Abstract
The bioemulsifier of Acinetobacter radioresistens KA53, referred to as alasan, is a high-molecular-weight complex of polysaccharide and protein. Recently, one of the alasan proteins, with an apparent molecular mass of 45 kDa, was purified and shown to constitute most of the emulsifying activity. The N-terminal sequence of the 45-kDa protein showed high homology to an OmpA-like protein from Acinetobacter spp. In the research described here the gene coding for the 45-kDa protein was cloned, sequenced, and expressed in Escherichia coli. Recombinant protein AlnA (35.77 kDa without the leader sequence) had an amino acid sequence homologous to that of E. coli OmpA and contained 70% of the specific (hydrocarbon-in-water) emulsifying activity of the native 45-kDa protein and 2.4 times that of the alasan complex. In addition to their emulsifying activity, both the native 45-kDa protein and the recombinant AlnA were highly effective in solubilizing phenanthrene, ca. 80 μg per mg of protein, corresponding to 15 to 19 molecules of phenanthrene per molecule of protein. E. coli OmpA had no significant emulsifying or phenanthrene-solubilizing activity. The production of a recombinant surface-active protein (emulsification and solubilization of hydrocarbons in water) from a defined gene makes possible for the first time structure-function studies of a bioemulsan.
Microorganisms synthesize a wide variety of high- and low-molecular-mass bioemulsifiers (21). The high-molecular-mass bioemulsifiers, referred to as bioemulsans (20), are amphipathic polysaccharides, proteins, lipopolysaccharides, lipoproteins, or complex mixtures of these biopolymers that stabilize oil-in-water emulsions. Although most research on bioemulsans has focused on potential industrial and environmental applications (1, 9, 16, 19, 22, 28, 32), there is also a growing interest in the natural role of bioemulsans for the producing microorganism (19). To better understand how bioemulsans function in the growth and survival of microorganisms, it is essential to elucidate their detailed chemical structures as well as the genes required for their biosynthesis.
The majority of Acinetobacter strains, including both hospital and environmental isolates, produce bioemulsans (27). The best studied are the bioemulsans of Acinetobacter calcoaceticus RAG-1 (22), A. calcoaceticus BD4 (7), and Acinetobacter radioresistens KA53 (12). RAG-1 emulsan is a complex of an anionic heteropolysaccharide and protein (24). Its surface activity is largely due to the presence of fatty acids, comprising 15% of the emulsan dry weight, which are attached to the polysaccharide backbone via O-ester and N-acyl linkages (4). The protein component of RAG-1 emulsan stimulates the emulsifying activity (34). A. calcoaceticus BD4, initially isolated by Taylor and Juni (30), produces a large polysaccharide capsule. Under certain growth conditions, the capsule is released together with the bound protein, producing a highly active emulsifier complex (7). The purified polysaccharide and protein components have no emulsifying activity by themselves. However, mixing the polysaccharide and protein led to reconstitution of the emulsifying activity (8).
The bioemulsan produced by A. radioresistens KA53, referred to as alasan, is a complex of an anionic polysaccharide containing covalently bound alanine (apoalasan) and protein (12). During exponential growth alasan is primarily bound to the cells, and during stationary phase it is released into the extracellular fluid (12). The protein fraction was essential for emulsifying activity (13). Three of the proteins were purified from the alasan complex by preparative sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and had apparent molecular masses of 16, 31, and 45 kDa (31). Essentially all of the emulsifying activity of the alasan complex was present in the 45-kDa protein.
The present study deals with the cloning, sequencing, and overexpression of the gene (alnA) responsible for the synthesis of the recombinant emulsifier protein (AlnA). Interestingly, the recombinant protein showed a sequence similarity to OmpA of Escherichia coli. Purified recombinant protein AlnA and the native 45-kDa protein showed similar, high emulsifying and phenanthrene (PHE)-solubilizing activities. With the potent tools of protein structure analysis, it should now be possible to examine structure-function relationships of the OmpA-like protein emulsifier of A. radioresistens KA53 with regard to its ability to stabilize hydrocarbon-in-water emulsions, solubilize hydrocarbons (2), and help the bacterium interact with interfaces (14). Although we show here that E. coli OmpA is not an effective bioemulsifier, the facts that the structure of the OmpA transmembrane domain of E. coli has been determined to a resolution of 1.65 Å and that hydrophobic binding sites have been defined (17, 18) should be useful for comparative studies.
MATERIALS AND METHODS
Bacterial strain and production of alasan, apoalasan, and the 45-kDa alasan protein.
