Abstract
We describe the further utilization of a genetic screen that identifies mutations defective in the assembly of proteins into the Escherichia coli cytoplasmic membrane. The screen yielded mutations in each of the known genes encoding components of the E. coli signal recognition particle pathway: ffh, ffs, and ftsY, which encode Ffh, 4.5S RNA, and FtsY, respectively. In addition, the screen yielded mutations in secM, which is involved in regulating levels of the SecA component of the bacterium’s protein export pathway. We used a sensitive assay involving biotinylation to show that all of the mutations caused defects in the membrane insertions of three topologically distinct membrane proteins, AcrB, MalF, and FtsQ. Among the mutations that resulted in membrane protein insertion defects, only the secM mutations also showed defects in the translocation of proteins into the E. coli periplasm. Genetic evidence suggests that the S382T alteration of Ffh affects the interaction between Ffh and 4.5S RNA.
In mammalian cells, much of protein targeting to the membrane of the endoplasmic reticulum depends on the signal recognition particle (SRP) and the membrane-bound SRP receptor (24, 36). The mammalian SRP complex consists of six proteins and an RNA (43, 44). As a nascent chain emerges from the ribosome, its amino-terminal signal sequence is recognized by the 54-kDa protein of the SRP (SRP54) (9, 13, 48). The SRP ribosome-nascent-chain complex is then delivered to the SRP receptor (6). The nascent chain is released to the membrane-embedded translocon and subsequently translocated into the endoplasmic reticulum (24).
The Escherichia coli SRP, first identified by searching for sequence homologs of the mammalian SRP subunits, is a simplified version of its mammalian counterpart (3, 21, 27, 35). Instead of six proteins and one RNA, the E. coli SRP consists only of a 48-kDa protein, Ffh, and the 4.5S RNA. Mature-form 4.5S RNA is 114 nucleotides long, much smaller than its mammalian homolog 7S L RNA (over 300 nucleotides long). The E. coli SRP receptor, FtsY, unlike the heterodimeric eukaryotic SRP receptor, consists of only one subunit. The ffh, ffs, and ftsY genes, which encode Ffh, 4.5S RNA, and FtsY, respectively, are all essential for cell viability (5, 14, 20).
Despite the presence of an SRP analog in E. coli and reports indicating its involvement in the translocation of several secreted proteins (14, 20, 21), the E. coli SRP does not appear to play a major role in the translocation of proteins into the periplasm and outer membrane. Instead, it appears to function mainly in the targeting and integration of cytoplasmic membrane proteins (7, 12, 15, 33, 39, 41). The SRP binds to the hydrophobic transmembrane segment of cytoplasmic membrane proteins (40, 41). The SRP ribosome-nascent-chain complex is targeted to the cytoplasmic membrane through interaction with FtsY, which is associated with the inner membrane (14, 22). Besides the SRP, the E. coli secretion machinery, including the membrane-bound SecYEG translocation channel as well as the ATPase SecA, has been demonstrated both genetically and biochemically to be important for the integration of membrane proteins (10, 38, 42, 45, 46). YidC, a homolog of the mitochondrial import protein Oxa1p (4), also appears to be required for the insertion process (28).
Evidence for E. coli SRPs in membrane protein assembly comes from in vitro studies and from in vivo studies examining the effects of depleting SRP components on the integration of membrane proteins. An in vivo genetic screen that yields E. coli mutants defective in membrane protein insertion has been described previously (37). The E. coli strain used expresses a hybrid protein, MalF-β-galactosidase 102, in which β-galactosidase is fused to the second periplasmic domain of the cytoplasmic membrane protein MalF. The insertion of MalF into the inner membrane and the export of its second periplasmic domain leads to the partial translocation of β-galactosidase, resulting in the loss of β-galactosidase activity (16). Mutations disrupting either the membrane protein insertion process or disulfide bond formation result in localization of β-galactosidase in the cytoplasm, where it folds into an active conformation (1, 37).
Colonies of the strain used for the screen appear white on minimal agar containing the substrate dye X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) due to the lack of active β-galactosidase. Colonies of a dsbA- or dsbB-null mutant appear dark blue due to high levels of β-galactosidase activity. Mutagenized cells were screened for those that exhibited a less intense blue color than that of the dsbA or dsbB phenotypically null cells, and 108 mutants with various degrees of β-galactosidase activity were found. Twenty-five of these mutants were the result of ffs mutations that cause defects in membrane protein insertion (37). In this study, we describe nine additional mutants with mutations in each component of the E. coli SRP-SRP receptor complex and mutations in secM, a gene involved in the regulation of SecA synthesis (8, 26, 30, 32). This study represents the first genetic approach that identifies mutations in all known components of the E. coli SRP. We further demonstrate that the ffs, ffh, ftsY, and secM mutants exhibit defects in the membrane assembly of three cytoplasmic membrane proteins: FtsQ, MalF, and AcrB. Among the mutations affecting membrane protein insertion, only secM mutations also cause defects in protein secretion.
MATERIALS AND METHODS
Strains, plasmids, media, and growth conditions.
