Abstract
A bovine plasminogen activator of atypical molecular mass (∼45 kDa) from Streptococcus uberis strain SK880 had been identified previously (L. B. Johnsen, K. Poulsen, M. Kilian, and T. E. Petersen. Infect. Immun. 67:1072–1078, 1999). The strain was isolated from a clinical case of bovine mastitis. The isolate was found not to secrete PauA, a bovine plasminogen activator expressed by the majority of S. uberis strains. Analysis of the locus normally occupied by pauA revealed an absence of the pauA open reading frame. However, an alternative open reading frame was identified within the same locus. Sequence analysis of the putative gene suggested limited but significant homology to other plasminogen activators. A candidate signal peptide sequence and cleavage site were also identified. Expression cloning of DNA encoding the predicted mature protein (lacking signal peptide) confirmed that the open reading frame encoded a plasminogen activator of the expected size, which we have named PauB. Both native and recombinant forms of PauB displayed an unexpectedly broad specificity profile for bovine, ovine, equine, caprine, porcine, rabbit, and human plasminogen. Clinical and nonclinical field isolates from nine United Kingdom sites were screened for the pauB gene and none were identified as carrying it. Similarly, clinical isolates from 20 Danish herds were all found to encode PauA and not PauB. Therefore, PauB represents a novel but rare bacterial plasminogen activator which displays very broad specificity.
Mammalian plasminogen activator and plasmin systems play a key role in numerous biological processes including fibrinolysis, degradation of extracellular matrix proteins, cellular migration, and cancer metastasis. Conversion of the plasma zymogen plasminogen to the serine protease plasmin represents the end point of a cascade of reactions culminating in fibrinolytic dissolution of blood clots (3). Plasmin also plays a role in tissue remodeling through its ability to hydrolyze extracellular matrix proteins (34) and can also initiate activation of further metalloproteinases (11). By commandeering elements of this system, a wide range of bacteria are able to bind plasmin(ogen) and thereby augment their pathogenesis over less able competitors (23). A smaller subset of bacterial species that are able to bind plasmin(ogen) also produce potent plasminogen activators. Staphylokinase (SAK), a 15.5-kDa protein secreted by Staphylococcus aureus, forms a complex in association with either fibrin-bound human plasmin or free plasmin in preference to plasminogen (reviewed recently in reference 9). The SAK-plasmin complex can in turn activate other plasminogen molecules to plasmin. Streptokinase (SK), a 47-kDa protein, was the first streptococcal plasminogen activator described for which a mechanism of action was determined (24). In contrast to SAK, SK can associate with either human plasminogen or plasmin to form an activator complex which can then interact with further plasminogen molecules to generate plasmin (29).
Once activated, plasmin can associate with the streptococcal cell surface (21, 22). Bound plasmin(ogen) has been proposed to confer the ability to access deep tissue sites upon certain streptococcal species through the action of plasmin upon extracellular matrix proteins (22). Furthermore, it has also been suggested that by gaining access to essential nutrients through the hydrolytic action of plasmin upon milk proteins, nutritionally fastidious microorganisms such as Streptococcus uberis could colonize environments such as the bovine mammary gland (16). It has been recognized that streptococci produce a diverse range of secreted plasminogen activators capable of converting plasminogen to plasmin and that this occurs in a host-specific manner. Most Lancefield group A, C, and G streptococci isolated from human sources secrete the human plasminogen activator SK. Further plasminogen activators identified in equine and porcine isolates of Streptococcus equisimilis which display little sequence similarity to streptokinase have been described (6). Interestingly, these plasminogen activators exhibit hierarchical preferences for mammalian plasminogens headed in each case by plasminogen obtained from the respective host animals (25, 27). Novel plasminogen activators have also been identified for isolates obtained from bovine streptococcal infections. A 32-kDa plasminogen activator isolated from S. uberis, PauA, displayed a preference for bovine, ovine, and equine plasminogen but was unable to activate porcine or human plasminogen (18). Similarly, a further unrelated 16-kDa plasminogen activator (designated PadA for plasminogen activator dysgalactiae A) was identified in bovine isolates of Streptococcus dysgalactiae which could activate bovine, ovine, equine, and rabbit but not human plasminogens (20). In the course of cloning pauA, which encodes the 32-kDa plasminogen activator from S. uberis, a single isolate (SK880) from a panel of 11 strains tested was reported to display plasminogen-dependent fibrinolysis due to a plasminogen activator of ∼45 kDa (14). This molecule failed to cross-react with antibody raised to PauA, suggesting that significant differences between the two S. uberis plasminogen activators existed (14).
We report here the molecular characterization of a second plasminogen activator (PauB) isolated from S. uberis (SK880) and discuss the significance of this molecule in terms of our understanding of the mechanism of action of streptococcal plasminogen activators and their role in disease.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bovine isolates of S. uberis 0140J and S. uberis SK880 were grown either on sheep blood agar containing esculin (1% [wt/vol]) or in Todd-Hewitt broth at 37°C. Escherichia coli XL1-Blue and BL21(DE3)pLysS (Stratagene Ltd.) were used as host strains for cloned DNA and recombinant expression studies, respectively, and were grown under standard conditions (32).
Extraction of genomic DNA.
