494, PH550, PH202, PH470, PH278, PH372, | lktC/F/1 | Forward | 5′-TTGGCTATGGATGAACTC-3′ | lktC/bp39 |
| PH588 | lktC/R/2 | Reverse | 5′-GCTGTAAGCCACGAATTT-3′ | lktA/bp56 |
| PH292, PH392, PH296, PH484, PH706, PH66, | lktC/F/1 | Forward | 5′-TTGGCTATGGATGAACTC-3′ | lktC/bp39 |
| PH344, PH498, PH240, PH496, PH574, PH290 | lktC/R/3 | Reverse | 5′-GCAGTCAACCAGGAACTT-3′ | lktA/bp56 |
| PH246, PH252, PH254, PH68 | lktC/F/2 | Forward | 5′-CACATGGCTATGGATGAA-3′ | lktC/bp36 |
| lktC/R/4 | Reverse | 5′-CGCCTCTTGTTGCAGTTA-3′ | lktA/bp70 | |
| lktB (all strains) | lktB/F/1 | Forward | 5′-CAATTTGCTAGAGCAGCT-3′ | lktA/bp2842 |
| lktB/R/1 | Reverse | 5′-TTTTCCATACTTCTRCCC-3′ | lktD/bp73 | |
| lktD (all strains) | lktD/F/1 | Forward | 5′-GCAAGCAYCACGAATTACTG-3′ | lktB/bp2058 |
| lktD/R/1 | Reverse | 5′-GCGTTCCCTTAAACTTTC-3′ | lktD/bp1434 | |
Nucleotide position corresponding to the first 5′ bp of the primer.
Analysis of nucleotide and protein sequence data.
Nucleotide sequence data were analyzed and edited with SEQED (version 1.0.3; Applied Biosystems) and the DNASTAR suite of programs (DNASTAR, Inc.). Statistical and phylogenetic analyses were carried out with MEGA (29) in conjunction with alignment programs written by T.S.W. Statistical analyses for clustering of polymorphic sites were carried out by the maximum chi-square method (49) with a computer program (MAXCHI) written by T.S.W. (45).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the lktCABD sequences obtained in this study are given in Table 1.
RESULTS
Nucleotide and amino acid sequence variation.
The complete nucleotide sequence of the lktB gene (2,127 nucleotides) and partial sequences of the lktC (447 of 504 [89%] nucleotides) and lktD (1,416 of 1,437 [98.5%] nucleotides) genes were determined in 23 M. haemolytica, 6 M. glucosida, and 4 P. trehalosi isolates (Table 1). We were unable to amplify the lktC gene in strain PH574. The nucleotide sequences of the lktA gene in these strains have been described previously (20), and lktA data are included here for comparative purposes. The noncoding intergenic regions, comprising 15 (lktC-A), 74 (lktA-B), and 11 (lktB-D) nucleotides, respectively, were omitted from the analyses. The total aligned length, including gaps required for multiple alignment, of the lktC (447 nucleotides), lktA (2,862 nucleotides), lktB (2,127 nucleotides), and lktD (1,416 nucleotides) genes was 6,861 nucleotides.
Nucleotide and amino acid sequence diversities of the lktC, lktA, lktB, and lktD genes were determined separately for each of the three species and also for the complete population (Table 3). With the exception of the lktC gene of M. glucosida, the nucleotide and amino acid substitution rates of each of the four genes are relatively low in M. glucosida and P. trehalosi, compared to M. haemolytica. However, as described below, the significantly higher substitution rates in M. haemolytica are due to the effects of horizontal gene transfer and recombination. Nucleotide and amino acid sequence diversities ranged from 2.0 to 4.7% and 0.4 to 1.9%, respectively, for M. glucosida and from 0.4 to 2.7% and 0 to 1.3%, respectively, for P. trehalosi. The lktC gene of M. glucosida has higher nucleotide and amino acid sequence diversities (7.4 and 6.8%, respectively) due to recombination (discussed below). In contrast, the lktA (22.0%), lktB (9.2%) and, to a lesser extent, lktC (6.3%) genes of M. haemolytica have higher nucleotide diversities than the same genes of M. glucosida and P. trehalosi (with the exception, as mentioned above, of the lktC gene of M. glucosida). The extensive nucleotide diversity of the lktA gene of M. haemolytica predicts a correspondingly high diversity (16%) at the amino acid level. Although amino acid diversity of lktC is also relatively high (7.4%), in M. haemolytica, amino acid diversity of the lktB gene is, in relation to its nucleotide diversity, disproportionately low at 2.3%. Although not as obvious, the amino acid diversity of the lktB gene is also comparatively low, in relation to nucleotide diversity, in M. glucosida and P. trehalosi. These findings suggest that amino acid replacement is subject to a higher degree of selective constraint in the lktB gene product than in the encoded proteins of the other three genes, particularly lktC and lktA. Nucleotide and amino acid diversities of lktD in M. haemolytica are very low (3.3 and 2.8%, respectively) and are comparable to the values obtained for M. glucosida and P. trehalosi.
TABLE 3.
Nucleotide and amino acid sequence diversity,dS and dN values, and dS/dN ratios for the lktA, lktC, lktB, and lktD genes of M. haemolytica, M. glucosida, and P. trehalosia
| Gene and organism | Diversity (%)
|
Mean ± SD
|
dS/dN | ||
|---|---|---|---|---|---|
| Nucleotide | Amino acid | dS | dN | ||
| lktC | |||||
| M. haemolytica | 6.3 | 7.4 | 7.37 ± 2.07 | 1.90 ± 0.51 | 3.9 |
| M. glucosida | 7.4 | 6.8 | 16.21 ± 3.74 | 2.12 ± .061 | 7.6 |
| P. trehalosi | 0.4 | 0.0 | 1.09 ± 0.78 | 0.00 ± 0.00 | |
| All strains | 19.0 | 14.9 | 25.90 ± 3.88 | 2.71 ± 0.52 | 9.6 |
| lktA | |||||
| M. haemolytica | 22.0 | 16.0 | 33.25 ± 1.78 | 3.76 ± 0.28 | 8.8 |
| M. glucosida | 2.0 | 1.5 | 2.45 ± 0.38 | 0.23 ± 0.06 | 10.7 |
| P. trehalosi | 0.7 | 0.0 | 0.94 ± 0.27 | 0.18 ± 0.06 | 5.2 |
| All strains | 25.8 | 18.5 | 39.47 ± 1.78 | 4.00 ± 0.26 | 9.9 |
| lktB | |||||
| M. haemolytica | 9.2 | 2.3 | 10.88 ± 0.97 | 0.28 ± 0.08 | 38.9 |
| M. glucosida | 2.7 | 0.4 | 5.31 ± 0.71 | 0.07 ± 0.04 | 75.9 |
| P. trehalosi | 1.3 | 0.1 | 2.76 ± 0.55 | 0.03 ± 0.03 | 92.0 |
| All strains | 12.9 | 3.0 | 18.61 ± 1.17 | 0.42 ± 0.09 | 44.3 |
| lktD | |||||
| M. haemolytica | 3.3 | 2.8 | 1.75 ± 0.31 | 0.19 ± 0.05 | 9.2 |
| M. glucosida | 4.7 | 1.9 | 8.25 ± 1.12 | 0.35 ± 0.12 | 23.5 |
| P. trehalosi | 2.7 | 1.3 | 5.42 ± 0.99 | 0.30 ± 0.12 | 18.1 |
| All strains | 15.7 | 8.3 | 16.21 ± 1.29 | 1.03 ± 0.16 | 15.7 |
dS and dN represent the number of synonymous substitutions per 100 synonymous sites and the number of nonsynonymous substitutions per 100 nonsynonymous sites, respectively.
