Abstract
There are two primary levels of control of the expression of the fructanase gene (fruA) of Streptococcus mutans: induction by levan, inulin, or sucrose and repression in the presence of glucose and other readily metabolized sugars. The goals of this study were to assess the functionality of putative cis-acting regulatory elements and to begin to identify the trans-acting factors involved in induction and catabolite repression of fruA. The fruA promoter and its derivatives generated by deletions and/or site-directed mutagenesis were fused to a promoterless chloramphenicol acetyltransferase (CAT) gene as a reporter, and strains carrying the transcriptional fusions were then analyzed for CAT activities in response to growth on various carbon sources. A dyadic sequence, ATGACA(TC)TGTCAT, located at −72 to −59 relative to the transcription initiation site was shown to be essential for expression of fruA. Inactivation of the genes that encode fructose-specific enzymes II resulted in elevated expression from the fruA promoter, suggesting negative regulation of fruA expression by the fructose phosphotransferase system. Mutagenesis of a terminator-like structure located in the 165-base 5′ untranslated region of the fruA mRNA or insertional inactivation of antiterminator genes revealed that antitermination was not a mechanism controlling induction or repression of fruA, although the untranslated leader mRNA may play a role in optimal expression of fructanase. Deletion or mutation of a consensus catabolite response element alleviated glucose repression of fruA, but interestingly, inactivation of the ccpA gene had no discernible effect on catabolite repression of fruA. Accumulating data suggest that expression of fruA is regulated by a mechanism that has several unique features that distinguish it from archetypical polysaccharide catabolic operons of other gram-positive bacteria.
Streptococcus mutans, the principal etiological agent of human dental caries, is able to synthesize and hydrolyze polymers of 𝒹-fructose, commonly known as fructans. The ability of this cariogenic bacterium to produce and degrade fructans enhances the survival of the organism during periods of nutrient starvation, results in prolonged production of organic acids through glycolysis, and has been shown to contribute directly to caries formation (4). Fructanase (FruA) is apparently the sole enzyme involved in utilization of β(2,1)- and β(2,6)-linked extracellular fructose polymers by S. mutans (5, 6). FruA is enzymatically similar to the levanase of Bacillus subtilis (11), attacking levans and inulins to liberate fructose as the only end product and cleaving sucrose into glucose and fructose (6). In S. mutans, fruA is cotranscribed with fruB, which lies immediately downstream of fruA and is predicted to encode a protein with strong similarity to bacterial β-fructosidases, although no function has yet been assigned to FruB (7). Previously, we reported that expression of fruA was inducible by growth on the homopolymers of fructose, levan, and inulin and was also exquisitely sensitive to carbon catabolite repression (CCR) (7; Z. T. Wen and R. A. Burne, Abstr. 100th Gen. Meet. Am. Soc. Microbiol., abstr. H66, p. 362, 2000).
In most cases, the genes for catabolism of nonpreferred carbohydrates and most polysaccharides are inducible by growth on the cognate substrate(s), and induction is frequently controlled by transcriptional activation or antitermination (35). For example, the levanase gene of B. subtilis requires the action of a sigma 54-like regulator that activates transcription when cells are growing with levans or low levels of fructose (22) Similarly, a variety of transcriptional antiterminator proteins with homology to BglG, the transcriptional antiterminator of the Escherichia coli bglGFB operon, are present in Bacillus (SacY, SacT, LicT, and GlcT), Lactobacillus casei (LacT), Staphylococcus aureus, Lactococcus lactis, and S. mutans (BglG) (9, 10, 12, 14, 17, 19, 31, 32, 37), where they have been shown to control the expression of carbohydrate catabolism by allowing transcription of various operons. The binding site of antiterminator proteins, a ribonucleic antiterminator (RAT) sequence, has been well characterized in B. subtilis (2, 20). In many cases, the activity of these antiterminators is modified by both the general and sugar-specific components of the phosphotransferase system (PTS) (36).
CCR of genes for catabolism of secondary carbohydrate sources in Bacillus and other gram-positive bacteria is mediated primarily through catabolite control protein A (CcpA), a member of the LacI-GalR family of bacterial transcriptional regulators (15). A palindromic sequence, the carbon catabolite response element (CRE), located in the promoter regions or coding sequences, is the target site for repression of genes exerted through CcpA (41). The histidine-containing phosphocarrier protein (HPr) of the phosphoenolpyruvate-dependent sugar:PTS, when phosphorylated by a fructose-1,6-bisphosphate-activated HPr kinase at serine residue 46, allosterically stimulates the binding of CcpA to CREs (25). Recently, CcpA-independent mechanisms for CCR that involve PTS-dependent phosphorylation of transcriptional activators and antiterminators have been reported for the levanase and licBCAH operons of B. subtilis and the lac operon of L. casei (14, 22, 38). For example, regulation of the lev operon of B. subtilis by CcpA was evident only when LevR was truncated (22). The carboxyl terminus of LevR was found to be homologous to the BglG-SacY-SacT family of antiterminator proteins, containing a PTS regulatory domain (PRD), and CCR was exerted by phosphorylation through enzyme I (EI) and HPr of the PTS.
Our previous analysis of the fruA promoter revealed the presence of a 165-base 5′ untranslated region (5′UTR) and two elements that may function as CcpA binding sites that flank or overlap the promoter (7). Additionally, we identified two potential transcription terminators, designated SL1 and SL2, in the 5′UTR mRNA, as well as a sequence with roughly 50% identity to RAT sequences that was correctly positioned with respect to the terminator-like sequences (Fig. 1). The purpose of this study was to assess the role of the putative regulatory elements and to begin to identify the trans-acting factors that are involved in catabolite repression and substrate induction of fruA.
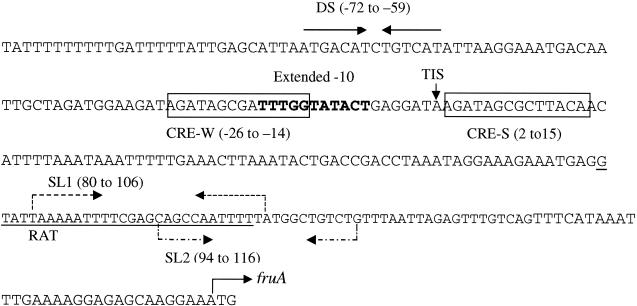
FIG. 1.