The E. coli strains and plasmids used in this study are listed in Table 1. Cells were grown at 37 or 30°C in NZY rich medium (10) or M63 minimal medium with appropriate supplements and antibiotics (37). Additional supplements used in this study included nicotinic acid (2 ng/ml), leucine, and phenylalanine (both at 50 μg/ml).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| HPT57 | MC1000 phoA+phoR leu+ λ102 (MalF-LacZ102 Ampr) | 37 |
| HPT129 | HPT57 zih::Tn10 | 37 |
| HPT130 | HPT57 fadR::Tn10 | 37 |
| HPT264 | JP313 leu::Tn10, arabinose sensitive | This study |
| HPT265 | JP313 secM73 leu::Tn10, arabinose sensitive | This study |
| HPT301 | JP313 leu::Tn10 secM93 | This study |
| HPT302 | JP313 leu::Tn10 secM98 | This study |
| HPT299 | JP313 zhg::Tn10 | This study |
| HPT300 | JP313 zhg::Tn10 ftsY70 | This study |
| HPT369 | HPT264, arabinose resistant | This study |
| HPT370 | HPT265, arabinose resistant | This study |
| HPT404 | JP313 pheA3141::Tn10Kan | This study |
| HPT405 | JP313 pheA3141::Tn10Kan ffh-77 | This study |
| HPT406 | JP313 pheA3141::Tn10Kan ffh-87 | This study |
| HPT407 | JP313 pheA3141::Tn10Kan ffh-103 | This study |
| HPT-UV23 | HPT57 ffs-23 | This study |
| HPT-UV46 | HPT57 ffs-46 | This study |
| HPT-UV70 | HPT57 fts Y70 | This study |
| HPT-UV73 | HPT57 secM73 | This study |
| HPT-UV77 | HPT57 ffh-77 | This study |
| HPT-UV87 | HPT57 ffh-87 | This study |
| HPT-UV89 | HPT57 ffh-89 | This study |
| HPT-UV93 | HPT57 secM93 | This study |
| HPT-UV98 | HPT57 secM98 | This study |
| HPT-UV103 | HPT57 ffh-103 | This study |
| CAG18608 | MG1655 pheA3141::Tn10Kan | 34 |
| CAG599 | F−zhg::Tn10 lacZ(Am) trp(Am) pho(Am) supC (Ts) mal(Am) rpsL | Harris D. Bernstein |
| EC294 | MG1655 leu::Tn10 | Lab collection |
| JP313 | MC4100 Δara714 | Lab collection |
| SG20253 | MG1655 zba-3054::Tn10 | 34 |
| Plasmids | ||
| CJ1 | pACYC184-secM Cmr | Don B. Oliver |
| pBAD42-secA | pBAD42-secA Cmr | Jan-Willem de Gier |
| pBE2 | gene X-secA in pACYC, Cmr | Don B. Oliver |
| pGJ78 series (J and K) | PSBT fusions at positions J (periplasmic) and K (cytoplasmic) of MalF in pBR origin plasmid under control of the IPTG-inducible promoter, Kanr | 10 |
| pHP5 | pAM238-ffs Spcr | 37 |
| pHP42 | pBAD18-ftsQ-PSBT Ampr | 37 |
| pHP44 | pBR322-acrR′ acrA acrB576-PSBT | 37 |
| pMS421 | placIq Spcr | Lab collection |
| pTRC-ftsY | pTRC-ftsY Cmr | 39 |
| RB11-ffh | plac-ffh, with lacIq on the same plasmid, Ampr | Harris D. Bernstein |
Mapping of ffs, secM, ffh, and ftsY mutations.
Possible mutations linked to ffs, ffh, ftsY, and secM were determined by P1 transduction using transposons linked to each of the genes; Tn10 in strain SG20253 was 50% linked to the wild-type ffs, Tn10 in strain EC294 was 50% linked to secM-secA, Tn10 Kan from strain CAG18608 was 60% linked to ffh, and Tn10 in strain CAG599 was 30% linked to ftsY.
Pulse-chase, immunoprecipitation, and steady-state protein level.
Pulse-chase and immunoprecipitation experiments were carried out as described previously (37). Anti-OmpA and anti-maltose binding protein (anti-MBP) antibodies (laboratory collection) were used to immunoprecipitate OmpA and MBP. Steady-state SecA, Ffh, and FtsY levels were examined by carrying out a standard Western blot analysis. Whole-cell proteins were precipitated by treating cells grown to an optical density at 600 nm (OD600) of 0.2 with 6% trichloroacetic acid. Anti-SecA (Jan-Willem de Gier), anti-Ffh, and anti-FtsY (both from Harris Bernstein) were used for the detection of the protein of interest.
Preparation and detection of total PSBT fusion proteins and biotinylated fusion proteins.
The expression and preparation of the MalF-Proprionibacterium shermanii transcarboxylase (PSBT), AcrB-PSBT, and FtsQ-PSBT fusion proteins from wild-type and mutant strains were carried out according to procedures described previously (37). Polyclonal anti-MalF PhoA J and anti-FtsQ antibodies (laboratory collection), as well as the anti-AcrB antibody (Hiroshi Nikaido), were used for the detection of total MalF-PSBT, FtsQ-PSBT, and AcrB576-PSBT fusion proteins. Streptavidin-horseradish peroxidase (HRP) (Amersham) was used to detect the biotinylated fusion proteins.
Spheroplast preparation and trypsin sensitivity assay.
E. coli strains carrying plasmids pGJ78-J or -K and pMS421 were grown overnight at 37°C in NZY medium containing spectinomycin (100 μg/ml) and kanamycin (40 μg/ml). After being diluted 1:100 in the same medium and grown to an OD600 of 0.3, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 0.5 mM to induce the fusion protein. After the culture reached an OD600 of 0.6, cells were chilled on ice for at least 20 min and 1.5 ml of cells was spun down and resuspended in ice-cold 18% sucrose-100 mM Tris-HCl (pH 8.0). After addition of EDTA (0.01 M) and lysozyme (100 μg/ml) to the resuspended cells and incubation on ice for 20 min, cells were split into three parts. To the first, trypsin was added at a final concentration of 10 μg/ml. To the second, only the sucrose buffer mentioned above was added. To the third, before addition of trypsin at 10 μg/ml, Triton X-100 was added at a final concentration of 1%. All three samples were incubated on ice for 15 min before the addition of phenylmethylsulfonyl fluoride to a final concentration of 35 mg/ml. Proteins were then precipitated with 10% trichloroacetic acid. After the protein pellets were washed with acetone and dried, sodium dodecyl sulfate sample buffer containing 0.7 M β-mercarptoethanol was added to resuspend the proteins. Whole-cell proteins were first separated by sodium dodecylsulfate-polyacrylamide gel electrophoresis before being transferred to a nitrocellulose membrane using a Semi-Dry protein transfer apparatus (Bio-Rad). After incubation of the membrane with streptavidin-HRP (Amersham) diluted at 1:1,500 and detection of the biotinylated proteins using an ECL kit (Amersham), the nitrocellulose membrane was incubated in 0.5 M NaOH for 5 min. The membrane was then washed several times before incubation with anti-MalF-PhoA J antibody (laboratory collection) for subsequent detection of the total MalF-PSBT fusion proteins.