Nucleotide substitution rates in lktC and lktD are substantially higher across all three species (19.0 and 15.7%, respectively) than for the individual species, indicating limited sharing of DNA in these genes among the three species. In contrast, nucleotide substitution rates in lktA and lktB are only slightly higher across all three species (25.8 and 12.9%, respectively) than the corresponding rates for M. haemolytica. This suggests that segments of DNA encoding the lktA and lktB genes have been recently exchanged among M. haemolytica, M. glucosida, and P. trehalosi.
Synonymous and nonsynonymous substitution rates.
The numbers of synonymous substitutions per 100 synonymous sites (dS) and nonsynonymous substitutions per 100 nonsynonymous sites (dN) were estimated (39) for the four genes, and the dS/dN ratios were calculated (Table 3). A high dS/dN ratio indicates that natural selection at the molecular level is purifying (conservative), acting against mutations resulting in amino acid replacements. The dS/dN ratios for the lktB gene in each species (38.9 to 92.0) and across all three species (44.3) are significantly greater than the corresponding ratios for lktC (3.9 to 7.6 and 9.6) and lktA (5.2 to 10.7 and 9.9); the dS/dN ratios for lktD (9.2 to 23.5 and 15.7) are approximately twofold higher than those for lktC and lktA. These findings confirm that amino acid divergence in LktB is subject to a higher degree of selective constraint in comparison to the proteins encoded by the other three genes, particularly lktC and lktA.
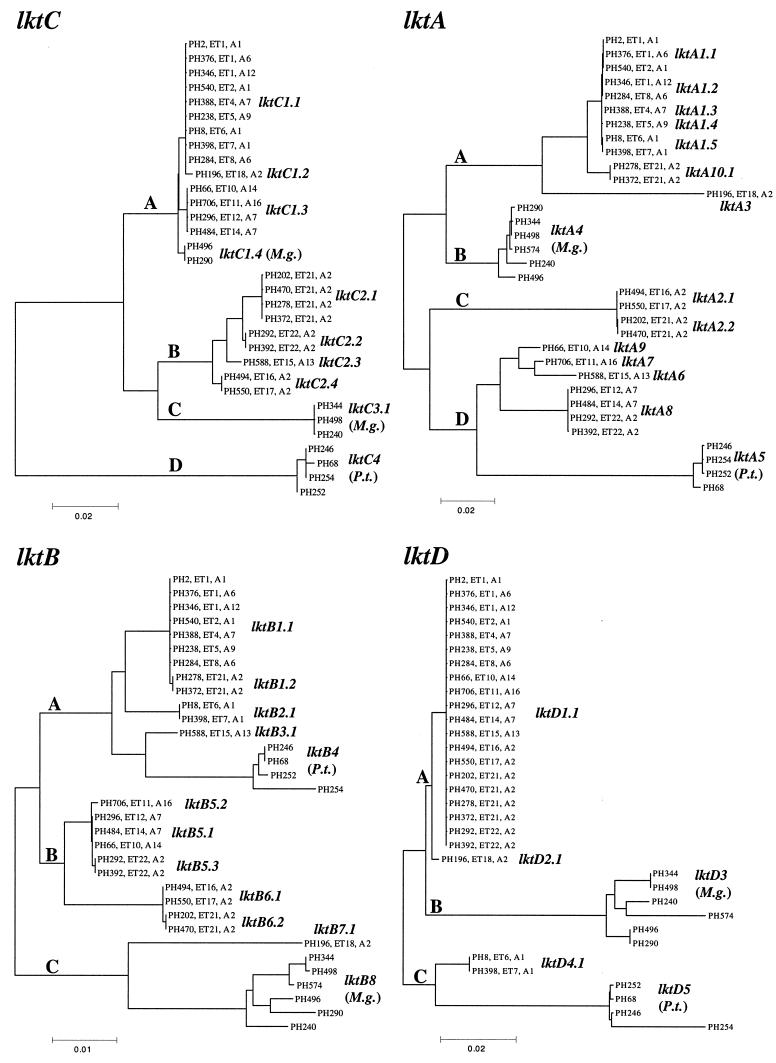
Allelic variation and phylogenetic relationships.
Nucleotide sequence comparison of the lktC, lktB, and lktD genes identified 12 different lktC sequences (Fig. 2), 19 lktB sequences (Fig. 3), and 11 lktD sequences (Fig. 4). Individual sequences represent distinct alleles which were classified as described below. Phylogenetic trees were constructed for the lktC, lktA, lktB, and lktD genes by using the neighbor-joining method with Jukes-Cantor correction for synonymous changes (29) (Fig. 5). Four major groups of alleles, lktC1-lktC4, representing lineages A to D, were identified among the 32 lktC sequences, and individual alleles within each group were classified as lktC1.1, lktC1.2, etc. (Fig. 5 and Table 1). Classification of the lktA alleles has been described previously (20), but their phylogenetic relationships are shown in Fig. 5 for comparison. Eight major groups of alleles, lktB1-lktB8, were identified among the 33 lktB sequences and these formed three major lineages, A to C, represented by lktB1-lktB4, lktB5 and lktB6, and lktB7 and lktB8, respectively. Distinct alleles within each group were classified in the same way as the lktC gene, i.e., lktB1.1, lktB1.2, etc. Five major groups of alleles, lktD1-lktD5, were identified among the 33 lktD sequences, and these formed three major lineages, A to C, represented by lktD1 and lktD2, lktD3, and lktD4 and lktD5, respectively. Individual alleles within each group were classified in the same way as the lktC and lktB genes.