Relevant nucleotide sequences and features of the 5′ region of fruA (positions −80 to 152 relative to the TIS). In boldface is the extended −10-like promoter that directs transcription of fruA. Boxed and labeled are two CREs, CRE-S (positions 2 to 15 relative to the TIS) and CRE-W (−26 to −14). Two inverted repeats, SL1 (positions 80 to 106) and SL2 (94 to 116), with potential to function as Rho-independent transcriptional terminators, are indicated with dashed arrows. Overlapping SL1 and SL2 and underlined is a sequence with 50% similarity to a RAT sequence (positions 76 to 104). A dyadic sequence (DS) located at −72 to −59 relative to the TIS is a putative binding site of a positive regulatory protein. The start codon of fruA is indicated with an arrow.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All E. coli strains were grown in Luria-Bertani medium with or without inclusion of ampicillin (100 μg/ml), kanamycin (40 μg/ml), tetracycline (10 μg/ml), erythromycin (500 μg/ml), chloramphenicol (20 μg/ml), kanamycin and chloramphenicol (16 μg/ml each), or erythromycin and tetracycline (10 μg/ml each), when necessary. S. mutans UA159 strains were maintained on brain heart infusion medium, and when necessary, kanamycin (1 mg/ml), tetracycline (10 μg/ml), and/or erythromycin (10 μg/ml) was added. Preparation of competent cells and transformation of S. mutans were done as previously described (7). For enzyme assays and growth studies, S. mutans strains were grown on tryptone-vitamin-based (TV) medium (7) with glucose, inulin (from Dahlia tubers; Sigma), or glucose plus inulin at 0.5% (wt/vol) each as the carbohydrate source(s). All agar media were prepared similarly, but agar (Difco Laboratories, Detroit, Mich.) was added at a concentration of 1.5% (wt/vol).
CAT assay.
For assay of chloramphenicol acetyltransferase (CAT) activity, S. mutans strains were grown in TV medium with the indicated carbohydrates, and early-log-phase cells (optical density at 600 nm ≅ 0.2 to 0.35) were harvested by centrifugation at 3,800 × g at 4°C for 10 min. The cells were then washed once with 10 mM Tris buffer (pH 7.8), and cell pellets either were quickly frozen in an ethanol-dry ice bath and stored at −86°C until assays were to be performed or were immediately homogenized as described previously (7). The cell lysates were assayed for CAT activity using the method of Shaw (33). CAT activity was expressed as nanomoles minute−1 milligram of protein−1.
DNA manipulations.
Unless otherwise stated, standard recombinant DNA techniques were performed as described previously (7, 30). All restriction and modifying enzymes and reagents were purchased from Gibco BRL Life Technologies, Inc. (Rockville, Md.), New England Biolabs (Beverly, Mass.), or MBI Fermentas (Amherst, N.Y.) and used as recommended by the supplier. For Southern hybridization analysis, 12 μg of genomic DNA was digested with enzymes and transferred to NewBond nylon membranes (NEN Life Science Products, Inc., Boston, Mass.). Probes were labeled with [α-32P]dATP using the Random Primers DNA Labeling System of Gibco BRL.
Strategy for dissecting cis-acting elements.
Mutations or deletions were generated by using regular PCR and recombinant PCR (16). All primers (Table 1) were designed in such a way as to introduce a BamHI site at the 5′ ends and a PstI site at the 3′ ends of the PCR products for subsequent cloning purpose. The fruA promoter (PfruA), obtained as a 232-bp fragment upstream of the fruA start codon, or its derivatives generated by site-directed mutagenesis and/or deletions were directly fused with a promoterless CAT gene (cat) in plasmid pU1 (7). The correct nucleotide sequences of all PCR products were confirmed by sequencing. The transcriptional fusions were then released by partial SmaI and HindIII digestions and inserted into EcoRV- and HindIII-digested pBluescript KS(+). The resulting plasmids were then digested with SmaI and HincII, and the DNA fragments with the gene fusions were gel purified and ligated into the integration vector pBGK (42) at the unique SmaI site. Plasmid pBGK is an insertion vector constructed from the S. mutans gtfA gene and allows insertion of foreign DNA immediately downstream, with respect to the direction of transcription of gtfA, of a kanamycin marker flanked by strong transcriptional terminators. Clones carrying the fusions were selected on kanamycin- and chloramphenicol-containing medium and analyzed by BamHI and EcoRI digestions to determine the orientations of the transcriptional fusions with respect to that of the gtfA gene on the vector. Although we have previously shown that orientation of the fusion had little effect on measured transcriptional activity, only those clones with orientations opposite to that of gtfA were selected for further study to ensure that readthrough from gtfA would not affect the results (42). The constructs were then used to transform S. mutans UA159 using procedures described previously (7). Transformants of S. mutans were selected on brain heart infusion containing kanamycin, and integration of the individual fusions into the chromosome as a result of double-crossover recombination was confirmed by PCRs (Table 1).
TABLE 1.
Primers used in this studya
| Primer | Sequence | Application |
|---|---|---|
| ccpa5′ | 5′-GACAGGATCTGAAAGGACAGCAGC-3′ | ccpA amplification |
| ccpa3′ | 5′-CTGGATTAGCTCATCAGTAAATG-3′ | ccpA amplification |
| fruA5′ | 5′-GAGCATTAATGAACTATGTCATATTAAGG-3′ | PfruA amplification |
| fruA3′ | 5′-TTTTCAAATTTATGAAACTGACAAACTC-3′ | PfruA amplification |
| cat3′ | 5′-CGGAAATCGTCGTGGTATTCACTC-3′ | Fusion confirmation |
| mtA | 5′-CTGCTCGAAACTCTGCTATACCTC-3′ | Mutations of SL1 |
| mtB | 5′-GAGGTATAGCAGAGTTTCGAGCAG-3′ | Mutations of SL1 |
| cw1 | 5′-GGAAGATAGACGATACTTTGGTATACTGAGG-3′ | Mutations of CRE-W |
| cw2 | 5′-CCTCAGTATACCAAAGTATCGTCTATCTTCC-3′ | Mutations of CRE-W |
| dCS | 5′-CTATCTTATCCTCAGTATACCAAATC-3′ | Deletion of CRE-S |
| dT | 5′-CTATTTAGGTCGGTCAGTATTTAAC-3′ | Deletion of SL1 and SL2 and their 3′ sequences |
| mcA | 5′-AGATGGTACCTAAAACATTTTAAATAAATTTTTGAAAC-3′ | Mutations of CRE-S |
| mcB | 5′-TTTAGGTACCATCTTATCCTCAGTATACCAAATCGCTAT-3′ | Mutations of CRE-S |
| dtA | 5′-CTCTAATTAAATACCTCATTTCTTTCCTATTTAGGTC-3′ | Deletion of SL1 and SL2 |
| dtB | 5′-GAAATGAGGTATTTAATTAGAGTTTGTCAGTTTC-3′ | Deletion of SL1 and SL2 |
| up1 | 5′-AGGAAATGACAATTGCTAGATG-3′ | Deletion of DS |
| mDS | 5′-GAGCATTAATTCGTTCTGTCATATTAAGG-3′ | Mutations of DS |
| gtfC3′ | 5′-AAAAATAGTTAGAGTTAGTG-3′ | Amplification of PgtfC |
| gtfC5′ | 5′-GATGCTAACTCTGGAGAACG-3′ | Amplification of PgtfC |
| DSR5′ | 5′-CTGACCGACCTAAATAGGAAAG-3′ | Amplification of 3′UTR |
Quantitative RNA analysis.