RESULTS
Three classes of mutational events might cause cytoplasmic localization of the β-galactosidase of the MalF-β-galactosidase 102 fusion protein: disruption of disulfide bond formation, interference with membrane protein insertion, and removal of portions of the MalF protein, altering the location of the attached β-galactosidase from the periplasm to the cytoplasm. Eighty-eight of 108 mutations obtained were characterized previously (37). Mutants with 4 of the 20 remaining mutations grew poorly and were difficult to analyze genetically. To determine to which category the other 16 mutations belonged, P1 transduction was carried out to detect linkage of the mutations to each of the genes involved in disulfide bond formation, to those genes required for membrane protein insertion, or to the malF-lacZ102 fusion (reference 37 and Materials and Methods). Four of these mutations were linked to ffh, one was linked to ftsY, two were linked to ffs, and two were linked to secM. An additional mutation that caused MalF membrane assembly defects was linked to none of these genes and was not studied further. The remainder of the mutations resulted in mutants that showed no defects in MalF assembly, being mutations either in dsbA or in none of the genes tested (Table 2).
TABLE 2.
Summary of 104 UV-induced mutants
| Class | Position(s) of the mutation(s) | No. of mutants identified |
|---|---|---|
| I (defective in disulfide bond formation) | dsbA and dsbB | 28 (dsbA) and 21 (dsbB) |
| II (defective in membrane protein insertion) | ffs | 27 |
| ffh | 4 | |
| ftsY | 1 | |
| secM | 3 | |
| Unmapped | 1 | |
| III | λ102 (malF-lacZ102) | 14 |
| IV | 90-min region | 1 |
| Unmapped | 4 |
The ffh mutations.
The four ffh mutants exhibited low levels of β-galactosidase activity (<10-fold higher than for the wild-type parent). The faint-blue-colony phenotype on X-Gal plates was fully complemented by plasmid RB11-ffh, carrying only ffh (results not shown). DNA sequencing revealed that ffh-77 has a single nucleotide change of G109C (counting from the ATG start site), resulting in an alanine 37-to-proline change (Table 3). ffh-87 and ffh-103 share the same T1144A nucleotide alteration, which results in a serine 382-to-threonine change. ffh-89 contains a single nucleotide change of A355G, leading to a lysine 119-to-glutamate change. Western blot analysis indicated that the steady-state Ffh levels in mutants containing the ffh-77, ffh-87, or ffh-103 mutation were similar to that in the wild type (results not shown). For some of the subsequent analyses, ffh mutations were moved to a JP313 strain background by P1 transduction.
TABLE 3.
Mutations in secM, ffs, ffh, and ftsY mutants
| Mutated gene(s) | Nucleotide changea | Amino acid changeb |
|---|---|---|
| secM73, secM93 | C181T | Q60 stop site |
| secM98 | Deletion of one T from TT at positions 170 and 171 | Alteration of amino acids at positions 58–88 before stop siteat position 89 |
| ffs-23 | TG to AA from −1 and +1 positionsc | |
| ffs-46 | GG to AA from +57 and +58 positionsc | |
| ffh-77 | G109C | A37P |
| ffh-87, ffh-103 | T1144A | S382T |
| ffh-89 | A355G | K119E |
| ftsY70 | A1241G | E414G |
Nucleotide numbering starts from the start codon.
Amino acid numbering starts from the first translated amino acid (including the signal sequence).
Positions are relative to the first base of the mature RNA.
The ftsY mutation.
The single ftsY mutant showed a faint-blue-colony phenotype with an intensity like that of the ffh mutants and was complemented by a plasmid (pTRC-ftsY) carrying only the ftsY gene. Sequencing analysis revealed a single A1241G nucleotide alteration (numbering starts from the ATG start site) causing a glutamate 414-to-glycine change (Table 3). Western blot analysis indicated no significant difference between the steady-state FtsY protein levels for this mutant and the wild type (results not shown). This ftsY mutant allele, ftsY70, was moved by P1 transduction to a new strain background for some of the subsequent analyses.
The ffs mutants.
Two mutations linked to the ffs region (Table 3) were complemented by introducing plasmid pHP5 carrying only ffs. The ffs-23 mutation converts a TG to an AA at the −1 and +1 positions relative to the starting point of the mature RNA. ffs-46 has GG-to-AA nucleotide changes at the 157 and 158 positions, corresponding to a region between the apical tetraloop and the symmetric internal loop of the 4.5S RNA. ffs-46 differs from our other ffs alleles in that it caused significantly slowed cell growth both on plates and in liquid culture.
The secM mutants.
The two new mutations linked to the secM-secA-mutT operon were complemented for the pale-blue-colony phenotype by plasmid pBE2 carrying secM and secA. secM93 shares the same nucleotide alteration with the previously identified secM73, which results in a C181T change (counting from the GTG start site), resulting in a stop codon at the amino acid residue at position 60 (with the signal sequence included in the numbering). secM98 has a frameshift mutation that deletes a T from two T’s at positions 170 and 171, resulting in a stop codon at the amino acid residue at position 89 (Table 2).
Initially termed gene X, secM regulates SecA and does not play a direct role in protein secretion (23). secM, secA, and mutT are cotranscribed; known nonsense mutations in secM have a strong polar effect on secA expression (8, 26, 30, 32). secM encodes a 170-amino-acid-long protein which, upon secretion to the periplasm, is processed to its final mature form of 133 amino acids (23, 29). In wild-type cells, SecM is rapidly degraded by the periplasmic-tail-specific protease (17). SecM is also subjected to a transient-translation pause at a position close to the carboxyl terminus, and this elongation arrest involves the nascent SecM itself. This pausing is found to be significantly enhanced when SecM translocation is retarded (17).