FIG. 2.
Distribution of polymorphic nucleotide sites among the lktC alleles. The numbers above the sequences represent the positions of polymorphic nucleotide sites from the 5′ end of the gene. The dots represent sites where the nucleotides match those of the first sequence (lktC1.1). The shaded areas highlight regions of sequence identity that represent proposed recombinant segments (see Fig. 6).
FIG. 3.
Distribution of polymorphic nucleotide sites among the lktB alleles. The numbers above the sequences represent the positions of polymorphic nucleotide sites from the 5′ end of the gene. The dots represent sites where the nucleotides match those of the first sequence (lktB1.1). The shaded areas highlight regions of sequence identity that represent proposed recombinant segments (see Fig. 6).
FIG. 4.
Distribution of polymorphic nucleotide sites among the lktD alleles. The numbers above the sequences represent the positions of polymorphic nucleotide sites from the 5′ end of the gene. The dots represent sites where the nucleotides match those of the first sequence (lktD1.1). The shaded areas highlight regions of sequence identity that represent proposed recombinant segments (see Fig. 6).
FIG. 5.
Neighbor-joining trees for the lktC, lktA, lktB, and lktD genes of M. haemolytica, M. glucosida, and P. trehalosi constructed with the Jukes-Cantor correction for synonymous changes. ETs and serotypes are given for M. haemolytica strains only. M.g., M. glucosida; P.t., P. trehalosi.
The degree of congruency of phylogenetic trees representing different genes provides an indication as to the relative importance of recombination and mutation in their evolution. Highly congruent trees indicate that the genes are evolving primarily by accumulating point mutations, whereas noncongruent trees provide evidence that recombination has disrupted the gene phylogenies. Clearly, the phylogenetic trees for the lktC, lktA, lktB, and lktD genes are noncongruent (Fig. 5), suggesting that intragenic recombination has played a role in their evolution and that of the complete operon. Noncongruency of the gene trees is most clearly seen by comparing the phylogenetic relationships of genes in strains representing M. haemolytica, M. glucosida, and P. trehalosi. Based on multilocus enzyme electrophoresis and 16S rRNA sequence data (15, 18), we know that M. glucosida has diverged from M. haemolytica and that P. trehalosi is further diverged from both species. This phylogenetic relationship is partially true for lktC (the lktC genes of the M. glucosida strains PH290 and PH496 do not follow this pattern) and lktD (the lktD genes of the M. haemolytica isolates PH8 and PH398 are exceptions) but not for lktA and lktB. In the case of lktA, the M. glucosida lktA4-type alleles represent a cluster (lineage B) that is more closely related to lktA1-type M. haemolytica alleles (lineage A) than are many of the other M. haemolyica lktA alleles (lineages C and D). For lktB, the P. trehalosi lktB4-type alleles are more closely related to the M. haemolytica lktB1-, lktB2-, and lktB3-type alleles (lineage A) than are the lktB5- and lktB6-type alleles of other M. haemolytica strains (lineage B), whereas the M. glucosida lktB8-type alleles, together with the lktB7.1 allele of isolate PH196, represent the most divergent lineage (lineage C). Further examples of phylogenetic noncongruency of the lktC, lktA, lktB, and lktD genes are seen in the M. haemolytica strains PH494 and PH550, PH278 and PH372, and PH292 and PH392. Alleles of these three pairs of strains represent separate but closely related lineages in lktC, three highly divergent lineages in lktA, two closely related lineages (lineage B) and a third more distantly related lineage (lineage A) in lktB, and a single lineage in lktD.
Distribution of alleles in relation to other strain characteristics.
The distribution of individual lktC, lktB, and lktD alleles among isolates indicate that, in general, alleles of the same type, e.g., lktC1.1, lktC1.2, etc., are associated with strains representing closely related ETs, but alleles of closely related types are often associated with distantly related ETs (Table 1). For example, lktC1.1 alleles are present in strains representing the closely related ETs 1, 2, 4, and 5 to 8, but the almost identical allele lktC1.2 occurs in strain PH196 of ET 18, lktC1.3 is associated with strains of ETs 10 to 12, and 14, and lktC1.4 is present in the M. glucosida strains PH290 and PH496. LktC2.1 alleles are present in serotype A2 strains of ET 21 and lktC2.2 alleles in serotype A2 strains of ET 22, but lktC2.3 occurs in the serotype A13 strain PH588 of ET 15 and lktC2.4 occurs in serotype A2 strains of ETs 16 and 17. LktC3-type alleles are associated with the M. glucosida strains PH240, PH344, and PH498 and lktC4-type alleles occur in P. trehalosi strains.
For lktB, in lineage A lktB1.1 alleles occur in strains representing ETs 1, 2, 4, 5, and 8, but lktB1.2 alleles are associated with ovine serotype A2 strains of ET 21; lktB2.1 alleles are present in ovine serotype A1 strains of ETs 6 and 7; lktB3.1 is represented by the serotype A13 strain PH588 of ET 15; and lktB4-type alleles occur in P. trehalosi strains. In lineage B, lktB5.1 alleles are present in strains of ETs 10, 12, and 14, and lktB5.2 occurs in strain PH706 of ET 11, but lktB5.3 alleles are associated with ovine serotype A2 strains of ET 22; lktB6.1 alleles occur in bovine and bovine-like serotype A2 strains of ETs 16 and 17, whereas lktB6.2 alleles are present in bovine serotype A2 strains of ET 21. In lineage C, lktB7.1 is associated with strain PH196 of ET 18, whereas lktB8-type alleles are associated with M. glucosida strains.
In the case of lktD, the lktD1.1 allele (lineage A) is present in all strains of M. haemolytica, except for PH196, PH8, and PH398. In lineage B, lktD3-type alleles are associated with M. glucosida isolates but, in lineage C, lktD4.1 alleles occur in ovine serotype A1 strains of ETs 6 and 7, whereas lktD5-type alleles are associated with P. trehalosi strains.
Intragenic recombination within the lktC, lktB, and lktD genes.
Intragenic recombination events lead to the formation of linked runs of nucleotides within a sequence whose ancestry is different from other nucleotides in the same sequence (50). If visual inspection of the polymorphic sites in a set of sequences suggests that one or more recombination events have occurred, the maximum chi-square method (49) will locate the most likely positions of the crossovers and test their statistical significance.