For slot blot analysis, total RNAs were prepared using a previously described procedure (7). Briefly, S. mutans UA159 and its derivatives were grown to mid-exponential phase (optical density at 600 nm ≅ 0.2 to 0.4) in TV medium supplemented with the carbohydrate(s) of interest at a concentration of 0.5% (wt/vol). Total RNAs were denatured and transferred to nylon membranes (GeneScreen Plus; NEN Life Science Products, Inc.) by using standard procedures (30). RNAs were UV cross-linked to the membranes, and the membranes were then hybridized with radioactively labeled probes.
RESULTS
Analysis of the 5′ UTR.
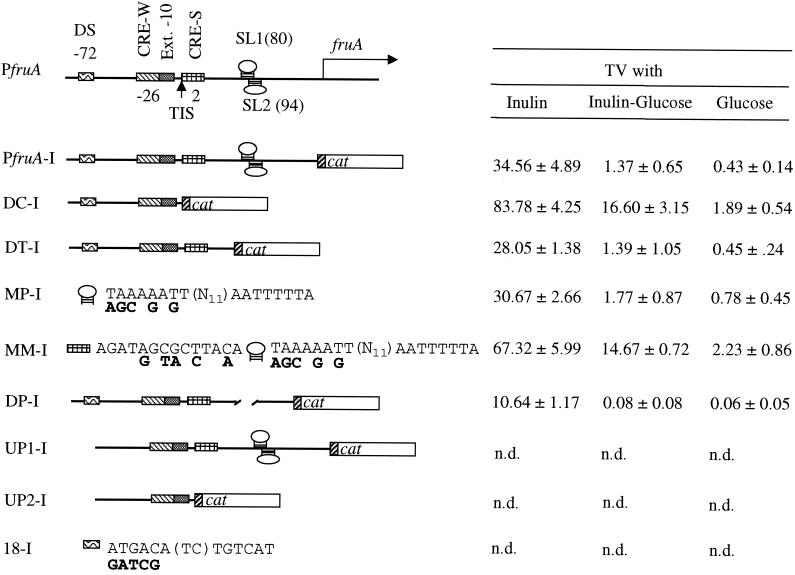
Substrate induction by transcriptional antitermination, in which the antiterminator binds to the upstream region of the RNA hairpin stem of the intrinsic terminator transcript, protecting the elongation complex from destabilization (43), is well documented in gram-positive bacteria (23, 27, 43). Previously, we identified in the 5′ UTR of the fruA transcript two overlapping stem-loops, designated SL1 at positions 80 to 106 (relative to the transcription initiation site [TIS]) and SL2 at positions 94 to 116, with the potential to function as transcriptional terminators (Fig. 1). Overlapping with SL1 (bases 76 to 104) was another dyadic sequence with 50% similarity to the binding site of the BglG family of transcriptional antiterminators, known as RAT sequences (2). CAT activity of the strain that carried the intact promoter-reporter fusion, PfruA-′cat, was 34.56 ± 4.89 nmol min−1 mg of protein−1 in cells grown on inulin, 1.37 ± 0.65 nmol min−1 mg of protein−1 in cells grown on glucose plus inulin, and 0.43 ± 0.14 nmol min−1 mg of protein−1 in cells grown on glucose alone (Fig. 2). Deletion of the two stem-loops (SL1 and SL2) and the 3′ region of the UTR (bases 78 to 152) did not affect the pattern of expression from the fruA promoter (DT-I [Fig. 2]). When SL1 was destabilized by mutating the sequence of the stem region from TAAAAATTTTCGAGCAGCCAATTTTTA to agcAgAgTTTCGAGCAGCCAATTTTTA (MP-I), no differences in expression from the fruA promoter were observed in cells grown on inulin, although there was a slight increase of CAT activity in cells grown on glucose plus inulin (Fig. 2) compared to the wild-type fusion. However, strain DP-I, carrying an internal deletion of SL1 and SL2 (bases 78 to 116), yielded only about one-third of the CAT activity of the strain that carried the PfruA-′cat fusion (10.64 versus 34.56 nmol min−1 mg of protein−1 for the strain with PfruA-′cat [Fig. 2]) when cells were growing on inulin as the sole carbohydrate source. CAT activity was barely detectable when glucose or glucose plus inulin was used to support growth.
FIG. 2.
The fruA promoter (PfruA) and its derivatives (left panel) and CAT activities expressed from the respective fusions after integration into the chromosome and growth of cells with the indicated carbohydrates(s) (right panel). Bar PfruA shows the intact fruA promoter and the putative cis elements SL1 (positions 80 to 106 relative to the TIS), SL2 (94 to 116), CRE-S (2 to 15), CRE-W (−26 to −14), extended −10-like promoter, and DS (−72 to −59) (see Fig. 1 for more detail). All constructs, PfruA and its derivatives generated from deletions and/or site-directed mutagenesis, were fused with a promoterless cat gene, which had its own ribosome binding site. The transcriptional fusions were then integrated into the S. mutans UA159 chromosome. PfruA-I, intact fruA promoter (−80 to 152) fusion integrated in the chromosome; DC-I, PfruA with deletions of CRE-S and the UTR 3′ to the promoter; DT-I, PfruA with deletions of SL1 and SL2 and their 3′UTR; MP-I, PfruA with base substitutions of SL1; MM-I, PfruA with mutations of both CRE-S and SL1; DP-I, PfruA with internal deletions of SL1 and SL2 (positions 78 to 116); UP1-I, PfruA with deletions of DS and its 5′ region; UP2-I, PfruA with deletions of both DS and CRE-S and its 3′UTR; and 18-I, base substitutions of DS. Nucleotides (in boldface) underneath each element indicate the substitutions made. Expression of the fusions were examined by CAT assays of cells grown on TV medium with the indicated sugar(s) as the sole carbohydrate source (see text for more details). CAT activity is expressed as nanomoles minute−1 milligram of protein−1, and the values presented here represent means ± standard deviations from no fewer than four separate experiments. n.d., not detectable.