Streptococcal genomic DNA was prepared by using a variation of the method of Hill and Leigh (13). Briefly, 1.5 ml of an overnight culture was centrifuged at 10,000 × g for 2 min, and the cell pellet was washed with 500 μl of 10 mM Tris-Cl-5 mM EDTA (pH 7.8). Bacterial cell walls were disrupted by resuspension in 375 μl of 10 mM Tris-Cl-5 mM EDTA (pH 7.8) containing 30 U of mutanolysin and 10 mg of lysozyme (both from Sigma) per ml and subsequent incubation at 37°C for 30 min. Total cell lysis was achieved by addition of 20 μl of sodium dodecyl sulfate (SDS) solution (20% [wt/vol] in 50 mM Tris-Cl, 20 mM EDTA [pH 7.8]) and proteinase K (Sigma) to a final concentration of 150 μg/ml and a further incubation at 37°C for 1 h. Cell wall material was removed by precipitation following the addition of 200 μl of saturated NaCl and subsequent centrifugation at 12,000 × g for 10 min. The supernatant was extracted with phenol chloroform and DNA precipitated by addition of 2 volumes of absolute ethanol. DNA pellets were washed with 70% aqueous ethanol and air dried prior to resuspension in Tris-EDTA (TE) buffer containing 20 μg of RNase A (Sigma) per ml.
DNA amplification and further analysis.
Amplification of DNA was performed using an Omn-E thermal cycler (Hybaid Ltd., Ashford, United Kingdom), with conditions and primers as listed in Table 1. Amplified DNA was purified with DNA Purification Kit II spin columns (Hybaid) and sequenced directly by Cambridge Bioscience, Cambridge, United Kingdom. Sequence analysis was performed using Wisconsin Package software version 10.1, Genetics Computer Group (GCG), Madison, Wis. Analysis of the putative signal peptide of PauB was performed with SignalP software available at http://genome.cbs.dtu.dk/services/SignalP/ (26). Digoxigenin-labeled probe DNA was generated by thermal cycling using a DIG DNA labeling mix (Roche Molecular Biochemicals).
TABLE 1.
Oligonucleotide primer sequences and their applications
| Designation | Sequence | Application (reference) | Template | Annealing temp (°C) |
|---|---|---|---|---|
| P73 | 5′-GAGAGTTTGATCCTGGCTCAGGA | 16S rRNA V2 region (4) | S. uberis genomic DNA | 55 |
| P74 | 5′-TTACCGCGGCTGCTGGCACGT | |||
| ER45 | 5′-GAGATTCCTCTCTAGATATCA | pauA locus/pauB sequencing template (31) | S. uberis genomic DNA | 50 |
| ER46 | 5′-GGGCTGCAGATCCGTTAAAAAATGACATTAATAT | |||
| P44 | 5′-GACGACGACAAGATAACCGGTTATGATTCCGAC | LIC pauA | S. uberis 0140J genomic DNA | 50 |
| P45 | 5′-GGAACAAGACCCGTATTTAATGGATACTTCCTTTA | |||
| P169 | 5′-GACGACGACAAGATCACTTCAAAAGAAGTTAATTACC | LIC pauB | S. uberis SK880 genomic DNA | 55 |
| P170 | 5′-GGAACAAGACCCGTCTTTATTTCAGTACCAATTATTGC | |||
| P38 | 5′-AATAACCGGTTATGATTCCGACTAC | pauA gene probe (31) | S. uberis 0140J genomic DNA | 50 |
| P39 | 5′-AAAATTTACTCGAGACTTCCTTTAAGG | |||
| P168 | 5′-CAAAGTAGAGGCCATGGCTTCAAAAGAAG | pauB gene probe | S. uberis SK880 genomic DNA | 50 |
| P167 | 5′-CACTTTATTTCGGATCCAATTATTGC |
Dot blot analysis of streptococcal field isolates was performed by using Immobilon-Ny+ transfer membrane and a 96-well vacuum manifold (both from Millipore Corp.) using standard techniques (2). Samples for dot blot analysis were prepared as follows. Five hundred microliters of a stationary-phase broth culture was centrifuged at 10,000 × g for 2 min, and the cell pellet was washed with 500 μl of 10 mM Tris-Cl-1 mM EDTA (pH 8). Samples were incubated at 100°C for 10 min and then transferred to ice. Once cool, the lysed cell debris was removed by centrifugation at 10,000 × g for 4 min. Two hundred microliters of each cleared lysate was then applied to the transfer manifold, and the samples were immobilized on nylon membrane held under vacuum. The filter was air dried for 2 h prior to prehybridization and hybridization at 65°C using digoxigenin-labeled probes amplified from S. uberis 0140J and SK880. Filters were washed down to 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS at 65°C, and hybridization signals were visualized according to the manufacturer’s instructions (Roche Molecular Biochemicals). The potential for nonspecific immunoglobulin binding by some candidate strains such as S. dysgalactiae was negated by introduction of 1% bovine serum (∼0.1 mg of immunoglobulin G per ml) prior to adding anti-digoxigenin Fab fragment during the visualization stage.
Expression cloning of plasminogen activators.