Visual inspection of the aligned lktC sequences identified runs of nucleotides representing recombinant segments in alleles lktC2.1-lktC2.4 (Fig. 2). Pairwise comparison of lktC sequences by the maximum chi-square method identified statistically significant partitions at nucleotide positions 163, 194, 272, and 342 (results not shown) which represent the end points of the recombinant segments. Similarly, visual inspection of the aligned lktB sequences identified blocks of nucleotides that represent recombinant segments (Fig. 3). Pairwise comparison of lktB sequences by the maximum chi-square method identified statistically significant partitions (representing the end points) for the majority of these segments (results not shown). In contrast, the lktD gene of M. haemolytica is highly conserved, and evidence of recombination occurs only in the lktD2.1 and lktD4.1 alleles (Fig. 4). However, visual inspection of the lktD3- and lktD5-type alleles of M. glucosida and P. trehalosi identified blocks of nucleotides that appear to represent recombinant segments. Pairwise comparison of these sequences with lktD1.1 by the maximum chi-square method identified statistically significant partitions that represent the end points of these segments (results not shown).
Certain strains, notably PH8, PH398, PH196, and PH588, are characterized by the presence of multiple small recombination events within the lktB and lktD genes. The lktB gene of strains PH8 and PH398 (lktB2.1) has a segment (nucleotides 318 to 1377) containing short runs of nucleotides (nucleotides 318 to 525, 744 to 768, and 1282 to 1377) that are identical, or almost identical, to the corresponding regions of the lktB gene in various other strains (Fig. 3). Similarly, the lktD gene of isolates PH8 and PH398 (lktD4.1) contains short runs of nucleotides that are identical to the corresponding regions of the lktD gene in strains of P. trehalosi (nucleotides 420 to 449 and 897 to 1002) and M. glucosida (nucleotides 1029 to 1038, 1086 to 1115, and 1203 to 1286) (Fig. 4). The lktB gene of strain PH588 (lktB3.1) also contains small runs of nucleotides that are identical to the corresponding regions of different strains (nucleotides 744 to 768, 795 to 958, and 1062 to 1377) (Fig. 3). Finally, the lktB gene of strain PH196 (lktB7.1) contains runs of nucleotides that have been derived from three different sources, namely, bovine A2 strains (nucleotides 624 to 906), M. glucosida (nucleotides 1389 to 1860), and P. trehalosi (nucleotides 1920 of lktB to 205 of lktD) (Fig. 3 and 4).
DISCUSSION
Mosaic structure of the lktCABD operon of M. haemolytica.
The lktA gene of M. haemolytica has previously been shown to have a complex mosaic structure that has been derived by a series of inter- and intraspecies horizontal DNA transfer and intragenic recombination events (20). Comparative sequence analysis of the lktC, lktB, and lktD genes has enabled us to build up a complete picture of the genetic organization of the lktCABD operon in M. haemolytica and its relatives M. glucosida and P. trehalosi. Based on the presence of well-defined recombinant segments within each gene (Fig. 2 to 4; see also reference 20) and noncongruent gene phylogenies (Fig. 5), it is clear that the lktCABD operon itself has a complex mosaic structure that has been derived by extensive horizontal DNA transfer and intragenic recombination. However, as described below, the pattern of recombination varies throughout the operon and among the different evolutionary lineages of M. haemolytica (15, 20).
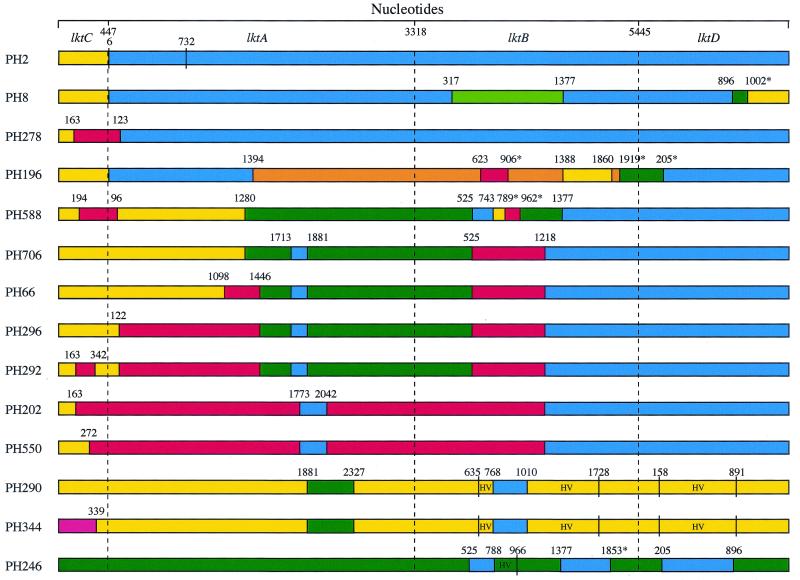
The mosaic structure of the operon is shown schematically in Fig. 6 and has greatest complexity in the lktA and lktB genes of certain M. haemolytica strains. Clearly, intragenic recombination has been most frequent within these two genes, which consist of recombinant segments derived from up to four different sources. These include M. haemolytica strains representing ETs 1 to 8 (the “A1/A6 group”); M. haemolytica isolates of ETs 16, 17, and 21 (the “bovine A2 group”) (15, 20); M. glucosida; and P. trehalosi. The lktC gene has a less complex mosaic structure because it contains recombinant segments derived from only two sources. In the majority of M. haemolytica strains the lktC gene is almost identical to that of the M. glucosida isolates PH290 and PH496 (Fig. 5 and 6). However, the lktC gene of strains PH278, PH588, and PH292 contains small recombinant segments derived from isolates of the bovine A2 group, e.g., PH202. The location of the partition at bp 163 in strains PH278, PH292, and PH202 (Fig. 6) and the complete identity of donor and recipient segments provides strong evidence of a common origin. In contrast to the lktC, lktA, and lktB genes, the lktD gene is, with the exception of strains PH8, PH398, and PH196, highly conserved among M. haemolytica strains and shows no evidence of recombination.
FIG. 6.
Schematic representation of the mosaic structures of the leukotoxin operon in M. haemolytica, M. glucosida, and P. trehalosi strains. The different colors indicate sequence identity and the likely origins of recombinant segments. All recombination sites were shown to be statistically significant by maximum chi-square analysis with the exception of those marked with an asterisk. Numbers above the proposed recombination sites indicate the positions of the last nucleotide at the downstream end of the recombinant segment. HV, hypervariable sequence.