As demonstrated by enzyme assays and reporter gene studies, FruA is strongly repressed in response to availability of rapidly metabolizable carbohydrates, including fructose and glucose (7) (Fig. 2). Consequently, effects of alterations in the DNA sequence of the 5′UTR on substrate induction may not have been apparent because of the dominant effect of CCR on fruA expression. To eliminate the effects of CCR on expression, the DNA for the 5′UTR beginning just 3′ to CRE-S, and with the same 3′ end as in the aforementioned constructs, was amplified by PCR and cloned behind the S. mutans gtfC promoter, which appears to be constitutively expressed (13). Plasmids carrying the intact fruA promoter PfruA and the constructs MP-I and DP-I (Fig. 2) were used as the templates in separate reactions, generating constructs that had deletions of the 5′ portion of the 5′UTR (46 to −80, relative to the TIS), including CRE-S and the fruA promoter, and that had the 3′ region of this 5′UTR intact (G19), SL1 mutated (G20), or SL1 and SL2 deleted (G21), respectively (Fig. 2). These constructs were then fused with the gtfC promoter (13), which was amplified from S. mutans UA159 by PCR. In general, CAT activities of the strains carrying the truncated 5′UTR or its derivatives were lower than those of strains carrying the fusions that had the 5′ region of the 5′UTR intact (data not shown), although the patterns of expression of cat were similar in all cases. Thus, the results obtained using fusions with mutations and deletions in the 5′UTR indicated that antitermination is not a primary mechanism for induction or repression of fruA. The significant decrease in CAT activity associated with deletion of SL1 and SL2 indicates that these structures may be involved in stability of the fruA transcript.
A dyadic sequence upstream of the fruA promoter is required for fruA expression.
Expression of fruA is inducible by substrate (7) (Fig. 2), with the highest activities measured when levans or inulins are the carbohydrate sources for growth. However, the 152-base 5′UTR of the fruA transcript does not seem to have any major role in induction of fruA in response to growth on inulins or levans, as demonstrated by use of deletions and/or base substitutions of the putative transcriptional terminators and RAT-like sequence (Fig. 1). Consequently, we examined the region 5′ to the promoter and identified an inverted repeat, ATGACA(TC)TGTCAT, designated DS (for dyadic sequence) and located at positions −72 to −59 relative to the initiation site of transcription. Deletion of this region (from −80 to −55) resulted in complete loss of expression of cat from the fruA promoter regardless of the sugar on which cells were grown (UP1-I and UP2-I [Fig. 2]). To verify the functionality of the dyadic sequence in fruA expression, DS was mutated to gatcgA(TC)TGTCAT by recombinant PCR. For that purpose, the intact fruA promoter, PfruA, was used as the template and oligonucleotide primers mDS and fruA3′, or the primers mDS and dCS, were used in PCRs to generate two constructs: one with the full-length fruA promoter and a mutated DS and the other with a mutated DS and deletion of CRE-S and regions 3′ to CRE-S (Fig. 1 and 2). These constructs were then fused with the promoterless cat gene and examined for CAT activity. No CAT activity could be measured with strains that carried the reporter gene fused with either one of the promoters containing mutations of DS (18-I [Fig. 2]), indicating that the dyadic sequence is required, probably as a binding site of a positive regulatory protein, for the transcription of fruA. Preliminary gel shift experiments with cell-free protein preparations from S. mutans UA159 showed some mobility shift of a radioactively labeled oligonucleotide containing the dyadic region (DS), indicating the presence of a factor(s) that interacts with the dyadic sequence (Z. T. Wen and R. A. Burne, unpublished data).
Involvement of the fructose-specific PTS in fruA regulation.
The PTS is not only central to uptake and phosphorylation of various sugars but also mediates transcriptional regulation of numerous genes (24, 29, 36). A class of transcriptional regulators contains two conserved homologous PRDs that function as the targets of PTS-mediated phosphorylation (36, 40). The activator proteins LevR of the lev operon, LicR of the licBCAH operon of B. subtilis, and members of BglG-SacY-SacT family of antiterminator proteins belong to the family of PRD-containing regulators (36). The activity of such regulators is HPr and EI dependent but is negatively regulated by EII proteins of the PTS. As a result of mutations affecting EIIA or EIIB, the PRD-containing regulators become active and the respective catabolic operons are then constitutively expressed.
We have previously characterized two fructose permease gene clusters containing the fruCD and fruI genes. The fruI gene codes for an IIABCFru that is inducible by fructose, and the fruCD genes code for IIBCFru and IIAFru proteins that are constitutively expressed (Z. T. Wen et al., submitted for publication). To examine the functionality of these two enzymes in regulation of fruA expression, strains that had fruI, fruCD, or fruI and fruCD inactivated by deletion of an internal portion of the genes and insertion of a selectable marker were introduced with the PfruA-′cat transcriptional fusion. The resulting transformants were grown on TV medium with glucose, inulin, or glucose plus inulin as the carbohydrate source(s) and analyzed for CAT activity. As shown in Table 2, inactivation of fruI resulted in a twofold increase of CAT activity when inulin was used as the sole carbohydrate source. CAT activity in the fruI wild-type background was barely detectable when the cells were grown on glucose-containing medium. However, CAT activity in the fruI mutant was 6.87 ± 1.41 and 4.71 ± 0.68 nmol min−1 mg of protein−1 when the cells were grown on glucose plus inulin or glucose alone, respectively. The CAT activity in the fruCD mutant background was elevated by 50% when inulin was used as the sole carbohydrate source, but no significant differences were observed between the wild type and the fruCD mutant when the cells were grown on glucose or glucose plus inulin. Clearly, the sensitivity of the operon to repression by glucose exerted through CRE-S makes it difficult to distinguish between induction and alleviation of CCR. Therefore, to exclude the impact of catabolite repression exerted through CRE-S, the fruI, fruCD, and fruI fruCD mutations were introduced into a strain carrying the PfruAΔCRE-′cat fusion, which had CRE-S and the 5′UTR deleted (DC-I) (Fig. 2). Similar to what was seen with the wild type (Fig. 2) following inactivation of fruI or fruCD, expression of the PfruAΔCRE-′cat fusion was increased by as much as fourfold when the cells were grown on inulin, compared to the PfruA-′cat fusion in the wild-type background (Table 2). Furthermore, cat expression under the direction of PfruAΔCRE-′cat in the fruI and fruCD mutants was increased by 7- and 25-fold, respectively, when the cells were grown on glucose plus inulin and glucose alone, compared to CAT expressed from the same promoter in a wild-type background. In the fruCD fruI mutant background, CAT activity expressed from PfruAΔCRE-′cat was further increased by 30 and 10% when the cells were grown on inulin-containing medium and glucose alone, respectively. Therefore, we can conclude that IIFru proteins act as negative regulators of fruA expression and likely control, at least in part, the induction of fruA by its substrates.