Genomic DNA encoding mature PauA and the predicted mature PauB open reading frame (ORF) was amplified using oligonucleotide primer pairs P44-P45 and P169-P170, respectively, under the conditions specified in Table 1. The primers were designed in accordance with the requirements for ligation-independent cloning (1) into the inducible expression vector pCAL-n-FLAG (Stratagene). Amplified DNA was prepared for cloning as described in the Affinity LIC Cloning and Protein Purification Kit manual (Stratagene), and recombinants were identified by sequence analysis of plasmid DNA prepared from E. coli SoloPack host cells using QIAprep spin columns (Qiagen). The pCAL-n-FLAG expression vector enabled production of fusion proteins composed of a 4-kDa calmodulin-binding protein (CBP) linked via a short polypeptide encoding the FLAG epitope to the mature plasminogen activators. Expression of recombinant fusion proteins was induced by growth of E. coli BL21(DE3)pLysS (Stratagene) harboring the CBP-FLAG-plasminogen activator constructs in Luria-Bertani medium supplemented with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 37°C.
Partial purification and N-terminal sequencing of PauB from S. uberis SK880.
Todd-Hewitt broth cultures of S. uberis SK880 were grown overnight at 37°C to stationary phase. Cleared culture supernatant was recovered following centrifugation at 10,000 × g for 2 min. PauB was selectively precipitated typically from 5 ml of broth culture using sequential 30 and 40% ammonium sulfate saturation at room temperature for 2 h. The precipitated material recovered following centrifugation at 10,000 × g for 5 min was redissolved in approximately 500 μl of phosphate-buffered saline. Samples were prepared for N-terminal sequence analysis according to the method of Coligan et al. (8). Briefly, samples were prepared for SDS-polyacrylamide gel electrophoresis (PAGE) in the concentration range of 100 to 200 pmol. Polyacrylamide gels (4% stacking; 8% resolving) were cast well in advance to allow complete polymerization according to the method of Laemmli (17). Gels were prerun with additional 2 mM mercaptoacetic acid present in the upper buffer reservoir to scavenge amino-terminal-blocking free radicals. Reduced and denatured samples were resolved at 15 V/cm and then transblotted to Immobilon-PSQ (polyvinylidene fluoride) transfer membrane (Millipore) with 10 mM cyclohexylamino propane sulfonic acid (pH 11)-1 mM dithiothreitol-10% (vol/vol) methanol as transfer buffer at 100 V for 1 h. Transferred protein was visualized on the membrane by rinsing with distilled water followed by immersion in 0.1% (wt/vol) amido black for 5 s and extensive destaining in distilled water. A heavily stained band at the appropriate molecular weight was excised and prepared for amino acid sequence analysis.
Zymographic analysis of S. uberis plasminogen activators.
Samples containing partially purified plasminogen activators PauA and PauB were prepared by ammonium sulfate precipitation from culture supernatants of S. uberis 0140J and SK880, respectively, as described above. Samples containing approximately 50 ng of plasminogen activator were denatured and reduced but not boiled prior to being resolved by SDS-PAGE alongside prestained molecular weight marker proteins (New England Biolabs, Inc.). SDS was removed by washing the gel twice in 2.5% Triton X-100 for 30 min at room temperature with gentle shaking. The acrylamide gel was overlaid upon a 1% agarose gel made with phosphate-buffered saline and containing 1% (wt/vol) Oxoid skim milk and 1 U of bovine plasminogen (Sigma) per ml and incubated at 37°C. Polaroid 667 film was used to record images (1/125 s, f11) showing proteolytic zones due to the activity of activated plasminogen.
Specificity profiling of S. uberis plasminogen activators.
The secreted (mature) forms of plasminogen activators from S. uberis 0140J and SK880 were partially purified from broth culture supernatants as described above. Expression of CBP-tagged recombinant versions of the mature forms of the PauA and PauB plasminogen activators described above was induced in overnight broth cultures. Crude cell lysates of the recombinant clones were prepared by three successive freeze-thaw cycles of bacterial cells resuspended in 0.5 volume of phosphate-buffered saline followed by centrifugation of cellular debris at 10,000 × g for 5 min. The relative amounts of plasminogen activators prepared from S. uberis broth cultures and the recombinant equivalents from E. coli clones were determined by SDS-PAGE and found to be of comparable concentration. Ten microliters of plasminogen activator preparation or equivalent control sample was incubated with 6 μl of human, rabbit, porcine, ovine, equine, or bovine plasminogen (Sigma, 1 U/ml) at 37°C for 1 h. Caprine plasminogen purified from goat serum (Harlan Sera-Lab Ltd. Loughborough, United Kingdom) at 0.75 mg/ml was kindly donated by Abu-Bakr Abu-Median (Institute for Animal Health, Compton, United Kingdom). A similar volume (6 μl) containing 4.5 μg of caprine plasminogen was incubated with the various plasminogen activator preparations. Following incubation, 15 μl of each sample was introduced into wells cut into 1% agarose gel made with phosphate-buffered saline and 1% Oxoid skim milk and incubated for up to 24 h at 37°C. Zones of clearance due to proteolytic cleavage of milk proteins were recorded photographically at suitable intervals as described above.
Nucleotide sequence accession number.
The nucleotide sequence for the region of S. uberis pauB in strain SK880 has been submitted to the EMBL nucleotide database under accession number AJ314852.
RESULTS
Confirmation of the identity of isolate SK880.