The complexity of the mosaic structure of the operon varies among the different lineages of M. haemolytica. The operon is highly conserved in strains representing ETs 1, 2, 4, 5, and 8 (Table 1 and Fig. 6), although there is strong evidence of recombination at the junction between the lktC and lktA genes (nucleotide position 6 of lktA), as well as at nucleotide position 732 in lktA (20). There are a small number of nucleotide substitutions in the lktA gene (20) but the lktC, lktB, and lktD genes are identical in all strains examined. Clearly, the leukotoxin operons of strains representing these ETs have a recent and common evolutionary origin and have not been disrupted by incoming “foreign” DNA. However, strains representing these ETs do appear to have acted as donors in the exchange of DNA to other strains, including M. glucosida and P. trehalosi isolates (Fig. 6). The operons of strains PH8 and PH398 of ETs 6 and 7 are unusual in that they have both been involved in DNA uptake and multiple small intragenic recombination events. The stability of the lktCABD operon in strains representing ETs 1 to 8 is in marked contrast to the variation that occurs in capsular polysaccharide; at least seven capsular serotypes are associated with these ETs (15). These differences suggest that the leukotoxin and capsular polysaccharide genetic loci are subject to different evolutionary processes in strains of ETs 1 to 8.
In contrast, the leukotoxin operon has greatest complexity in strains representing ETs 10 to 12, 14, 15, 18, and 22 (PH66, PH706, PH296, PH484, PH588, PH196, PH292, and PH392) (Table 1 and Fig. 6). The operons in strains of these ETs have a common evolutionary origin and have been derived by a sequential series of horizontal DNA transfer and intragenic recombination events (20). Presumably, strains representing these lineages are more susceptible to horizontal transfer of the operon and intragenic recombination than are strains of ETs 1, 2, 4, 5, and 8. The operons of strains of ETs 16, 17, and 21 (PH494, PH550, PH202, PH470, PH278, and PH372) have less complex mosaic structures that have been derived independently. However, the region between nucleotides 163 and 272 of lktC and 1218 of lktB in the bovine A2 strains PH202 and PH550 (Fig. 6) is more divergent from other M. haemolytica strains than is the same region of M. glucosida, and as divergent as P. trehalosi (results not shown). This strongly suggests that the ancestral bovine operon has been acquired by horizontal gene transfer from a more distantly related species. M. glucosida and P. trehalosi strains have clearly acted as donors in the horizontal transfer of DNA to M. haemolytica strains, but they have also received DNA from M. haemolytica strains of the A1/A6 group. Although DNA from the bovine A2 group has been widely transferred to other M. haemolytica strains, there is no evidence that DNA from this source has been acquired by M. glucosida and P. trehalosi strains.
Recent evolutionary origin of the lktCABD operon of M. haemolytica and the influence of host switching.
Homologous recombinant segments in donor and recipient lktA alleles were previously shown to be identical, or nearly identical, suggesting that the mosaic lktA alleles evolved relatively recently (20). Our data for the lktC, lktB, and lktD genes have extended these findings and demonstrate that homologous donor and recipient segments within the lktCABD operon have identical, or nearly identical, nucleotide sequences (Fig. 2 to 4 and 6). For example, the entire 4,968-bp region downstream of position 1446 in the lktA gene of strains PH706, PH66, PH296, and PH292 is identical, except for two base changes at nucleotide positions 1446 and 1918 in the lktB gene of PH706 and one base change at nucleotide position 1402 in the lktB gene of PH292. Clearly, there have been very few point mutations throughout the operon since the recombination events took place. These findings provide strong evidence that the recombination events responsible for the mosaic structure of the lktCABD operon occurred very recently, in evolutionary terms.
It has previously been shown that the recombinant lktA alleles of ovine M. haemolytica strains contain DNA segments derived from bovine serotype A2 strains (20). Evidence was also provided to support the view that these segments could only have become incorporated into the lktA gene of ovine strains as a consequence of host switching of the bovine strains from cattle to sheep (20). In the present study we have shown that DNA segments derived from the lktCABD operon of bovine or bovine-like serotype A2 strains (e.g., PH202, PH470, PH494, and PH550) have become incorporated into the lktC, lktA, and lktB genes of ovine M. haemolytica strains representing every ET except for those belonging to the A1/A6 group (Table 1 and Fig. 6). These findings not only demonstrate that bovine A2 strains have played a central role in the evolution of the lktCABD operon of M. haemolytica but also confirm the view that host switching of these strains from cattle to sheep has been the major factor responsible for driving these evolutionary events.
Conserved mechanism of horizontal transfer.
The extreme 5′ and 3′ ends of the leukotoxin operon are highly conserved in all strains of M. haemolytica, with the exception of the 3′ ends in isolates PH8 and PH398 (Fig. 6). The 5′ end of the operon consists of segments of various length that extend into the lktC or lktA genes and have identical, or nearly identical, nucleotide sequences (Fig. 2 and 6). These conserved segments are present in strains representing every lineage of M. haemolytica, as well as in the M. glucosida strains PH290 and PH496, and clearly have a common origin. The 3′ end of the operon is highly conserved over the entire length of the lktD gene and for almost half the length of the lktB gene in every M. haemolytica strain except PH8, PH398, and PH196 (Fig. 3, 4, and 6). The identical, or nearly identical, nucleotide sequences of the lktB/lktD 3′ region of the operon in strains representing every major lineage of M. haemolytica strongly suggests a common origin which is most likely to be M. haemolytica strains of the A1/A6 group.
The remarkably high degree of sequence similarity at the 5′ and 3′ ends of the leukotoxin operon in M. haemolytica strains of divergent lineages was, after the diversity previously observed in lktA (20), an unexpected finding and has the following implications. First, it provides additional evidence that the horizontal transfer and recombination events responsible for the mosaic structure of the operon occurred relatively recently. Second, it suggests that the complete operon, rather than parts of it, has been horizontally transferred between divergent lineages of M. haemolytica. Evidence for this is provided by the similarity of the operon in isolates PH66, PH706, PH296, PH484, PH292, and PH392 that represent ETs 10, 11, 12, 14, and 22, respectively (Table 1 and Fig. 6). Horizontal transfer of the complete operon is consistent with the reasoning behind the selfish operon model of Lawrence and Roth (32). This model suggests that the horizontal transfer of adjacent genes with coordinate function is a formative force in the evolution of operon structure. According to the model, the cotransfer of genes encoding novel metabolic functions allow new hosts to exploit novel ecological niches. Clearly, the transfer of only parts of the operon would provide no evolutionary benefit to the recipient. Third, it provides evidence that a common and highly conserved mechanism has been responsible for the recent horizontal transfer of the operon between strains of divergent lineages. It is interesting to speculate that the mechanism of DNA transfer is transduction because various toxin genes are known to be located within the genomes of temperate bacteriophages (9, 42, 59) and lysogenic bacteriophages have been identified in M. haemolytica (46). However, conjugation and transformation cannot be ruled out as possible mechanisms of DNA transfer.