The ffh, ftsY, ffs, and secM mutants are all defective in membrane protein insertion.
We considered it likely that the ffh, ftsY, and ffs mutations caused the cytoplasmic localization of β-galactosidase in the fusion as a result of their effects on membrane protein insertion; secM mutations might cause the same defects due to their polar effect on the downstream secA. We used a biotinylation assay, which was previously employed for detecting membrane protein insertion defects in the other ffs mutants, to test this possibility (37). In our assay system, a biotinylatable domain (80 amino acids in size) from the 1.3S subunit of PSBT was fused to the periplasmic domains of MalF, AcrB, and FtsQ (37). If the fusion proteins failed to be inserted into the cytoplasmic membrane, the PSBT domain was biotinylated by biotin ligase, an enzyme found only in the cytoplasm in E. coli. However, if efficiently inserted, the PSBT domain was translocated into the periplasm rapidly enough so that it would remain unbiotinylated.
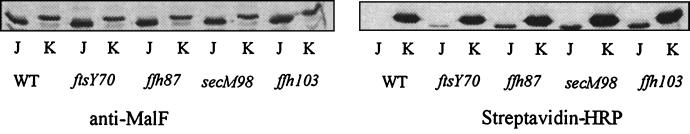
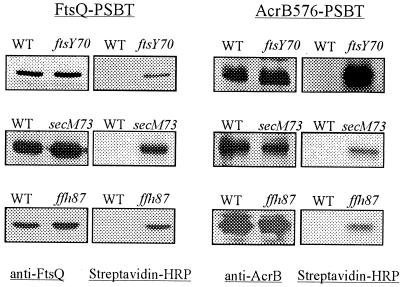
ffh, ftsY, ffs, and secM mutants are all defective in the insertion of MalF into the cytoplasmic membrane, as indicated by the biotinylation of a fraction of the MalF-PSBT periplasmic fusion J (results for ffh-87, ffh-103, ftsY70, and secM98 mutants are shown in Fig. 1; other results are not shown). These mutants also exhibited defects in the insertion of AcrB and FtsQ (results for ffh-87, ftsY70, and secM73 are shown in Fig. 2; other results are not shown). In the wild-type parent strain, we observed no biotinylated signal for the PSBT fused to the periplasmic domains of MalF, AcrB, and FtsQ (Fig. 1 and 2). A biotin carboxyl carrier protein, which resides in the cytoplasm, showed the same levels of biotinylation in all strain backgrounds (results not shown).
FIG. 1.
ffh, ftsY, and secM mutants are defective in the membrane insertion of MalF. The MalF-PSBT periplasmic J fusion and cytoplasmic K fusion were expressed from pGJ78-J and pGJ78-K in HPT57 (wild type) and HPT-UV70, HPT-UV87, HPT-UV98, and HPT-UV103 containing the ftsY70, ffh-87, secM98, and ffh-103 mutations, respectively. Anti-MalF antibody was used to detect total fusion proteins, and streptavidin-HRP was used to detect the biotinylated fusion proteins. WT, wild type.
FIG. 2.
ffh, ftsY, and secM mutants are defective in the membrane insertion of AcrB and FtsQ. AcrB576-PSBT and FtsQ-PSBT were expressed from pHP44 and pHP42 in HPT404 (wild-type ffh), HPT406 (ffh-87), HPT299 (wild-type ftsY), and HPT300 (ftsY70). AcrB576-PSBT was expressed from pHP44 in HPT264 (wild-type secM) and HPT265 (secM73). FtsQ-PSBT was expressed from pHP42 in HPT369 (wild-type secM) and HPT370 (secM73). Anti-AcrB and anti-FtsQ antibodies were used to detect AcrB576-PSBT and FtsQ-PSBT, respectively, and streptavidin-HRP was used to detect the biotinylated fusion proteins. Blots on different rows were exposed for different lengths of time, and the band intensities, therefore, should not be compared. WT, wild type.
Only secM mutations cause defects in the secretion of periplasmic and outer membrane proteins.
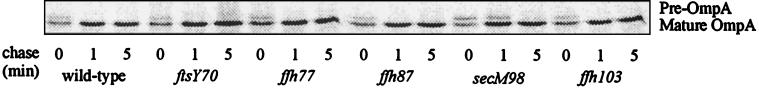
We asked whether the mutations affecting membrane protein assembly also exhibited defects in protein translocation across the cytoplasmic membrane by monitoring OmpA signal sequence processing in pulse-chase experiments (37). After a 1-min chase with cold methionine, while the wild-type cells and ffs, ffh, and ftsY mutant cells all showed only mature OmpA, the secM98 mutant still had significant amounts of precursor OmpA (Fig. 3). From this experiment, we conclude that while the secM mutations cause a defect in protein translocation into the periplasm, the same process remains unaffected by the ffh, ftsY, and ffs mutations.
FIG. 3.
Protein secretion is defective in cells containing mutations in secM but not in ffh or ftsY. Pulse-chase and immunoprecipitation of OmpA were carried out to examine the efficiency of OmpA translocation into the periplasm in HPT57 (wild type) and in HPT-UV98, HPT-UV77, HPT-UV87, HPT-UV103, and HPT-UV70, containing the secM98, ffh-77, ffh-87, ffh-103, and ftsY70 mutations, respectively.
The secM mutants produce apparently normal amounts of SecA.
Because the secM mutations all result in stop codons early in the protein coding region, a possible explanation for the defects we observed is that SecA levels were reduced. In fact, the faint-blue-mutant-colony phenotype of all three secM mutants was complemented by plasmid pBAD42-secA, carrying secA alone, but not CJ1, carrying only secM. However, Western blot analysis and pulse-chase-immunoprecipitation for both the wild type and the secM73 mutant showed no observable differences in SecA levels between the two strains (results not shown).
The biotinylated fusion proteins in ftsY and secM mutants stay in the cytoplasm.