When grown on sheep blood agar containing esculin, strain SK880 presented a morphology typical of S. uberis. In addition, when grown in the presence of esculin and viewed under UV light, strain SK880 was clearly surrounded by a darker, nonfluorescent zone indicative of esculin hydrolysis (10) in a manner typical of S. uberis. Sequence analysis of PCR-amplified DNA from SK880, corresponding to that from the highly variable V2 region of 16S ribosomal RNA exactly matched the sequence obtained for S. uberis 0140J and other S. uberis isolates (data not shown) (4).
Zymographic analysis of S. uberis SK880.
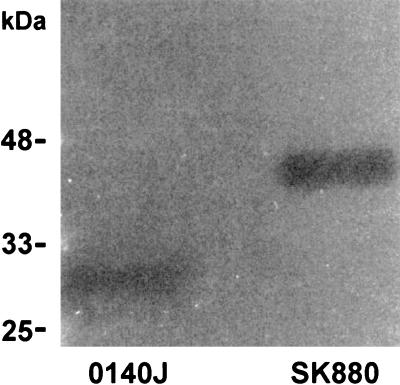
Concentrated cell-free culture supernatant from broth cultures of S. uberis SK880 was shown to generate zones of proteolytic clearance in agarose overlay containing bovine plasminogen and skim milk (Fig. 1). This activity was demonstrated to be plasminogen specific through an absence of proteolysis when bovine plasminogen was omitted from the overlay (data not shown). Comparisons of mobility in SDS-PAGE and subsequent zymography confirmed the unusually high molecular mass of the plasminogen activator from S. uberis SK880 (43 to 45 kDa), in agreement with the previously reported estimate of approximately 45 kDa (14). In addition, zymographic analysis showed that S. uberis SK880 lacked a plasminogen activator of a size corresponding to that of PauA (∼30 kDa).
FIG. 1.
Zymographic analysis of culture supernatants from S. uberis 0140J and SK880. Zones of proteolysis due to activated bovine plasmin are shown in a skim milk-agarose overlay.
Investigation of the pauA locus in S. uberis SK880.
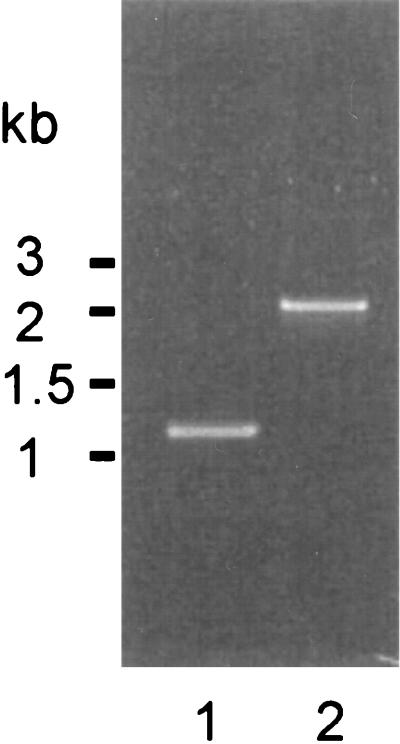
Primers ER45 and ER46 complimentary to the ORFs identified on either flank of pauA in strain 0140J (31) were used to amplify chromosomal DNA from the corresponding locus in S. uberis SK880. A single amplification product of approximately 2.1 kbp was generated, compared to a 1.2-kbp product isolated from S. uberis 0140J (Fig. 2). Amplification with primers complimentary to either strand of the coding sequence of pauA in conjunction with primers anchored within the flanking ORFs was attempted and in all cases yielded no product from S. uberis SK880, suggesting the absence of PauA coding sequence at this locus (data not shown).
FIG. 2.
Amplification products from the hexA-orf1 locus of S. uberis. Lane 1, S. uberis 0140J; lane 2, S. uberis SK880.
Identification of PauB ORF.
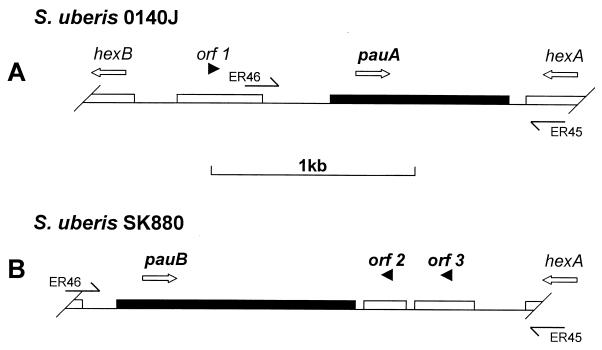
Sequence analysis of the 2.1-kbp amplification product generated from S. uberis SK880 with pauA flanking primers identified one large ORF of approximately 1.1 kbp (Fig. 3). Analysis of this ORF suggested only limited homology at the genetic level to other known streptococcal plasminogen activators. The highest score achieved was 58% identity over a 231-nucleotide segment of S. equisimilis ESK from strain 87-542-W, isolated from an equine host (6). The molecular mass of the theoretical translation product of 377 amino acids was estimated to be 43.8 kDa, which agreed closely with the apparent size of the plasminogen activator under zymographic analysis. Blast searches of protein databases using this sequence identified homology to SAK (31% identity and 54% conservation over 160 residues) and S. equisimilis ESK (27% identity and 47% conservation over 359 residues). Lower scores were obtained to classical SK-type plasminogen activators (i.e., those from isolates of groups A, C, and G streptococci of human origin) and also to the S. uberis plasminogen activator PauA. In the light of such low levels of sequence homology, the plasminogen activator identified in S. uberis SK880 was considered novel and was named PauB (for plasminogen activator uberis B). A putative leader peptide sequence of 25 residues that could direct export of the pauB gene product was identified by comparison to the precursor N-terminal sequences of other streptococcal plasminogen activators (data not shown). Additional use of SignalP software designed to facilitate prediction of leader peptides and their likely processing sites (26) also predicted a cleavage site located between Ala25 and Ile26. Furthermore, a hydrophobicity plot generated from the theoretical gene product showed a clear change from a hydrophobic to a hydrophilic nature at this point (data not shown).