TABLE 2.
CAT activities of the S. mutans UA159 fructose-specific EII mutantsa
| Genotype | CAT activity (nmol min−1 mg of protein−1)b of strain grown on TV medium with:
|
||
|---|---|---|---|
| Inulin | Inulin and glucose | Glucose | |
| fruCD+frul+/PfruA-I | 18.50 ± 3.25 | 0.02 ± 0.01 | NDc |
| fruI/PfruA-I | 35.36 ± 4.52 | 6.87 ± 1.41 | 4.71 ± 0.68 |
| fruCD/PfruA-I | 27.41 ± 1.19 | 0.06 ± 0.04 | 0.03 ± 0.03 |
| fruI fruCD/PfruA-I | 45.13 ± 2.89 | 8.82 ± 1.03 | 6.07 ± 0.14 |
| fruCD+fruI+/DC-I | 83.78 ± 4.25 | 16.60 ± 3.15 | 1.89 ± .54 |
| fruI/DC-I | 77.63 ± 1.67 | 118.80 ± 5.02 | 52.11 ± 6.57 |
| fruCD/DC-I | 86.58 ± 2.27 | 116.06 ± 22.04 | 62.44 ± 13.5 |
| fruI fruCD/DC-I | 102.16 ± 9.86 | 140.82 ± 12.8 | 65.17 ± 12.21 |
S. mutans UA159 and its fructose-specific EII mutants, carrying fruCD, fruI, and fruCD fruI, with either PfruA-′cat (PfruA-I) or PfruAΔCRE-′cat (DC-I, PfruA with CRE-S and its 3′UTR deleted) integrated in the chromosome were grown on TV medium with the indicated sugar(s) as the sole carbohydrate source.
CAT activities were measured as described in the text. The data are means ± standard deviations from three separate experiments.
ND, not detectable.
Involvement of CRE sequences in CCR of fruA.
Two sequences, called CRE-S and CRE-W, for strong and weaker homology to the consensus sequence for CREs, were identified at positions 2 to 15 and −26 to −13 with respect to the TIS, respectively (7) (Fig. 1). Previously, by using plasmid-borne transcriptional fusions, we found that deletion of CRE-S and the entire 5′UTR largely alleviated glucose repression of fruA expression (7). To confirm the potential role of CRE-S in CCR and to determine if CRE-W was also involved, the fusion PfruAΔCRE-′cat was cloned into the integration vector pBGK (42) and integrated into the S. mutans chromosome in single copy. As shown in Fig. 2, deletion of CRE-S and the 5′UTR (DC-I) resulted in about a 6-fold increase of CAT activity when the cells were grown on glucose, a 12-fold increase when they were grown on glucose plus inulin, and a 2.5-fold increase when they were grown on inulin, compared to the strain that carried the intact PfruA-′cat fusion grown under identical conditions, which further supports the notion that CRE-S is at least partially responsible for glucose-induced CCR of fruA.
According to Weickert and Chambliss (41), a palindrome with the consensus sequence TGWNANCGNTNWCA is the target site for CCR of many genes in B. subtilis. Some bases are more important than others, but it is the dyad symmetry that determines the strength of this element. The nucleotide sequence of CRE-S, which differs from the consensus at a single base pair, was mutated from AGATAGCGCTTACA to AGATgGtaCcTAaA; base changes in the selected positions have been shown to give maximal derepression of amyE in B. subtilis (41). CAT activity of the strain that had the construct with mutations of both CRE-S and SL1 (MM-I) was increased by sevenfold when the strain was grown on glucose, eightfold when it was on glucose plus inulin, and twofold when it was grown on inulin, but mutations of SL1 alone (MP-I) had no major impact on cat expression (Fig. 2). These data further support the notion that CRE-S is a dominant element involved in CCR of fruA.
CRE-W, positioned at −26 to −13 relative to the TIS, differs from the CRE consensus sequence at positions 1, 13, and 14. Because it overlaps with the probable extended −10 promoter for fruA (7), the feasibility of assessing the functionality of this element without perturbing promoter recognition was uncertain. Notwithstanding this, specific primers were designed for recombinant PCR to introduce point mutations of the CRE-W element without altering key bases in the extended −10 promoter (28) (Fig. 1; Table 1). The resulting PCR product had the CRE-W sequence altered from AGATAGCGATTTGG to AGAcgatacTTTGG. Compared to that in the strain that carried the PfruA-′cat fusion, CAT activity in the strain carrying the mutated CRE-W was decreased by 50% when inulin was the sole carbohydrate source and was undetectable when the cells were grown on glucose-containing medium (data not shown). Although base substitutions were designed so as not to physically alter the putative promoter sequence, it seems that the imposed mutations did in fact interfere with transcription, resulting in a decrease in CAT activity. Importantly, though, no alleviation of CCR was observed, indicating that CRE-W has either a secondary role or no role in CCR of fruA.
Inactivation of ccpA and its impact on glucose repression of fruA..
In Bacillus, CcpA is found to bind CREs and to be primarily responsible for CCR (15, 41). We previously cloned a gene from S. mutans UA159 of 999 bp (Wen and Burne, Abstr. 100th Gen. Meet. Am. Soc. Microbiol.) and with a deduced amino acid sequence that had 52, 59, and 55% identity to the CcpA proteins of B. subtilis, Lactobacillus pentosus, and L. lactis, respectively (3, 15, 21). To assess the impact of the CcpA protein on expression of fruA, mutants were generated by using a spectinomycin resistance element to replace a large portion (nucleotides 56 to 687 relative to the TIS) of the ccpA coding sequences. Replacement of the wild-type ccpA gene was achieved by double-crossover integration of the interrupted ccpA gene, and successful inactivation of the wild-type copy of ccpA was confirmed by Southern blot and Western blot analyses using antibodies that were raised against a purified histidine-tagged CcpA protein from S. mutans GS-5 (E. F. Albone and R. A. Burne, Abstr. 97th Gen. Meet. Am. Soc. Microbiol., abstr. D-166, p. 237, 1997) (data not shown). Several mutant strains were isolated and selected for further experiments.