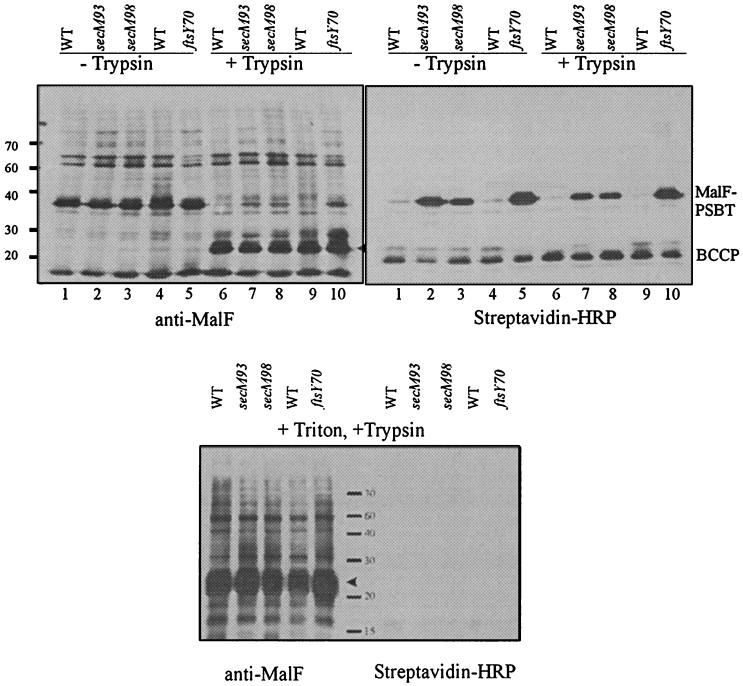
In a mutant defective only in the kinetics of protein translocation, biotinylation of a MalE-PSBT fusion does not block its translocation into the periplasm (10, 25). We had assumed that, similarly, our SRP mutants might be defecFStive only in the kinetics of membrane protein insertion; the membrane proteins would likely still be inserted into the cytoplasmic membrane, although at a lower rate than that for the wild-type (37). We tested this hypothesis directly by determining whether the biotinylated domain of the periplasmic MalF-PSBT fusion in our mutants was exposed to the periplasm. To do so, we added trypsin to the spheroplasts prepared from mutants expressing the fusion protein. A trypsin site in the MalF-PSBT fusion is located in the second periplasmic domain in MalF. If properly inserted into the membrane, a trypsin digest may generate a 25-kDa band, replacing the full-length MalF-PSBT fusion band that was seen prior to the addition of trypsin. If the protein fails to be inserted into the membrane or is inserted in a different topology such that the trypsin-sensitive site is not exposed to the periplasm, the full-length fusion band will remain.
In a wild-type background, when the spheroplasts expressing the fusion protein were treated with trypsin, a 25-kDa band replaced the full-length MalF-PSBT fusion band as we expected (top left panel of Fig. 4, lanes 6 and 9), indicating the proper membrane insertion of this fusion protein. In the secM93, secM98, and ftsY70 mutant spheroplasts, although the same 25-kDa band replaced most of the full-length fusion signals when trypsin was added, there was still a full-length MalF-PSBT signal remaining (top left panel of Fig. 4, lanes 7, 8, and 10), indicating that a proportion of the fusion protein was insensitive to trypsin and, therefore, unlikely to be inserted properly in all three mutants. When the same membranes were incubated with streptavidin-HRP to reveal the biotinylated signal, the lanes corresponding to mutant samples treated or not treated with trypsin all revealed a biotinylated band running at the same position as that of the full-length MalF-PSBT protein and the intensities of the biotinylated bands remained the same under both conditions (top right panel of Fig. 4, lanes 2, 3, and 5, and lanes 7, 8, and 10). These results indicated that the biotinylated fusion proteins in all the mutants were protected from trypsin digest. When the spheroplasts from the mutants were treated with Triton X-100, first to disrupt the membrane and then with trypsin, all the full-length fusion proteins disappeared, indicating that the inaccessibility of trypsin to the fusion protein was due to the protection provided by the cytoplasmic membrane (Fig. 4, lower panel). We could also confirm that the spheroplasts used for our trypsin digest (without Triton X-100) were intact based on the presence of the biotinylated cytoplasmic carrier protein, which disappeared when Triton X-100 was added to dissolve the membrane prior to the addition of trypsin.
FIG. 4.
The biotinylated PSBT domain is located in the cytoplasm. Spheroplasts were prepared from HPT264 (wild-type secM), HPT301 (secM93), HPT302 (secM98), HPT299 (wild-type ftsY), and HPT300 (ftsY70), each of which expresses the MalF-PSBT periplasmic fusion J from plasmid pGJ78-J. (Upper panels) Trypsin was added to the indicated samples. (Lower panel) Triton X-100 was added before trypsin. Anti-MalF PhoA J antibody was used to visualize the expressed fusion proteins, and streptavidin-HRP was used to visualize the biotinylated fusion protein. Arrowheads point to the 25-kDa band of the trypsin-digested product. WT, wild type.
DISCUSSION
A genetic screen for membrane protein defects has yielded point mutations in the E. coli genes coding for 4.5S RNA, Ffh, and FtsY; in all components of the E. coli SRP and SRP receptors and in the secM gene, a regulator of SecA. Using the biotinylation assay, we demonstrated that all mutations lead to a defect in the proper insertion of the cytoplasmic membrane proteins MalF, AcrB, and FtsQ. SecM mutations, however, also caused slowed translocation of the outer membrane protein OmpA and the periplasmic protein MBP (37).
The ffs mutations.
We suspect that the ffs-23 and ffs-46 mutations affect the 4.5S RNA differently from the way the ffs-29 and ffs-69 mutations described previously affect 4.5S RNA (37). ffs-23, which alters the base composition in the region corresponding to the start site of the mature 4.5S RNA, may cause a defect in the proper 5′-end processing of the precursor RNA. ffs-46 alters two bases in the most internal stem region, and this region has been shown to be important for the interaction between 4.5S RNA and Ffh in vitro (47). Although the bases altered by the mutations do not directly contact Ffh according to the crystal structure of the Ffh M domain-4.5S RNA complex (2), they disrupt base pairing in this region, which can subsequently alter the conserved symmetric and asymmetric loops surrounding it and lead to the disruption of 4.5S RNA Ffh binding.