Amino acid sequence diversity is related to mosaic structure and gene function.
Nucleotide sequence diversities of the lktA, lktB and, to a lesser extent, lktC genes of M. haemolytica are considerably higher than the corresponding genes of M. glucosida and P. trehalosi because they have mosaic structures that have been generated by intragenic recombination. Although the inferred amino acid sequence diversities of the lktC and lktA genes are relatively high, the amino acid diversity of lktB is very low in relation to the corresponding nucleotide diversities. Furthermore, there is a strong correlation between amino acid replacement and ratios of synonymous and nonsynonymous nucleotide substitution rates (dS/dN) for these genes (Table 3). The lktA and lktC genes have high amino acid diversities and low dS/dN ratios, whereas the lktB gene has low amino acid diversity and a high dS/dN ratio. These data clearly indicate that amino acid replacement in the lktB gene product is subject to a higher degree of selective constraint in comparison to the proteins encoded by the other three genes, particularly lktC and lktA.
The variation in amino acid replacement rates may be accounted for in terms of the different functions of the lktC, lktA, lktB, and lktD gene products and is perhaps best illustrated by comparison of the lktA and lktB genes. LktA has high amino acid diversity generated by intragenic recombination, but the majority of amino acid replacements are associated with surface-exposed, hydrophilic domains (20). It is likely that mosaic lktA genes derived by recombination between lktA alleles provide an adaptive advantage against the host antibody response by generating novel antigenic variation at surface-exposed sites (20). Recombinational exchange is thought to confer a selective advantage on various pathogens by generating antigenic variation in cell surface antigens (11, 21, 34, 44, 51).
Intragenic recombination is also responsible for the high nucleotide diversity of lktB but, unlike LktA, amino acid replacement within LktB is highly constrained. LktB is a cytoplasmic membrane protein that is involved in transport of LktA into the periplasm. The protein has a transmembrane organization with up to eight membrane-spanning regions (12), and amino acid replacement is highly constrained to maintain the proteins’ structural integrity and function. The importance of amino acid conservation in this protein is also demonstrated by the fact that LktB exhibits 90.5% similarity with the corresponding HlyB protein of the hemolysin determinant of E. coli (55); in contrast, the LktA and HlyA proteins are only 36.4% similar (54). We suggest that the high degree of nucleotide diversity of lktB is primarily a result of its close proximity to the lktA gene and not because of positive selection acting on LktB. Similarly, the high rate of recombination at the gnd locus of E. coli is thought to be due to its close linkage with genes of the rfb region (5, 41). The latter mediate biosynthesis of the highly antigenic polysaccharide domain (O antigen) of somatic lipopolysaccharide and are believed to be subject to strong diversifying selection in connection with avoidance of the host immune system (44). Thus, amino acid diversity of individual genes within the lktCABD operon depends not only on the degree of nucleotide diversity generated by intragenic recombination but also on evolutionary constraints imposed by the function of the encoded gene product.
In conclusion, this study has shown that the leukotoxin operon of M. haemolytica has a complex mosaic structure and has been derived by relatively recent horizontal DNA transfer and intragenic recombination events. The lktA and lktB genes have the most complex structures with segments derived from up to four different sources, including the related species M. glucosida and P. trehalosi. In contrast, the lktD gene is highly conserved in M. haemolytica. The lktA and lktB genes both have high nucleotide diversities due to their mosaic structures, but the amino acid diversity of LktB is significantly lower than that of LktA due to evolutionary contraint against amino acid replacement. These differences reflect the very different functions of the LktA and LktB proteins. The 5′ and 3′ ends of the operon are highly conserved in M. haemolytica, which strongly suggests that multiple horizontal exchanges of the complete operon have been mediated by a common mechanism such as transduction. The different patterns of recombination in the various evolutionary lineages of M. haemolytica could be explained by differences in bacteriophage host range. Finally, our findings for the leukotoxin operon support the previous conclusion that host switching of bovine A2 strains from cattle to sheep, probably postdating the domestication of these species, has played a major role in the evolution of the operon in ovine M. haemolytica strains.
Acknowledgments
This study was supported by a Wellcome Trust University Award to R.L.D. (053669/Z/98/Z). T.S.W. is supported by grants from the National Institutes of Health.
REFERENCES
- 1.Ambagala, T. C., A. P. N. Ambagala, and S. Srikumaran. 1999. The leukotoxin of Pasteurella haemolytica binds to β2 integrins on bovine leukocytes. FEMS Microbiol. Lett. 179:161–167. [DOI] [PubMed] [Google Scholar]
- 2.Angen, O., R. Mutters, D. A. Caugant, J. E. Olsen, and M. Bisgaard. 1999. Taxonomic relationships of the [Pasteurella] haemolytica complex as evaluated by DNA-DNA hybridizations and 16S rRNA sequencing with proposal of Mannheimia haemolytica gen. nov., comb. nov., Mannheimia granulomatis comb. nov., Mannheimia glucosida sp. nov., Mannheimia ruminalis sp. nov. and Mannheimia varigena sp. nov. Int. J. Syst. Bacteriol. 49:67–86. [DOI] [PubMed] [Google Scholar]
- 3.Baluyut, C. S., R. R. Simonson, W. J. Bemrick, and S. K. Maheswaran. 1981. Interaction of Pasteurella haemolytica with bovine neutrophils: identification and partial characterization of a cytotoxin. Am. J. Vet. Res. 42:1920–1926. [PubMed] [Google Scholar]
- 4.Berggren, C. A., C. S. Baluyut, R. R. Simonson, W. J. Bemrick, and S. K. Maheswaran. 1981. Cytotoxic effects of Pasteurella haemolytica on bovine neutrophils. Am. J. Vet. Res. 42:1383–1388. [PubMed] [Google Scholar]