FIG. 3.
Schematic representation of pauA in S. uberis 0140J (A) and pauB in S. uberis SK880 (B). Open arrows represent the orientation of plasminogen activator and flanking genes. Filled arrowheads indicate the orientation of putative ORFs. Partial and horizontal arrows indicate oligonucleotide primer locations (Table 1).
Two smaller nonoverlapping ORFs running antiparallel to the pauB ORF were also identified (Fig. 3). These ORFs spanned a region of genomic DNA exhibiting homology to part of a single but larger negative regulator for transcription of fibrillar surface protein antigen (pag) of Streptococcus sobrinus (accession number D13323). Corresponding Blast homology searches using the theoretical translation products of both ORFs identified regions homologous to part of the S. sobrinus regulatory element and also to the Listeria monocytogenes surface protein negative regulator (accession number CAC13964).
The GC contents of the 2.1-kbp amplification product from S. uberis SK880 and the corresponding 1.1-kbp product from S. uberis 0140J were both calculated to be 33.1%. No evidence of homology to bacteriophage-like elements or inverted and direct repeat sequences suggestive of transposon insertion was found at either end of the 2.1-kbp segment of S. uberis SK880 DNA.
Confirmation of N-terminal processing of PauB.
Preparations of native PauB isolated from cell-free culture supernatant were subjected to N-terminal sequence analysis to confirm the point where cleavage of the putative leader peptide occurred. Three determinations were made due to the presence of additional contaminating proteins in the samples. The first eight residues of the PauB sequence were clearly identified and confirmed the predicted cleavage point between Ala25 and Ile26, yielding a mature protein commencing at the NH2 end with Ile, Thr, Ser, Lys, Glu, Val, Asn, Tyr.
Expression cloning of the mature pauB gene.
Genomic DNA encoding mature PauB commencing at Ile26 was amplified from S. uberis SK880. The mature pauB coding sequence was cloned into the expression vector pCAL-n-FLAG (Stratagene) and introduced into a suitable E. coli host strain. Expression of a functional fusion protein comprising a calmodulin-binding moiety linked to the N terminus of the mature PauB molecule was verified by microtiter tray assay using clarified cell lysates, bovine plasminogen (Sigma), and plasmin-specific chromogenic substrate H-D-Val-Leu-Lys-pNA as described previously (15).
Specificity profiling of PauB.
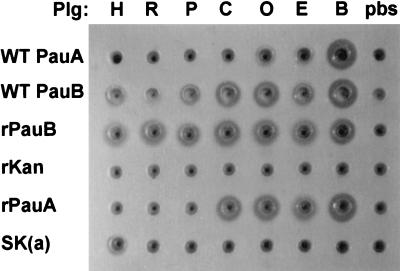
The abilities of preparations of PauB isolated from S. uberis SK880 culture supernatant and recombinant PauB (rPauB) to activate a variety of mammalian plasminogens were determined alongside those of PauA and SK (Fig. 4). PauA and PauB preparations from S. uberis 0140J and SK880, respectively, activated bovine, ovine, and equine plasminogen. Native and recombinant PauB were able to activate caprine plasminogen. The recombinant form of PauA also appeared to activate caprine plasminogen, whereas native PauA did not. Both native and recombinant PauA were unable to activate porcine, rabbit, and human plasminogen. Surprisingly, both native and recombinant preparations of PauB were shown to activate porcine, rabbit, and human plasminogen.
FIG. 4.
Specificity profiles of native and recombinant plasminogen activators from S. uberis. Wells were cut in phosphate-buffered saline-agarose containing 1% skim milk. To these were added preincubated mixtures of plasminogen activator preparations such as WT PauA and WT PauB, wild-type plasminogen activators from culture supernatants of S. uberis 0140J and SK880, respectively; rPauA and rPauB, cleared whole-cell lysates from recombinant clones of PauA and PauB, respectively; SK(a), culture supernatant containing streptokinase from Group A streptococcus strain 0358) or a control recombinant lysate (rKan, encoding a kanamycin resistance gene) with mammalian plasminogen preparations designated as follows: H, human; R, rabbit; P, porcine; C, caprine; O, ovine; E, equine; B, bovine or a buffered saline control (pbs).
Prevalence of PauB in field isolates obtained from bovine milk.