A. radioresistens KA53 (NCIMB-40692), isolated previously on acetate medium (12), was maintained on brain heart infusion agar (Difco Laboratories, Detroit, Mich.). After incubation for 3 days, the plates were stored at 4°C. For emulsifier production, inocula were prepared in ethanol medium (EM) containing (per liter of deionized water) 5 ml of ethanol, 1.8 g of urea, 13.7 g of Na2HPO4, 7.26 g of KH2PO4, 0.4 g of MgSO4 · 7H2O, and 1 ml of trace elements (25). Alasan production was carried out at 30°C in a 2.5-liter fermentor (Multigen; New Brunswick Scientific Co. Inc.) containing 1.4 liters of EM. Bacterial growth was initiated by introducing a 0.1% inoculum. After 80 h of batch-fed fermentation, as described previously (12), the culture reached a turbidity of 30 at 600 nm. The cell-free culture broth was then obtained by centrifugation, followed by filtration through a 0.45-μm-pore-size membrane filter. After addition of ammonium sulfate to 65% saturation, the turbid suspension was allowed to stand overnight at 4°C and then centrifuged at 10,000 × g for 20 min. The pellets were dissolved in water, dialyzed extensively against deionized water, and lyophilized, yielding 3.5 g of alasan. Apoalasan (deproteinized alasan) was obtained by the hot-phenol method (33). Stock solutions of alasan and apoalasan were prepared fresh by hydration at 0°C in 20 mM Tris-HCl, pH 8.5. The 45-kDa alasan protein was obtained from alasan by preparative SDS-PAGE as described previously (31).
Peptide analysis.
SDS-PAGE was carried out on 12% gels (Bio-Rad Co., Hercules, Calif.). The gels were stained either with Coomassie blue (11) or anti-45-kDa monoclonal antibodies (10). Edman degradation analysis of the N-terminal sequence was carried out by the University of California at Davis after resolved proteins were transferred to polyvinylidene difluoride paper. Eleven N-terminal amino acids were sequenced from the 45-kDa protein isolated from alasan (GVTITPLMLGY). In addition, tryptic digests of the 45-kDa protein were separated by high-performance liquid chromatography and two additional N-terminal fragments were sequenced (VFFDTNK and LTAQGFAWDQ). The sequenced fragments were used to design reverse and forward degenerate PCR primers. The primers produced a 900-bp fragment from genomic DNA that was cloned into pGEM-T (Promega Corp., Madison, Wis.). This fragment was labeled using the DIG PCR labeling kit (Roche Diagnostics, Mannheim, Germany) for screening a mini-genomic library.
Southern hybridization.
DNA of A. radioresistens KA53 was restricted using different combinations of enzymes (New England Biolabs, Beverly, Mass.) and run on 0.8% agarose gels. DNA was transferred to a positively charged nylon membrane (Roche), as described in the manufacturer’s instructions. PCR products were first obtained using Takara Taq DNA polymerase and genomic DNA as a template following 10 min of initial denaturation at 94°C; 30 cycles of 45 s at 94°C, 45 s at 56°C, and 60 s at 72°C; and a final elongation step of 10 min at 72°C. Products were visualized with ethidium bromide after running on a 1% agarose gel; DNA was extracted from the gels using Elutip-D columns (Schleicher & Schuell). PCR products were diluted 1:50 and used as a template for digoxigenin (DIG)-labeled probes obtained by PCR incorporation under the same PCR conditions, using DIG PCR labeling mixture (Roche). Following high-stringency washes, an anti-DIG antibody was added and the probes were visualized using CDPstar chemiluminescent substrate. A 2.2-kb fragment generated by BamHI/HindIII digestion hybridized to this probe and was extracted from the gel and cloned into pUC-18 (New England Biolabs). E. coli JM-109 (Promega) colonies were transformed with the plasmid, and the resulting colonies were screened using the colony PCR technique. Nine different positive colonies that yielded the appropriate band size were sequenced, resulting in the full-length open reading frame (ORF) of the gene coding for the 45-kDa protein. Automated DNA sequencing was performed on double-stranded DNA templates by the dideoxynucleotide chain termination method using an Applied Biosystems (Foster City, Calif.) model 373A sequencer.
Production of recombinant protein AlnA.
To produce recombinant 45-kDa protein, the full-length ORF (minus the coding sequence for the putative signal peptide) was amplified using a forward primer with an NdeI restriction site and a reverse primer with a BamHI restriction site. Initial denaturation was at 94°C for 10 min. The Taq DNA polymerase reaction was carried out at 72°C for 45 s (30 cycles). The annealing temperature was 52°C. The resulting fragment was cloned into pET15b (Novagen Inc., Madison, Wis.) and transformed into E. coli ER2566 (New England Biolabs). The resulting fragment was then sequenced to determine that the right reading frame was maintained. Synthesis of the recombinant protein was induced at 30°C with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 2 h, and then the recombinant protein was purified from the bacterial pellet according to the method developed by Qiagen (Chatsworth, Calif.). Briefly, the bacterial pellet was lysed with sample buffer (0.25 M Tris [pH 6.8] and 0.8% SDS in 40% glycerol), and the lysate was incubated for 30 min with nickel beads (Qiagen) and loaded on a column. The beads were then washed with increasing amounts of imidazole (Sigma Chemical Co., St. Louis, Mo.) in order to wash out nonspecifically bound proteins. The elution of the induced protein, AlnA, was accomplished using 250 mM imidazole at a rate of 0.5 ml/min. For immunoblotting, monoclonal antibodies were produced according to a modified method of Koehler and Milstein (10).