Ffh mutations in the N and G domains.
The three ffh mutations each altered a different structural domain of Ffh: ffh-77 resulted in an A37P change in the amino-terminal N domain; ffh-89 resulted in a K119E change in the GTPase G domain; and ffh-87 and ffh-103 resulted in the same S382T change in the methionine-rich M domain. The A37P and S382T mutations did not alter the stability of the protein and, therefore, must affect some aspect of Ffh function. Alanine 37, which was changed to proline as a result of the ffh-77 mutation, is located in a stretch of amino acid residues in the N domain that are especially well conserved among distantly related species (ALLEADV in E. coli). The N domain of mammalian SRP54 was proposed to promote efficient signal sequence binding in an in vitro study that examined the binding by using SRP54 variants with mutations in this stretch of amino acid residues (18). Although A37 was not among the mutated residues, mutations altering LL and DV from the ALLEADV stretch all led to the decreased binding of signal sequences by SRP54. If the same function exists for the N domain of Ffh, we might expect the ffh-77 mutation to affect its binding to the hydrophobic transmembrane segment of inner membrane proteins.
Lysine 119, which was converted to glutamate as a result of the ffh-89 mutation, is completely conserved across bacterial species. It resides between the first and the second GTPase consensus elements (there are a total of four such elements in Ffh). It is not obvious from the current structural information how this mutation affects the function of Ffh.
The Ffh M domain S382T mutation likely affects Ffh-4.5S RNA interaction.
The methionine-rich M domain, where the S382T mutations resulting from ffh-87 and ffh-103 reside, contains both the signal sequence recognition surface and the contact interface with 4.5S RNA (2). In the SRP core complex crystal, M382 forms hydrogen bonds with C62 from the symmetric loop and G48 from the asymmetric loop in conserved domain IV of the 4.5S RNA (2). When a plasmid carrying ffs (pHP5) was introduced into ffh-87 and ffh-103 mutants, the faint-blue-colony phenotype disappeared (results not shown). The same complementation was not observed when pHP5 was introduced into ffh-77. This implies that in the ffh-87 and ffh-103 mutants, because of the change of S382T, Ffh no longer interacts with the 4.5S RNA with 100% efficiency, and this results in fewer functional 4.5S RNA-Ffh complexes in the mutants than in the wild type. Introducing extra copies of the 4.5S RNA complements the defect by increasing its local concentration, thus promoting 4.5S RNA-Ffh complex formation. This complementation experiment also implies that the S382T mutant Ffh proteins remain intact in other functions such as FtsY binding.
FtsY mutation.
ftsY70 has a single amino acid change of E414G in the GTPase domain of FtsY. This glutamate residue, however, is not among the most conserved residues across different species. Since the ftsY70 mutation does not cause decreased stability of the protein, the E414G amino acid change likely affects some aspect of FtsY function.
SecM mutations.
It is striking that all three mutations in the chromosomal region where secA resides were in the secM gene, resulting in two truncated versions of SecM protein. They each caused defects in membrane protein insertion as well as presecretory protein translocation. Since secM does not play a direct role in protein secretion (23), we initially speculated that the secM mutations caused the observed defects by reducing expression of the downstream secA gene. This inference was supported by the finding that a plasmid expressing secA alone, but not one expressing secM alone, complemented the defects in secM. However, Western blot analysis revealed that the SecA protein levels in the secM mutants and the wild-type cells were similar. Previously, an amber mutation in secM at amino acid position 132 resulted in the repression of SecA translation, possibly due to the blocking of secA translational initiation by base pairing of the secA Shine-Dalgarno sequence with the secM terminus (19, 31, 32). Normally, this base pairing is disrupted upon translation of secM into this region, thus allowing translation of SecA (11). The fact that we observed normal amounts of SecA protein in our secM early-termination mutants seems to contradict this earlier observation. It may be that there is a second translational start site later in secM which in these mutants initiates the translation of a protein internal to wild-type SecM, allowing downstream SecA translation. Alternatively, the normal SecA levels in the secM mutants could be a result of the derepression of secA expression. However, neither explanation allows a clear explanation for the phenotype of our secM mutants. Therefore, we leave open the possibility that secM may play a role in membrane protein insertion as well as in secA expression.
Biotinylated fusion proteins remain in the cytoplasm in the ftsY70 and secM mutants.
n the ffh, ftsY, and secM mutants, the biotinylated derivatives of the MalF-PSBT periplasmic fusion proteins were not accessible to trypsin added from the outside of the spheroplasts, indicating a cytoplasmic location for the biotinylated PSBT domain. We offer two possible explanations for our results. First, in the ffs, ffh, ftsY, and secM mutants, a portion of the MalF-PSBT protein failed to be inserted into the membrane and was thus biotinylated in the cytoplasm. Second, the mutations only slowed the kinetics of membrane protein insertion, allowing biotinylation of some of the MalF-PSBT, which then could no longer be inserted into the membrane. Previously, we observed that in a sec mutant defective in the kinetics of translocation, biotinylation of the PSBT fused to the last amino acid residue of MalE did not block its further translocation into the periplasm. Our results here may be explained by differences between the mechanisms by which secreted proteins are translocated into the periplasm and inner membrane proteins are assembled into the inner membrane.
Our results strengthen the conclusion that the SRP pathway plays an important role in the process of membrane protein insertion (7, 12, 15, 33, 39, 41). Our mutants provide an advantage for in vivo studies of membrane proteins in mutant strains, as our mutants exhibit relatively normal growth rates. Previous in vivo studies had either depleted the ffh, ftsY, and ffs gene products or used dominant lethal variants in which the basis of the dominant lethality was not well worked out. Although the majority of mutations in ffs affect membrane protein insertion as a result of the decreased amount of 4.5S RNA (37), the ffh and ftsY mutations likely affect some functional aspects of the Ffh and FtsY proteins. This genetic approach to the understanding of membrane protein insertion can be exploited further to yield mutations that help to dissect the individual steps of the pathway.