The prevalence of the gene encoding PauB in isolates from the United Kingdom herd was determined by using allele-specific amplification from genomic DNA or full-length gene probes to conduct dot blot analysis of immobilized nucleic acid prepared from bacteria (predominantly S. uberis) isolated from bovine milk. Isolates tested were gathered from nine different farms located across the South of the United Kingdom. They corresponded to a variety of time points during the lactation cycle and also represented clinical and subclinical cases of mastitis. Growth characteristics on blood-esculin media were used initially to select predominantly S. uberis for this analysis. One hundred twelve isolates were tested, of which none were positive for the pauB allele; however, 88 of these were shown to carry the pauA allele (data not shown). Of the 24 isolates that carried neither pauA nor pauB, 6 of 6 were found to be species other than S. uberis when subjected to 16S rRNA analysis (data not shown). Similarly, Southern analysis of 20 clinical mastitis isolates, each gathered from a different Danish herd, was unable to detect pauB; however, pauA was present in all cases (data not shown).
DISCUSSION
Effector molecules that facilitate host-pathogen interactions involving pathogenic veterinary streptococci have come under increasing scrutiny in recent years (7, 12, 19, 28). Correspondingly, a number of novel streptococcal plasminogen activators distinct from classical SK have been identified over this period. These include ESK and PSK, identified from equine and porcine isolates of S. equisimilis (6, 27), PadA, identified in S. dysgalactiae (20), and PauA, the well-characterized plasminogen activator of S. uberis (14, 18, 31, 33). Intriguingly, each of these plasminogen activators exhibits activity towards mammalian plasminogen in a species-specific manner. In all examples to date, the host animal’s plasminogen and that of a limited number of additional species have been shown to interact with these molecules, leading to the conclusion that the pathogenesis of these streptococci is in some way dependent upon activation of host plasminogen.
PauB represents a second plasminogen activator identified from S. uberis. Homology scores derived from pairwise amino acid sequence alignments of mature plasminogen activators clearly illustrate the sequence diversity displayed by this expanding group of bacterial proteins (Table 2). The level of homology shown by PauB to other bacterial plasminogen activators may reflect the functional requirements of these molecules, namely, specific interaction with plasminogen leading to the formation of an activator complex. There is, however, little evidence from sequence analysis to suggest an evolutionary link between PauB and any of the other bacterial plasminogen activators.
TABLE 2.
Pairwise homology scoresa of streptococcal and staphylococcal bacterial plasminogen activators
| Plasminogen activators GenBank accession no) | % identity (% similarity) with:
|
||||||
|---|---|---|---|---|---|---|---|
| PauA | SK(a) | SK(c) | SK(g) | ESK(c) | PSK(c) | SAK (X00127) | |
| PauB (AJ314852) | 24.7 (38.3) | 26.1 (36.3) | 24.6 (34.9) | 24.6 (34.9) | 30.6 (42.0) | 26.0 (36.9) | 36.4 (53.7) |
| PauA (AJ012549) | 30.4 (42.8) | 33.2 (45.1) | 31.9 (44.9) | 25.1 (35.9) | 37.2 (45.7) | 24.5 (35.8) | |
| SK(a) (P10520) | 88.4 (90.3) | 88.9 (90.6) | 25.7 (33.9) | 33.3 (43.2) | 23.5 (35.3) | ||
| SK(c) (P00779) | 98.1 (98.3) | 19.2 (28.6) | 34.7 (45.4) | 25.0 (37.5) | |||
| SK(g) (X13400) | 19.6 (28.6) | 34.2 (45.2) | 25.0 (37.5) | ||||
| ESK(c) (AF104301) | 24.1 (34.0) | 28.7 (44.2) | |||||
| PSK(c) (AF104300) | 24.5 (32.6) | ||||||
Scores for pairwise alignment of mature amino acid sequences were generated with the gap program (GCG).
In attempting to define the mode of action of SK, the observation was made that the extreme N-terminal amino acids Ile1, Ala2, and Gly3 were similar in nature to the N-terminal residues of host-activated human plasmin (Val562, Val563, and Gly564) (35). Under the influence of host plasminogen activators (urokinase or tissue-type plasminogen activator), proteolytic cleavage of the activation loop of plasminogen between Arg561 and Val562 represents a key stage in the conversion of plasminogen. The new N-terminal amino acid (Val562) is thought to form a salt bridge with Asp740 adjacent to Ser741, one of three active site residues thereby helping to form the active center of plasmin using a mechanism similar to the activation of trypsinogen to trypsin (30). In the absence of proteolysis, bacterial plasminogen activators of the SK family interact with plasminogen to form activator complexes within which the active site of plasmin is generated. It was suggested that the N terminus of SK might form a surrogate salt bridge with Asp740 to achieve the specific interaction and conformational adjustments required to generate the active center (5). Further investigation of the role of the extreme N-terminal residues of mature SK supported this proposal when the essential nature of Ile1 in the activation of human plasminogen was demonstrated (35). Comparison of the N termini of other bacterial plasminogen activators PauA and PSK appears to support these findings; indeed, the predicted mature N terminus of PauB also conforms to this pattern (Table 3). One exception to this pattern is apparent in ESK, where the reported mature N terminus is biochemically quite different, leading to speculation as to whether an alternative mechanism might be employed to trigger the formation of plasmin in this case.
TABLE 3.