To determine whether loss of CcpA affected the utilization of fructans, growth of the wild-type strain on glucose, fructose, glucose plus inulin, or fructose plus inulin (at 0.5%, wt/vol) was compared to that of the ccpA mutant. The ccpA mutant grew as well as the wild type on all sugars tested, with the exception that there was a very slight reduction of the growth rate on inulin as the sole carbohydrate source (data not shown). Both the wild type and the ccpA mutant displayed diauxic growth on medium containing 0.1% (wt/vol) glucose and 0.25% inulin, indicating that the utilization of inulin was still sensitive to CCR (data not shown). The ccpA mutation was introduced into the strains carrying PfruA-′cat (PfruA-I), PfruAΔCRE-′cat (DC-I), and PfruAΔSL1-2-′cat (DT-I) fusions, and the transformants were analyzed for CAT activity in response to growth on different carbohydrates. As can be seen in Table 3, there were no major differences between the wild type and the ccpA mutant strains in terms of CAT activities when the cells were grown on glucose, inulin, or glucose plus inulin (Table 3), suggesting that CcpA may not play a role in CCR of fruA under the conditions studied or that there is redundancy in control of CCR of fruA.
TABLE 3.
CAT activity of ccpA mutant strains of S. mutans UA159a
| Strain | CAT activity (nmol min−1 mg of protein−1)b of strain grown on TV medium with:
|
||
|---|---|---|---|
| Inulin | Inulin and glucose | Glucose | |
| S. mutans ccpA+/PfruA-I | 31.23 ± 1.42 | 0.96 ± 0.41 | 0.25 ± 0.15 |
| S. mutans ccpA/PfruA-I | 32.45 ± 3.01 | 1.25 ± 0.45 | 0.5 ± 0.35 |
| S. mutans ccpA/DC-I | 82.06 ± 6.78 | 17.62 ± 3.02 | 0.45 ± 0.36 |
| S. mutans ccpA/DT-I | 28.73 ± 2.48 | 0.94 ± 0.53 | NDc |
The ccpA mutant strain was introduced with transcriptional fusions of the intact fruA promoter (PfruA-I) and its derivatives PfruAΔCRE (DC-I, PfruA with CRE-S and its 3′UTR deleted) and PfruAΔSL1-2 (DT-I, PfruA with SL1 and SL2 and their 3′UTR deleted) (see Fig. 1 and 2 for more detail).
CAT activities were measured as described in the text. The data represent means ± standard deviations from no fewer than three separate experiments.
ND, not detectable.
DISCUSSION
Substrate induction of carbohydrate catabolism in B. subtilis is mediated primarily by transcriptional activators and antitermination (35). Neither the putative transcriptional terminators (SL1 and SL2) nor the RAT-like sequence appears to play a significant role in regulation of fruA in response to carbohydrate source or availability. Further supporting this idea, in a search of the S. mutans UA159 genome database (www.genome.ou.edu/smutans.html) we have identified a gene that we have designated smaT. The smaT gene is predicted to encode a polypeptide of 281 amino acid residues with 44, 33, 30, and 30% identity to LicT of B. subtilis, BglG of E. coli, and SacY and SacT of B. subtilis, respectively. Inactivation of this gene by allelic exchange had no significant effect on growth on fructans or expression of PfruA-′cat fusion under all conditions tested (Wen and Burne, unpublished data). Inactivation of other transcriptional antiterminators in the genome also had no influence on fruA expression, further confirming that the role of the 5′UTR in the fruA mRNA is unrelated to attenuating transcription in response to carbohydrate source or availability.
Considering the fact that the 5′UTR does not have any role in regulation of fruA, it was logical to explore whether induction of fruA could be mediated through a target site located 5′ to the promoter. The data presented in this communication clearly show that a dyadic sequence positioned at −72 to −59 relative to the TIS is required for fruA expression and is probably the target for a transcriptional activator. In some ways, then, the fruA operon of S. mutans is regulated similarly to the levanase (sacC) operon of B. subtilis (22), albeit with some important differences. First, sacC induction requires LevR, which is a sigma 54-like regulator that is genetically linked to sacC. No regulatory genes are linked to the fruA operon, nor is fruA transcribed from a −12/−24-like promoter. Also, there are no genes encoding PTS-like components in the fruA operon, as there are in the B. subtilis levanase operon, and there is no gene in S. mutans that appears to be homologous to levR by use of computer algorithms or Southern hybridization. Further, the target sequence for binding of LevR and the dyadic sequence required for fruA show no similarity. On the other hand, the transcriptional activator LicR of the B. subtilis licBCAH operon binds to an inverted repeat just upstream of the promoter (38), similar to fruA. However, there is no apparent homology between the binding site of LicR and the region upstream of the fruA promoter (data not shown).
Inactivation of the fruI and/or fruCD genes resulted in constitutive elevation of expression of the PfruA-′cat fusion. In some aspects, control of fruA expression via the fructose PTS makes it again similar to levanase expression in B. subtilis (22). Levanase induction in B. subtilis is controlled by a phosphorelay circuit involving four gene products encoded in the levanase operon that phosphorylate LevR when fructose is absent from the growth medium. When low levels of fructose are added, a preferential transfer of phosphate to the incoming fructose leaves the PRD domain of LevR in an unphosphorylated state, which renders the protein competent for activation of sacC transcription. Unlike for the B. subtilis levanase operon, there are no genes in the fruA operon for EII-like gene products that regulate transcription of the operon, yet an apparently similar phosphorelay circuit involving IIFru proteins exists for induction and repression of fructanase in S. mutans. Notably, growth with fructose alone does not induce fruA expression, but we believe that this is a technical anomaly and that the genetic evidence for fructose induction through fructose EIIs provides a more credible picture of the fruA regulatory pathway. Specifically, the reason that there is no apparent induction of fruA in cells grown on fructose is that for the cells to grow well, it is necessary to provide relatively high concentrations of the hexose, which results in catabolite repression of fruA. In contrast, cells growing on inulin or levan, which optimally induce fruA expression, are exposed to much lower steady-state levels of fructose because the hexose is liberated from the fructans at a rate lower than the optimal rate for fructose transport.
Utilization of secondary carbon sources in B. subtilis and other gram-positive bacteria is governed primarily by CcpA, which binds to CREs (1, 15, 39), although CcpA-independent mechanisms have also been reported (14, 22, 38). As observed with other catabolite-repressible systems, deletion or mutation of a promoter-proximal CRE, in this case CRE-S, resulted in a dramatic decrease in CCR of fruA, indicative of the central role of this element in CCR of fruA. Interestingly, inactivation of ccpA in S. mutans had little impact on CCR of fruA (Table 3), consistent with other studies that explored the role of CcpA in CCR in streptococci. For example, in S. mutans GS-5, disruption of the ccpA homologue regM had no effect on diauxic growth when the strain was grown on a variety of nonpreferred carbohydrate sources and glucose. In fact, ccpA inactivation resulted in increased glucose repression of α-galactosidase, mannitol-1-phosphate dehydrogenase, and phospho-β-galactosidase (34). Similarly, sucrose-mediated repression of α-galactosidase (aga) expression by Streptococcus pneumoniae was not affected by mutation of a gene that encodes an apparent CcpA homologue (26). Nevertheless, to our knowledge, this communication provides the first evidence that a putative CRE actually functions in CCR of an operon and yet a CcpA deficiency does not have an impact on expression of the operon containing that CRE. Thus, there is the distinct possibility that CcpA, which we have shown is in fact expressed in UA159, fulfills some other function in the cells and that some other repressor(s) acts at the CRE to exert CCR.