Acknowledgments
We thank Harris D. Bernstein, Jan-Willem de Gier, Hiroshi Nikaido, and Don B. Oliver for providing strains, plasmids, and antibodies. We gratefully acknowledge advice given by members of the Beckwith Lab.
This work was supported by National Institutes of Health grant GM38922 to J.B. J.B. is an American Cancer Society research professor.
REFERENCES
- 1.Bardwell, J. C., K. McGovern, and J. Beckwith. 1991. Identification of a protein required for disulfide bond formation in vivo. Cell 67:581–589. [DOI] [PubMed] [Google Scholar]
- 2.Batey, R. T., R. P. Rambo, L. Lucast, B. Rha, and J. A. Doudna. 2000. Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science 287:1232–1239. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein, H. D., M. A. Poritz, K. Strub, P. J. Hoben, S. Brenner, and P. Walter. 1989. Model for signal sequence recognition from amino-acid sequence of 54K subunit of signal recognition particle. Nature 340:482–486. [DOI] [PubMed] [Google Scholar]
- 4.Bonnefoy, N., F. Chalvet, P. Hamel, P. P. Slonimski, and G. Dujardin. 1994. OXA1, a Saccharomyces cerevisiae nuclear gene whose sequence is conserved from prokaryotes to eukaryotes controls cytochrome oxidase biogenesis. J. Mol. Biol. 239:201–212. [DOI] [PubMed] [Google Scholar]
- 5.Brown, S., and M. J. Fournier. 1984. The 4.5 S RNA gene of Escherichia coli is essential for cell growth. J. Mol. Biol. 178:533–550. [DOI] [PubMed] [Google Scholar]
- 6.Connolly, T., and R. Gilmore. 1989. The signal recognition particle receptor mediates the GTP-dependent displacement of SRP from the signal sequence of the nascent polypeptide. Cell 57:599–610. [DOI] [PubMed] [Google Scholar]
- 7.de Gier, J.-W., P. Mansournia, Q. A. Valent, G. J. Phillips, J. Luirink, and G. von Heijne. 1996. Assembly of a cytoplasmic membrane protein in Escherichia coli is dependent on the signal recognition particle. FEBS Lett. 399:307–309. [DOI] [PubMed] [Google Scholar]
- 8.Fikes, J. D., and P. J. Bassford, Jr. 1989. Novel secA alleles improve export of maltose-binding protein synthesized with a defective signal peptide. J. Bacteriol. 171:402–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.High, S., and B. Dobberstein. 1991. The signal sequence interacts with the methionine-rich domain of the 54-kD protein of signal recognition particle. J. Cell Biol. 113:229–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jander, G., J. E. Cronan, Jr., and J. Beckwith. 1996. Biotinylation in vivo as a sensitive indicator of protein secretion and membrane protein insertion. J. Bacteriol. 178:3049–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiser, K. B., and M. G. Schmidt. 1999. Regulation of the Escherichia coli secA gene is mediated by two distinct RNA structural conformations. Curr. Microbiol. 38:113–121. [DOI] [PubMed] [Google Scholar]
- 12.Koch, H. G., T. Hengelage, C. Neumann-Haefelin, J. MacFarlane, H. K. Hoffschulte, K. L. Schimz, B. Mechler, and M. Muller. 1999. In vitro studies with purified components reveal signal recognition particle (SRP) and SecA/SecB as constituents of two independent protein-targeting pathways of Escherichia coli. Mol. Biol. Cell 10:2163–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krieg, U. C., P. Walter, and A. E. Johnson. 1986. Photocrosslinking of the signal sequence of nascent preprolactin to the 54-kilodalton polypeptide of the signal recognition particle. Proc. Natl. Acad. Sci. USA 83:8604–8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luirink, J., C. M. ten Hagen-Jongman, C. C. van der Weijden, B. Oudega, S. High, B. Dobberstein, and R. Kusters. 1994. An alternative protein targeting pathway in Escherichia coli: studies on the role of FtsY. EMBO J. 13:2289–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacFarlane, J., and M. Muller. 1995. Functional integration of a polytopic membrane protein of E. coli requires the bacterial signal recognition particle. Biochem. Soc. Trans. 23:560S. [DOI] [PubMed] [Google Scholar]
- 16.McGovern, K., and J. Beckwith. 1991. Membrane insertion of the Escherichia coli MalF protein in cells with impaired secretion machinery. J. Biol. Chem. 266:20870–20876. [PubMed] [Google Scholar]
- 17.Nakatogawa, H., and K. Ito. 2001. Secretion monitor, SecM, undergoes self-translation arrest in the cytosol. Mol. Cell 7:185–192. [DOI] [PubMed] [Google Scholar]
- 18.Newitt, J. A., and H. D. Bernstein. 1997. The N-domain of the signal recognition particle 54-kDa subunit promotes efficient signal sequence binding. Eur. J. Biochem. 245:720–729. [DOI] [PubMed] [Google Scholar]
- 19.Oliver, D. B., and J. Beckwith. 1982. Regulation of a membrane component required for protein secretion in Escherichia coli. Cell 30:311–319. [DOI] [PubMed] [Google Scholar]
- 20.Phillips, G. J., and T. J. Silhavy. 1992. The E. coli ffh gene is necessary for viability and efficient protein export. Nature 359:744–746. [DOI] [PubMed] [Google Scholar]
- 21.Poritz, M. A., H. D. Bernstein, K. Strub, D. Zopf, H. Wilhelm, and P. Walter. 1990. An E. coli ribonucleoprotein containing 4.5S RNA resembles mammalian signal recognition particle. Science 250:1111–1117. [DOI] [PubMed] [Google Scholar]
- 22.Powers, T., and P. Walter. 1997. Co-translational protein targeting catalyzed by the Escherichia coli signal recognition particle and its receptor. EMBO J. 16:4880–4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajapandi, T., K. M. Dolan, and D. B. Oliver. 1991. The first gene in the Escherichia coli secA operon, gene X, encodes a nonessential secretory protein. J. Bacteriol. 173:7092–7097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rapoport, T. A., B. Jungnickel, and U. Kutay. 1996. Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu. Rev. Biochem. 65:271–303. [DOI] [PubMed] [Google Scholar]