- 5.Bisercic, M., J. Y. Feutrier, and P. R. Reeves. 1991. Nucleotide sequences of the gnd genes from nine natural isolates of Escherichia coli: evidence of intragenic recombination as a contributing factor in the evolution of the polymorphic gnd locus. J. Bacteriol. 173:3894–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, J. F., F. Leite, and C. J. Czuprynski. 1997. Binding of Pasteurella haemolytica leukotoxin to bovine leukocytes. Infect. Immun. 65:3719–3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burrows, L. L., E. Olah-Winfield, and R. Y. C. Lo. 1993. Molecular analysis of the leukotoxin determinants from Pasteurella haemolytica serotypes 1 to 16. Infect. Immun. 61:5001–5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, Y. F., R. Young, D. Post, and D. K. Struck. 1987. Identification and characterization of the Pasteurella haemolytica leukotoxin. Infect. Immun. 55:2348–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheetham, B. F., and M. E. Katz. 1995. A role for bacteriophages in the evolution and transfer of bacterial virulence determinants. Mol. Microbiol. 18:201–208. [DOI] [PubMed] [Google Scholar]
- 10.Clinkenbeard, K. D., D. A. Mosier, A. L. Timko, and A. W. Confer. 1989. Effects of Pasteurella haemolytica leukotoxin on cultured bovine lymphoma cells. Am. J. Vet. Res. 50:271–275. [PubMed] [Google Scholar]
- 11.Coffey, T. J., M. C. Enright, M. Daniels, J. K. Morona, R. Morona, W. Hryniewicz, J. C. Paton, and B. G. Spratt. 1998. Recombinational exchanges at the capsular polysaccharide biosynthetic locus lead to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol. Microbiol. 27:73–83. [DOI] [PubMed] [Google Scholar]
- 12.Coote, J. G. 1992. Structural and functional relationships among the RTX toxin determinants of gram-negative bacteria. FEMS Microbiol. Rev. 88:137–162. [DOI] [PubMed] [Google Scholar]
- 13.Cruz, W. T., R. Young, Y. F. Chang, and D. K. Struck. 1990. Deletion analysis resolves cell-binding and lytic domains of the Pasteurella leukotoxin. Mol. Microbiol. 4:1933–1939. [DOI] [PubMed] [Google Scholar]
- 14.Czuprynski, C. J., E. J. Noel, O. Ortiz-Carranza, and S. Srikumaran. 1991. Activation of bovine neutrophils by partially purified Pasteurella haemolytica leukotoxin. Infect. Immun. 59:3126–3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies, R. L., S. Arkinsaw, and R. K. Selander. 1997. Evolutionary genetics of Pasteurella haemolytica isolates recovered from cattle and sheep. Infect. Immun. 65:3585–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davies, R. L., S. Arkinsaw, and R. K. Selander. 1997. Genetic relationships among Pasteurella trehalosi isolates based on multilocus enzyme electrophoresis. Microbiology 143:2841–2849. [DOI] [PubMed] [Google Scholar]
- 17.Davies, R. L., and W. Donachie. 1996. Intra-specific diversity and host specificity within Pasteurella haemolytica based on variation of capsular polysaccharide, lipopolysaccharide and outer-membrane proteins. Microbiology 142:1895–1907. [DOI] [PubMed] [Google Scholar]
- 18.Davies, R. L., B. J. Paster, and F. E. Dewhirst. 1996. Phylogenetic relationships and diversity within the Pasteurella haemolytica complex based on 16S rRNA sequence comparison and outer membrane protein and lipopolysaccharide analysis. Int. J. Syst. Bacteriol. 46:736–744. [DOI] [PubMed] [Google Scholar]
- 19.Davies, R. L., and M. Quirie. 1996. Intra-specific diversity within Pasteurella trehalosi based on variation of capsular polysaccharide, lipopolysaccharide and outer-membrane proteins. Microbiology 142:551–560. [DOI] [PubMed] [Google Scholar]
- 20.Davies, R. L., T. S. Whittam, and R. K. Selander. 2001. Sequence diversity and molecular evolution of the leukotoxin (lktA) gene in bovine and ovine strains of Mannheimia (Pasteurella) haemolytica. J. Bacteriol. 183:1394–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feavers, I. M., A. B. Heath, J. A. Bygraves, and M. C. J. Maiden. 1992. Role of horizontal genetic exchange in the antigenic variation of the class 1 outer membrane protein of Neisseria meningitidis. Mol. Microbiol. 6:489–495. [DOI] [PubMed] [Google Scholar]
- 22.Frank, G. H. 1989. Pasteurellosis of cattle, p.197–222. In C. F. Adlam and J. M. Rutter (ed.), Pasteurella and pasteurellosis. Academic Press, London, England.
- 23.Gerbig, D. G., Jr., M. R. Cameron, D. K. Struck, and R. N. Moore. 1992. Characterization of a neutralizing monoclonal antibody to Pasteurella haemolytica leukotoxin. Infect. Immun. 60:1734–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilmour, N. J. L., and J. S. Gilmour. 1989. Pasteurellosis of sheep, p.223–261. In C. F. Adlam and J. M. Rutter (ed.), Pasteurella and pasteurellosis. Academic Press, London, England.
- 25.Highlander, S. K., M. Chidambaram, M. J. Engler, and G. M. Weinstock. 1989. DNA sequence of the Pasteurella haemolytica leukotoxin gene cluster. DNA 8:15–28. [DOI] [PubMed] [Google Scholar]
- 26.Highlander, S. K., M. J. Engler, and G. M. Weinstock. 1990. Secretion and expression of the Pasteurella haemolytica leukotoxin. J. Bacteriol. 172:2343–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeyaseelan, S., S. L. Hsuan, M. S. Kannan, B. Walcheck, J. F. Wang, M. E. Kehrli, E. T. Lally, G. C. Sieck, and S. K. Maheswaran. 2000. Lymphocyte function-associated antigen 1 is a receptor for Pasteurella haemolytica leukotoxin in bovine leukocytes. Infect. Immun. 68:72–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaehler, K. L., R. J. F. Markham, C. C. Muscoplat, and D. W. Johnson. 1980. Evidence of species specificity in the cytocidal effects of Pasteurella haemolytica. Infect. Immun. 30:615–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar, S., K. Tamura, and M. Nei. 1993. MEGA: Molecular evolutionary genetics analysis software for microcomputers. Comput. Appl. Biosci. 10:189–191. [DOI] [PubMed] [Google Scholar]
- 30.Lan, R., and P. R. Reeves. 1996. Gene transfer is a major factor in bacterial evolution. Mol. Biol. Evol. 13:47–55. [DOI] [PubMed] [Google Scholar]