In attempting to reconcile the lack of involvement of CcpA in catabolite repression of an operon with a functional CRE, we identified an open reading frame corresponding to a product with a significant degree of similarity to the CcpB protein of B. subtilis (8). The B. subtilis CcpB protein has 30% identity to CcpA of the same organism and is responsible for CCR when cells are grown on solid medium or in liquid medium with little agitation (8). A homologue of CcpB was identified in S. mutans UA159, and the gene was inactivated using a strategy similar to that for ccpA, but mutation of ccpB alone, or of both ccpA and ccpB, had no effect on expression of the PfruA-′cat fusion (Wen and Burne, unpublished data). Yet another protein in B. subtilis, CcpC, is a member of the LysR family of transcriptional regulators that mediates repression of citB expression by glucose and sources of 2-ketoglutarate (18). However, we could not identify a CcpC paralogue in the genome database. Therefore, if there is another repressor controlling CCR of fruA, and possibly other catabolite-sensitive operons in S. mutans, it is probably not a CcpA, -B, or -C homologue.
We also believe that, like for lev, licBCAH, and some other catabolic operons, EI and HPr may modulate the transcription of the fruA operon by influencing the activity of the transcriptional activator, which could explain the residual CCR in strains carrying gene fusions to the fruA promoter with mutations or deletion of CRE-S (22, 38) (Fig. 2) or in the ccpA mutant of S. mutans. Such a phenomenon, called CcpA-independent CCR, occurs in regulation of licS, licBCAH, and lev operons of B. subtilis and the lac operon of L. casei (14, 31). Isolation of the fruA regulator of transcription will allow us to investigate this hypothesis in more detail.
In summary, expression of fruA in S. mutans UA159 is inducible by inulin and subject to catabolite repression. We present evidence here that expression of fruA requires a transcriptional activator that is probably negatively regulated by components of a fructose-specific PTS and that CCR of fruA occurs through a CRE, as well as through yet-undisclosed mechanisms. Thus, there are fundamental differences between substrate induction and catabolite repression of fruA of S. mutans and other genes encoding polysaccharide-degrading enzymes of eubacteria.
Acknowledgments
We acknowledge J. Lemos and S. Bhagwat for their critical evaluation of the manuscript. We also acknowledge B. A. Roe, R. Y. Tian, H. G. Jia, Y. D. Qian, S. P. Linn, L. Song, R. E. McLaughlin, M. McShan, and J. Ferretti for their immensely valuable contribution to this project through the Streptococcus mutans Genome Sequencing Project.
The Streptococcus mutans Genome Sequencing Project is supported by a grant from the National Institute of Dental and Craniofacial Research. This work was supported by NIDCR grant DE12236.
REFERENCES
- 1.Arnaud, M., M. Debarbouille, G. Rapoport, M. H. Saier, and J. Reizer. 1996. In vitro reconstitution of transcriptional antitermination by the SacT and SacY proteins of Bacillus subtilis. J. Biol. Chem. 271:18966–18972. [DOI] [PubMed] [Google Scholar]
- 2.Aymerich, S., and M. Steinmetz. 1992. Specificity determinants and structural features in the RNA target of the bacterial antiterminator proteins of the BglG/SacY family. Proc. Natl. Acad. Sci. USA 89:10410–10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burne, R. A., Y. Y. Chen, D. L. Wexler, H. Kuramitsu, and W. H. Bowen. 1996. Cariogenicity of Streptococcus mutans strains with defects in fructan metabolism assessed in a program-fed specific-pathogen-free rat model. J. Dent. Res. 75:1572–1577. [DOI] [PubMed] [Google Scholar]
- 5.Burne, R. A., and J. E. Penders. 1994. Differential localization of the Streptococcus mutans GS-5 fructan hydrolase enzyme, FruA. FEMS Microbiol. Lett. 121:243–249. [DOI] [PubMed] [Google Scholar]
- 6.Burne, R. A., K. Schilling, W. H. Bowen, and R. E. Yasbin. 1987. Expression, purification, and characterization of an exo-β-d-fructosidase of Streptococcus mutans. J. Bacteriol. 169:4507–4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burne, R. A., Z. T. Wen, Y. M. Chen, and J. E. C. Penders. 1999. Regulation of expression of the fructan hydrolase gene of Streptococcus mutans GS-5 by induction and carbon catabolite repression. J. Bacteriol. 181:2863–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chauvaux, S., I. T. Paulsen, and M. H. Saier, Jr. 1998. CcpB, a novel transcription factor implicated in catabolite repression in Bacillus subtilis. J. Bacteriol. 180:491–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cote, C. K., D. Cvitkovitch, A. S. Bleiweis, and A. L. Honeyman. 2000. A novel β-glucoside-specific PTS locus from Streptococcus mutans that is not inhibited by glucose. Microbiology 146:1555–1563. [DOI] [PubMed] [Google Scholar]
- 10.Debarbouille, M., M. Arnaud, A. Fouet, A. Klier, and G. Rapoport. 1990. The sacT gene regulating the sacPA operon in Bacillus subtilis shares strong homology with transcriptional antiterminators. J. Bacteriol. 172:3966–3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Debarbouille, M., I. Martin-Verstraete, M. Arnaud, A. Klier, and G. Rapoport. 1991. Positive and negative regulation controlling expression of the sac genes in Bacillus subtilis. Res. Microbiol. 142:757–764. [DOI] [PubMed] [Google Scholar]
- 12.Frenkiel, H., J. Bardowski, S. D. Ehrlich, and A. Chopin. 1998. Transcription of the trp operon in Lactococcus lactis is controlled by antitermination in the leader region. Microbiology 144:2103–2111. [DOI] [PubMed] [Google Scholar]
- 13.Goodman, S. D., and Q. Gao. 2000. Characterization of the gtfB and gtfC promoters from Streptococcus mutans GS-5. Plasmid 43:85–98. [DOI] [PubMed] [Google Scholar]
- 14.Gosalbes, M. J., V. Monedero, and G. Perez-Martinez. 1999. Elements involved in catabolite repression and substrate induction of the lactose operon in Lactobacillus casei. J. Bacteriol. 181:3928–3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henkin, T. M., F. J. Grundy, W. L. Nicholson, and G. H. Chambliss. 1991. Catabolite repression of alpha-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacI and galR repressors. Mol. Microbiol. 5:575–584. [DOI] [PubMed] [Google Scholar]
- 16.Higuchi, R. 1990. Recombinant PCR, p.177–183. In M. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, Inc., New York, N.Y.