- 25.Reed, K. E., and J. E. Cronan. 1991. Escherichia coli exports previously folded and biotinated protein domains. J. Biol. Chem. 266:11425–11428. [PubMed] [Google Scholar]
- 26.Riggs, P. D., A. I. Derman, and J. Beckwith. 1988. A mutation affecting the regulation of a secA-lacZ fusion defines a new sec gene. Genetics 118:571–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romisch, K., J. Webb, J. Herz, S. Prehn, R. Frank, M. Vingron, and B. Dobberstein. 1989. Homology of 54K protein of signal-recognition particle, docking protein and two E. coli proteins with putative GTP-binding domains. Nature 340:478–482. [DOI] [PubMed] [Google Scholar]
- 28.Samuelson, J. C., M. Chen, F. Jiang, I. Moller, M. Wiedmann, A. Kuhn, G. J. Phillips, and R. E. Dalbey. 2000. YidC mediates membrane protein insertion in bacteria. Nature 406:637–641. [DOI] [PubMed] [Google Scholar]
- 29.Sarker, S., K. E. Rudd, and D. Oliver. 2000. Revised translation start site for secM defines an atypical signal peptide that regulates Escherichia coli secA expression. J. Bacteriol. 182:5592–5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmidt, M. G., K. M. Dolan, and D. B. Oliver. 1991. Regulation of Escherichia coli secA mRNA translation by a secretion-responsive element. J. Bacteriol. 173:6605–6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt, M. G., and D. B. Oliver. 1989. SecA protein autogenously represses its own translation during normal protein secretion in Escherichia coli. J. Bacteriol. 171:643–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmidt, M. G., E. E. Rollo, J. Grodberg, and D. B. Oliver. 1988. Nucleotide sequence of the secA gene and secA(Ts) mutations preventing protein export in Escherichia coli. J. Bacteriol. 170:3404–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seluanov, A., and E. Bibi. 1997. FtsY, the prokaryotic signal recognition particle receptor homologue, is essential for biogenesis of membrane proteins. J. Biol. Chem. 272:2053–2055. [DOI] [PubMed] [Google Scholar]
- 34.Singer, M., T. A. Baker, G. Schnitzler, S. M. Deischel, M. Goel, W. Dove, K. J. Jaacks, A. D. Grossman, J. W. Erickson, and C. A. Gross. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol. Rev. 53:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Struck, J. C., H. Y. Toschka, T. Specht, and V. A. Erdmann. 1988. Common structural features between eukaryotic 7SL RNAs, eubacterial 4.5S RNA and scRNA and archaebacterial 7S RNA. Nucleic Acids Res. 16:7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tajima, S., L. Lauffer, V. L. Rath, and P. Walter. 1986. The signal recognition particle receptor is a complex that contains two distinct polypeptide chains. J. Cell Biol. 103:1167–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tian, H., D. Boyd, and J. Beckwith. 2000. A mutant hunt for defects in membrane protein assembly yields mutations affecting the bacterial signal recognition particle and Sec machinery. Proc. Natl. Acad. Sci. USA 97:4730–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Traxler, B., and C. Murphy. 1996. Insertion of the polytopic membrane protein MalF is dependent on the bacterial secretion machinery. J. Biol. Chem. 271:12394–12400. [DOI] [PubMed] [Google Scholar]
- 39.Ulbrandt, N. D., J. A. Newitt, and H. D. Bernstein. 1997. The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell 88:187–196. [DOI] [PubMed] [Google Scholar]
- 40.Valent, Q. A., D. A. Kendall, S. High, R. Kusters, B. Oudega, and J. Luirink. 1995. Early events in preprotein recognition in E. coli: interaction of SRP and trigger factor with nascent polypeptides. EMBO J. 14:5494–5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valent, Q. A., P. A. Scotti, S. High, J. W. de Gier, G. von Heijne, G. Lentzen, W. Wintermeyer, B. Oudega, and J. Luirink. 1998. The Escherichia coli SRP and SecB targeting pathways converge at the translocon. EMBO J. 17:2504–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.von Heijne, G. 1989. Control of topology and mode of assembly of a polytopic membrane protein by positively charged residues. Nature 341:456–458. [DOI] [PubMed] [Google Scholar]
- 43.Walter, P., and G. Blobel. 1982. Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum. Nature 299:691–698. [DOI] [PubMed] [Google Scholar]
- 44.Walter, P., and V. R. Lingappa. 1986. Mechanism of protein translocation across the endoplasmic reticulum membrane. Annu. Rev. Cell Biol. 2:499–516. [DOI] [PubMed] [Google Scholar]
- 45.Werner, P. K., M. H. Saier, and M. Muller. 1992. Membrane insertion of the mannitol permease of Escherichia coli occurs under conditions of impaired SecA function. J. Biol. Chem. 267:24523–24532. [PubMed] [Google Scholar]
- 46.Wolfe, P. B., M. Rice, and W. Wickner. 1985. Effects of two sec genes on protein assembly into the plasma membrane of Escherichia coli. J. Biol. Chem. 260:1836–1841. [PubMed] [Google Scholar]
- 47.Wood, H., J. Luirink, and D. Tollervey. 1992. Evolutionary conserved nucleotides within the E. coli 4.5S RNA are required for association with P48 in vitro and for optimal function in vivo. Nucleic Acids Res. 20:5919–5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zopf, D., H. D. Bernstein, A. E. Johnson, and P. Walter. 1990. The methionine-rich domain of the 54 kd protein subunit of the signal recognition particle contains an RNA binding site and can be crosslinked to a signal sequence. EMBO J. 9:4511–4517. [DOI] [PMC free article] [PubMed] [Google Scholar]