- 31.Lawrence, J. G., and H. Ochman. 1998. Molecular archaeology of the Escherichia coli genome. Proc. Natl. Acad. Sci. USA 95:9413–9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lawrence, J. G., and J. R. Roth. 1996. Selfish operons: Horizontal transfer may drive the evolution of gene clusters. Genetics 143:1843–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, J., K. D. Clinkenbeard, and J. W. Ritchey. 1999. Bovine CD18 identified as a species specific receptor for Pasteurella haemolytica leukotoxin. Vet. Microbiol. 67:91–97. [DOI] [PubMed] [Google Scholar]
- 34.Li, J., K. Nelson, A. C. McWhorter, T. S. Whitam, and R. K. Selander. 1994. Recombinational basis of serovar diversity in Salmonella enterica. Proc. Natl. Acad. Sci. USA 91:2552–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lo, R. Y. C., C. A. Strathdee, and P. E. Shewen. 1987. Nucleotide sequence of the leukotoxin genes of Pasteurella haemolytica A1. Infect. Immun. 55:1987–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maheswaran, S. K., M. S. Kannan, D. J. Weiss, K. R. Reddy, E. L. Townsend, H. S. Yoo, B. W. Lee, and L. O. Whiteley. 1993. Enhancement of neutrophil-mediated injury to bovine pulmonary endothelial cells by Pasteurella haemolytica leukotoxin. Infect. Immun. 61:2618–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maheswaran, S. K., D. J. Weiss, M. S. Kannan, E. L. Townsend, K. R. Reddy, L. O. Whiteley, and S. Srikumaran. 1992. Effects of Pasteurella haemolytica A1 leukotoxin on bovine neutrophils: degranulation and generation of oxygen-derived free radicals. Vet. Immunol. Immunopathol. 33:51–68. [DOI] [PubMed] [Google Scholar]
- 38.Mdurvwa, E. G., and C. J. Brunner. 1994. Bovine neutrophil activation by culture fluid from Pasteurella haemolytica serotypes A1 and A11. Vet. Microbiol. 41:311–319. [DOI] [PubMed] [Google Scholar]
- 39.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418–426. [DOI] [PubMed] [Google Scholar]
- 40.Nelson, K., and R. K. Selander. 1992. Evolutionary genetics of the proline permease gene (putP) and the control region of the proline utilization operon in populations of Salmonella and Escherichia coli. J. Bacteriol. 174:6886–6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nelson, K., and R. K. Selander. 1994. Intergeneric transfer and recombination of the 6-phosphogluconate dehydrogenase gene (gnd) in enteric bacteria. Proc. Natl. Acad. Sci. USA 91:10227–10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ochman, H., J. G. Lawrence, and E. A. Grolsman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299–304. [DOI] [PubMed] [Google Scholar]
- 43.Petras, S. F., M. Chidambaram, E. F. Illyes, S. Froshauer, G. M. Weinstock, and C. P. Reese. 1995. Antigenic and virulence properties of Pasteurella haemolytica leukotoxin mutants. Infect. Immun. 63:1033–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reeves, P. 1993. Evolution of Salmonella O antigen variation by interspecific gene transfer on a large scale. Trends Genet. 9:17–22. [DOI] [PubMed] [Google Scholar]
- 45.Reid, S. D., R. K. Selander, and T. S. Whittam. 1999. Sequence diversity of flagellin (fliC) alleles in pathogenic Escherichia coli. J. Bacteriol. 181:153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richards, A. B., H. W. Renshaw, and L. W. Sneed. 1985. Pasteurella haemolytica bacteriophage: identification, partial characterization, and relationship of temperate bacteriophages from isolates of Pasteurella haemolytica (biotype A, serotype 1). Am. J. Vet. Res. 46:1215–1220. [PubMed] [Google Scholar]
- 47.Saadati, M., H. A. Gibbs, R. Parton, and J. G. Coote. 1997. Characterisation of the leukotoxin produced by different strains of Pasteurella haemolytica. J. Med. Microbiol. 46:276–284. [DOI] [PubMed] [Google Scholar]
- 48.Shewen, P. E., and B. N. Wilkie. 1982. Cytotoxin of Pasteurella haemolytica acting on bovine leukocytes. Infect. Immun. 35:91–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith, J. M. 1992. Analyzing the mosaic structure of genes. J. Mol. Evol. 34:126–129. [DOI] [PubMed] [Google Scholar]
- 50.Smith, J. M. 1999. The detection and measurement of recombination from sequence data. Genetics 153:1021–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith, N. H., P. Beltran, and R. K. Selander. 1990. Recombination of Salmonella phase 1 flagellin genes generates new serovars. J. Bacteriol. 172:2209–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sneath, P. H. A., and M. Stevens. 1990. Actinobacillus rossii sp. nov., Actinobacillus seminis sp. nov., nom. rev., Pasteurella bettii sp. nov., Pasteurella lymphangitidis sp. nov., Pasteurella mairi sp. nov., and Pasteurella trehalosi sp. nov. Int. J. Syst. Bacteriol. 40:148–153. [DOI] [PubMed] [Google Scholar]
- 53.Stevens, P. K., and C. J. Czuprynski. 1996. Pasteurella haemolytica leukotoxin induces bovine leukocytes to undergo morphologic changes consistent with apoptosis in vitro. Infect. Immun. 64:2687–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Strathdee, C. A., and R. Y. C. Lo. 1987. Extensive homology between the leukotoxin of Pasteurella haemolytica A1 and the alpha-hemolysin of Escherichia coli. Infect. Immun. 55:3233–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strathdee, C. A., and R. Y. C. Lo. 1989. Cloning, nucleotide sequence, and characterization of genes encoding the secretion function of the Pasteurella haemolytica leukotoxin determinant. J. Bacteriol. 171:916–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sutherland, A. D. 1985. Effects of Pasteurella haemolytica cytotoxin on ovine peripheral blood leucocytes and lymphocytes obtained from gastric lymph. Vet. Microbiol. 10:431–438. [DOI] [PubMed] [Google Scholar]
- 57.Sutherland, A. D., and W. Donachie. 1986. Cytotoxic effect of serotypes of Pasteurella haemolytica on sheep bronchoalveolar macrophages. Vet. Microbiol. 11:331–336. [DOI] [PubMed] [Google Scholar]
- 58.Welch, R. A. 1991. Pore-forming cytolysins of gram-negative bacteria. Mol. Microbiol. 5:521–528. [DOI] [PubMed] [Google Scholar]
- 59.Yamaguchi, T., T. Hayashi, H. Takami, K. Nakasone, M. Ohnishi, K. Nakayama, S. Yamada, H. Komatsuzawa, and M. Sugai. 2000. Phage conversion of exfoliative toxin A production in Staphylococcus aureus. Mol. Microbiol. 38:694–705. [DOI] [PubMed] [Google Scholar]
- 60.Yoo, H. S., B. S. Rajagopal, S. K. Maheswaran, and T. R. Ames. 1995. Purified Pasteurella haemolytica leukotoxin induces expression of inflammatory cytokines from bovine alveolar macrophages. Microb. Pathog. 18:237–252. [DOI] [PubMed] [Google Scholar]