- 17.Idelson, M., and O. Amster-Choder. 1998. SacY, a transcriptional antiterminator from Bacillus subtilis, is regulated by phosphorylation in vivo. J. Bacteriol. 180:660–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jourlin-Castelli, C., N. Mani, M. M. Nakano, and A. L. Sonenshein. 2000. CcpC, a novel regulator of the LysR family required for glucose repression of the citB gene in Bacillus subtilis. J. Mol. Biol. 295:865–878. [DOI] [PubMed] [Google Scholar]
- 19.Knezevic, I., S. Bachem, A. Sickmann, H. E. Meyer, J. Stulke, and W. Hengstenberg. 2000. Regulation of the glucose-specific phosphotransferase system (PTS) of Staphylococcus carnosus by the antiterminator protein GlcT. Microbiology 146:2333–2342. [DOI] [PubMed] [Google Scholar]
- 20.Langbein, I., S. Bachem, and J. Stulke. 1999. Specific interaction of the RNA-binding domain of the Bacillus subtilis transcriptional antiterminator GlcT with its RNA target, RAT. J. Mol. Biol. 293:795–805. [DOI] [PubMed] [Google Scholar]
- 21.Mahr, K., W. Hillen, and F. Titgemeyer. 2000. Carbon catabolite repression in Lactobacillus pentosus: analysis of the ccpA region. Appl. Environ. Microbiol. 66:277–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin-Verstraete, I., J. Stulke, A. Klier, and G. Rapoport. 1995. Two different mechanisms mediate catabolite repression of the Bacillus subtilis levanase operon. J. Bacteriol. 177:6919–6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nussbaum-Shochat, A., and O. Amster-Choder. 1999. BglG, the transcriptional antiterminator of the bgl system, interacts with the β′ subunit of the Escherichia coli RNA polymerase. Proc. Natl. Acad. Sci. USA 96:4336–4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase system of bacteria. Microbiol. Rev. 57:534–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reizer, J., A. H. Romano, and J. Deutscher. 1993. The role of phosphorylation of HPr, a phosphocarrier protein of the phosphotransferase system, in the regulation of carbon metabolism in Gram-positive bacteria. J. Cell. Biochem. 51:19–24. [DOI] [PubMed] [Google Scholar]
- 26.Rosenow, C., M. Maniar, and J. Trias. 1999. Regulation of the α-galactosidase activity in Streptococcus pneumoniae: characterization of the raffinose utilization system. Genome Res. 9:1189–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rutberg, B. 1997. Antitermination of transcription of catabolic operons. Mol. Microbiol. 23:413–421. [DOI] [PubMed] [Google Scholar]
- 28.Sabelnikov, A. G., B. Greenberg, and S. A. Lacks. 1995. An extended −10 promoter alone directs transcription of the DpnII operon of Streptococcus pneumoniae. J. Mol. Biol. 250:144–155. [DOI] [PubMed] [Google Scholar]
- 29.Saier, M. H., Jr., and J. Reizer. 1994. The bacterial phosphotransferase system: new frontiers 30 years later. Mol. Microbiol. 13:755–764. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 31.Schnetz, K., J. Stulke, S. Gertz, S. Kruger, M. Krieg, M. Hecker, and B. Rak. 1996. LicT, a Bacillus subtilis transcriptional antiterminator protein of the BglG family. J. Bacteriol. 178:1971–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schnetz, K., C. Toloczyki, and B. Rak. 1987. β-Glucoside (bgl) operon of Escherichia coli K-12: nucleotide sequence, genetic organization, and possible evolutionary relationship to regulatory components of two Bacillus subtilis genes. J. Bacteriol. 169:2579–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaw, W. V. 1979. Chloramphenicol acetyltransferase activity from chloramphenicol-resistant bacteria. Methods Enzymol. 43:737–755. [DOI] [PubMed] [Google Scholar]
- 34.Simpson, C. L., and R. R. Russell. 1998. Identification of a homolog of CcpA catabolite repressor protein in Streptococcus mutans. Infect. Immun. 66:2085–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steinmetz, M. 1993. Carbohydrate metabolism: pathways, enzymes, genetic regulation, and evolution., p.157–170. In A. L. Sonenshein and R. Losick (ed.), Bacillus and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D. C.
- 36.Stulke, J., M. Arnaud, G. Rapoport, and Martin-Verstraete. 1999. PRD—a protein domain involved in PTS dependent induction and carbon catabolite repression of catabolic operons in bacteria. Mol. Microbiol. 28:865–874. [DOI] [PubMed] [Google Scholar]
- 37.Stulke, J., I. Martin-Verstraete, M. Zagorec, M. Rose, A. Klier, and G. Rapoport. 1997. Induction of the Bacillus subtilis ptsGHI operon by glucose is controlled by a novel antiterminator, GlcT. Mol. Microbiol. 25:65–78. [DOI] [PubMed] [Google Scholar]
- 38.Tobisch, S., J. Stulke, and M. Hecker. 1999. Regulation of the lic operon of Bacillus subtilis and characterization of potential phosphorylation sites of the LicR regulator protein by site-directed mutagenesis. J. Bacteriol. 181:4995–5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tobisch, S., D. Zuhlke, J. Bernhardt, J. Stulke, and M. Hecker. 1999. Role of CcpA in regulation of the central pathways of carbon catabolism in Bacillus subtilis. J. Bacteriol. 181:6996–7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tortosa, P., S. Aymerich, C. Lindner, M. H. Saier, Jr., J. Reizer, and D. Le Coq. 1997. Multiple phosphorylation of SacY, a Bacillus subtilis transcriptional antiterminator negatively controlled by the phosphotransferase system. J. Biol. Chem. 272:17230–17237. [DOI] [PubMed] [Google Scholar]
- 41.Weickert, M. J., and G. H. Chambliss. 1990. Site-directed mutagenesis of a catabolite repression operator sequence in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 87:6238–6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wen, Z. T., and R. A. Burne. 2001. Construction of a new integration vector for use in Streptococcus mutans. Plasmid 45:31–36. [DOI] [PubMed] [Google Scholar]
- 43.Yarnell, W. S., and J. W. Roberts. 1999. Mechanism of intrinsic transcription termination and antitermination. Science 284:611–615. [DOI] [PubMed] [Google Scholar]