Alignment of the extreme N termini of host-activated plasminogen and mature bacterial plasminogen activators
| Protein | N-terminal amino acid sequence | ||
|---|---|---|---|
| Human plasminogen | Val562 | Val563 | Gly564 |
| Bovine plasminogen | Ile562 | Val563 | Gly564 |
| Classical SK | Ile1 | Ala2 | Gly3 |
| PauA | Ile1 | Thr2 | Gly3 |
| PSK | Ile1 | Gly2 | Gly3 |
| PauB | Ile1 | Thr2 | Ser3 |
| ESK | Asn1 | Asn2 | Tyr3 |
Prior to this communication, all streptococcal plasminogen activators were reported to display distinct levels of species specificity. Classical SK isolated from human streptococcal pathogens is highly specific for human plasminogen, whereas PauA activates bovine, ovine, and equine, but not human plasminogen. The finding that PauB appeared to activate every mammalian plasminogen tested was surprising. No evidence of proteolytic degradation of milk proteins was evident from preparations of PauB (native or recombinant) when assayed in the absence of plasminogen. It remains to be determined whether the apparent lack of species specificity displayed by PauB is due to its primary sequence or perhaps results from an alternative mechanism of activation.
In the course of cloning the pauB gene from S. uberis SK880 it became apparent that this novel plasminogen activator occupied a locus normally filled by pauA. This finding clearly explained why no caseinolytic activity due to PauA was apparent on zymographic analysis; however, it did raise questions as to the origin and significance of the presence of pauB in this locus. The pauB gene was flanked by sequence homologous to a regulatory element from an oral streptococcus of the mutans group; however, no evidence to suggest horizontal transmission was identified. On the basis of the high level of sequence conservation observed for PauA (31) and its flanking sequences, it is tempting to speculate that this locus has served as a repository for determinants essential to the pathogenesis of S. uberis.
Evidence of pauB was not found in S. uberis isolated from the United Kingdom dairy herd. The occurrence of pauB in Danish isolates was reported once from a screen of 11 strains tested (14). A further investigation in this study found no further evidence of pauB in isolates from 20 different Danish herds. If the distribution of pauB within S. uberis is as low as these studies have suggested, the possibility that the acquisition of pauB by SK880 occurred recently is raised. Conversely, it could be argued that there was little advantage to be gained by S. uberis expressing PauB rather than PauA, thus limiting the spread of a low-abundance determinant. The finding of bovine plasminogen activators in the overwhelming majority of S. uberis isolates tested does suggest a role for these molecules in the pathogenesis of the bacterium. It remains to be seen whether the abundance of PauB increases within S. uberis or whether indeed it is identified in other streptococci, resulting from the absence of species specificity displayed by PauB.
Acknowledgments
We are most grateful to Laust Johnsen (Protein Chemistry Laboratory, Department of Molecular and Structural Biology, University of Aarhus, Denmark) for the kind gift of S. uberis SK880. In addition, the generous gifts of field isolates from the United Kingdom (Elizabeth Berry, Institute for Animal Health, Compton, United Kingdom) and Denmark (Lars Holst Pedersen, The Royal Veterinary and Agricultural University, Institute for Veterinary Microbiology, Stigbøjlen, Denmark) are acknowledged. N-terminal amino acid sequencing was performed by Lawrence Hunt (IAH, Compton) and Patrick Barker (Microchemical Facility, The Babraham Institute, Cambridge, United Kingdom).
We acknowledge funding from the Ministry of Agriculture, Fisheries and Food.
REFERENCES
- 1.Aslanidis, C., and P. J. de Jong. 1990. Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res. 18:6069–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 2001. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Bachman, F. 1994. Fibrinolysis, p. 549–625. In A. L. Bloom, C. D. Forbes, D. P. Thomas, and E. G. D. Tuddenham (ed.), Haemostasis and thrombosis. Churchill Livingstone, London, United Kingdom.
- 4.Bentley, R. W., and J. A. Leigh. 1995. Development of PCR-based hybridization protocol for identification of streptococcal species. J. Clin. Microbiol. 33:1296–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bode, W., and R. Huber. 1976. Induction of the bovine trypsinogen-trypsin transition by peptides sequentially similar to the N-terminus of trypsin. FEBS Lett. 68:231–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caballero, A. R., R. Lottenberg, and K. H. Johnston. 1999. Cloning, expression, sequence analysis, and characterization of streptokinases secreted by porcine and equine isolates of Streptococcus equisimilis. Infect. Immun. 67:6478–6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calvinho, L. F., R. A. Almeida, and S. P. Oliver. 1998. Potential virulence factors of Streptococcus dysgalactiae associated with bovine mastitis. Vet. Microbiol. 61:93–110. [DOI] [PubMed] [Google Scholar]
- 8.Coligan, J. E., B. M. Dunn, H. L. Ploegh, D. W. Speicher, and P. T. Wingfield (ed.). 2001. Current protocols in protein science. John Wiley & Sons, Inc., New York, N.Y.
- 9.Collen, D. 1998. Staphylokinase: a potent, uniquely fibrin-selective thrombolytic agent. Nat. Med. 4:279–284. [DOI] [PubMed] [Google Scholar]
- 10.Edberg, S. C., K. Gam, C. J. Bottenbley, and J. M. Singer. 1976. Rapid spot test for the determination of esculin hydrolysis. J. Clin. Microbiol. 4:180–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldberg, G. I., S. M. Frisch, C. He, S. M. Wilhelm, R. Reich, and I. E. Collier. 1990. Secreted proteases. Regulation of their activity and their possible role in metastasis. Ann. N. Y. Acad. Sci. 580:375–384. [DOI] [PubMed] [Google Scholar]
- 12.Gottschalk, M., and M. Segura. 2000. The pathogenesis of the meningitis caused by Streptococcus suis: the unresolved questions. Vet. Microbiol. 76:259–272. [DOI] [PubMed] [Google Scholar]
- 13.Hill, A. W., and J. A. Leigh. 1989. DNA fingerprinting of Streptococcus uberis: a useful tool for epidemiology of bovine mastitis. Epidemiol. Infect. 103:165–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnsen, L. B., K. Poulsen, M. Kilian, and T. E. Petersen. 1999. Purification and cloning of a streptokinase from Streptococcus uberis. Infect. Immun. 67:1072–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnston, K. H. 1993. Solid and fluid phase assays for bacterial plasminogen activators. J. Microbiol. Methods 18:267–274. [Google Scholar]
- 16.Kitt, A. J., and J. A. Leigh. 1997. The auxotrophic nature of Streptococcus uberis. The acquisition of essential acids from plasmin derived casein peptides. Adv. Exp. Med. Biol. 418:647–650. [PubMed] [Google Scholar]
- 17.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. [DOI] [PubMed] [Google Scholar]
- 18.Leigh, J. A. 1994. Purification of a plasminogen activator from Streptococcus uberis. FEMS Microbiol. Lett. 118:153–158. [DOI] [PubMed] [Google Scholar]
- 19.Leigh, J. A. 1999. Streptococcus uberis: a permanent barrier to the control of bovine mastitis? Vet. J. 157:225–238. [DOI] [PubMed] [Google Scholar]
- 20.Leigh, J. A., S. M. Hodgkinson, and R. A. Lincoln. 1998. The interaction of Streptococcus dysgalactiae with plasmin and plasminogen. Vet. Microbiol. 61:121–135. [DOI] [PubMed] [Google Scholar]
- 21.Leigh, J. A., and R. A. Lincoln. 1997. Streptococcus uberis acquires plasmin activity following growth in the presence of bovine plasminogen through the action of its specific plasminogen activator. FEMS. Microbiol. Lett. 154:123–129. [DOI] [PubMed] [Google Scholar]
- 22.Lottenberg, R., L. E. DesJardin, H. Wang, and M. D. Boyle. 1992. Streptokinase-producing streptococci grown in human plasma acquire unregulated cell-associated plasmin activity. J. Infect. Dis. 166:436–440. [DOI] [PubMed] [Google Scholar]
- 23.Lottenberg, R., D. Minning-Wenz, and M. D. Boyle. 1994. Capturing host plasmin(ogen): a common mechanism for invasive pathogens? Trends Microbiol. 2:20–24. [DOI] [PubMed] [Google Scholar]
- 24.McClintock, D. K., and P. H. Bell. 1971. The mechanism of activation of human plasminogen by streptokinase. Biochem. Biophys. Res. Commun. 43:694–702. [DOI] [PubMed] [Google Scholar]
- 25.McCoy, H. E., C. C. Broder, and R. Lottenberg. 1991. Streptokinases produced by pathogenic group C streptococci demonstrate species-specific plasminogen activation. J. Infect. Dis. 164:515–521. [DOI] [PubMed] [Google Scholar]
- 26.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1–6. [DOI] [PubMed] [Google Scholar]
- 27.Nowicki, S. T., D. Minning-Wenz, K. H. Johnston, and R. Lottenberg. 1994. Characterization of a novel streptokinase produced by Streptococcus equisimilis of non-human origin. Thromb. Haemost. 72:595–603. [PubMed] [Google Scholar]
- 28.Oliver, S. P., R. A. Almeida, and L. F. Calvinho. 1998. Virulence factors of Streptococcus uberis isolated from cows with mastitis. Zentbl. Vetmed. Reihe B 45:461–471. [DOI] [PubMed] [Google Scholar]
- 29.Reddy, K. N., and G. Markus. 1972. Mechanism of activation of human plasminogen by streptokinase. Presence of active center in streptokinase-plasminogen complex. J. Biol. Chem. 247:1683–1691. [PubMed] [Google Scholar]
- 30.Robbins, K. C., L. Summaria, B. Hsieh, and R. J. Shah. 1967. The peptide chains of human plasmin. Mechanism of activation of human plasminogen to plasmin. J. Biol. Chem. 242:2333–2342. [PubMed] [Google Scholar]
- 31.Rosey, E. L., R. A. Lincoln, P. N. Ward, R. J. Yancey, Jr., and J. A. Leigh. 1999. PauA: a novel plasminogen activator from Streptococcus uberis. FEMS Microbiol. Lett. 178:27–33. (Erratum, 180:353.) [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, New York, N.Y.
- 33.Sazonova, I. Y., A. K. Houng, S. A. Chowdhry, B. R. Robinson, L. Hedstrom, and G. L. Reed. 2001. The mechanism of a bacterial plasminogen activator intermediate between streptokinase and staphylokinase. J. Biol. Chem. 276:12609–12613. [DOI] [PubMed] [Google Scholar]
- 34.Vassalli, J. D., A. P. Sappino, and D. Belin. 1991. The plasminogen activator/plasmin system. J. Clin. Investig. 88:1067–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, S., G. L. Reed, and L. Hedstrom. 1999. Deletion of Ile1 changes the mechanism of streptokinase: evidence for the molecular sexuality hypothesis. Biochemistry 38:5232–5240. [DOI] [PubMed] [Google Scholar]