Solubilization of PHE.
To determine the kinetics of solubilization, 100 μg of PHE was crystallized in the bottom of 1-ml quartz cuvettes. The cuvettes were placed in a six-compartment holder of an Ultraspec 2000 spectrophotometer (Pharmacia, Uppsala, Sweden), and 1 ml of assay buffer containing different proteins was added to each cuvette. The amount of protein was determined by the Bradford reaction (Bio-Rad). Solubilization was performed without shaking. A252 measurements were taken at room temperature every 30 s for 3.5 h with the parallel nonsynchronized mode of the kinetics software package provided by the manufacturer. Assay buffer (20 mM Tris, pH 8.0) was used as the control. Absorbance readings were converted to concentration of PHE from a standard calibration curve of PHE in hexane.
Determination of emulsifying activity.
A micromodification of the standard emulsification assay (26) was used to measure emulsifying activity. Samples to be tested were introduced into 10-ml glass tubes containing TM buffer (20 mM Tris-HCl buffer [pH 7.0], 10 mM MgSO4) to a final volume of 1.5 ml, and then 0.02 ml of a 1:1 (vol/vol) mixture of hexadecane and 2-methylnaphthalene was added. The tubes were then vortexed at room temperature for 30 min. Turbidity was determined using a Gilford spectrophotometer, model 240. One unit of emulsifying activity was defined as the amount of biopolymer that yielded an A600 of 1.0 in the assay.
Nucleotide sequence accession number.
The GenBank accession number of the A. radioresistens KA53 alnA (OmpA) genomic sequence is AY033946.
RESULTS
Cloning and sequencing of the (alnA) gene coding for the 45-kDa alasan protein.
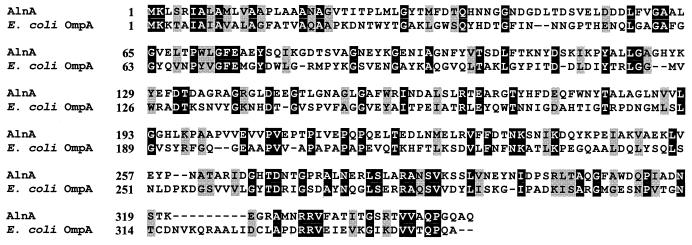
Most of the emulsifying activity of the alasan complex can be attributed to the 45-kDa protein (31). Thus, its N-terminal amino acid sequence was determined. The first 19 amino acids showed 85% identity to the outer membrane protein A (OmpA)-like protein from Acinetobacter spp. (15). Trypsin cleavage of the 45-kDa protein produced several internal peptides, two of which were sequenced. Each of the peptides showed a high similarity to the OmpA-like protein of Acinetobacter spp., one of them being near the C terminus. Degenerate PCR primers were designed from the N-terminal and C-terminal peptides and produced an 890-bp amplimer with genomic DNA isolated from A. radioresistens KA53. The resulting fragment was cloned and sequenced. The deduced amino acid sequence contained the three determined peptide sequences of the native 45-kDa alasan protein. This 890-bp fragment was then used to screen a mini-genomic library of strain KA53. A 2.2-kb BamHI/HindIII fragment that hybridized with the probe was extracted from the gel, cloned into pUC-18, and transformed into E. coli JM-109. Colonies were screened with the PCR primers, and several positive clones were obtained. Sequencing these clones demonstrated that the 2.2-kb fragment contained the full 1.1-kb coding sequence of the 36-kDa protein. The sequence of the first 21 amino acids upstream from the native 45-kDa protein, beginning with GVTIT, was 90% identical to the Acinetobacter sp. OmpA and 45% identical to the E. coli OmpA signal sequence. Based on the DNA sequence, the translated recombinant protein (AlnA) would contain 349 amino acids, corresponding to 37.85 kDa (35.77 kDa without the N-terminal leader sequence) with a predicted pI of 4.71. The difference in molecular mass between the native 45-kDa protein and AlnA is probably due to covalent modifications of the 45-kDa protein, as discussed later. There were no cysteine codons present within the entire (1,044-nucleotide) ORF. These features are consistent with the abundant, heat-modifiable OmpA group of proteins, found in most gram-negative bacteria (3). Figure 1 shows the deduced amino acid sequence of the AlnA protein compared with that of E. coli OmpA (27% identity). The putative ATG start codon (Fig. 1) of alnA was preceded by a perfect Shine-Dalgarno sequence (GGAGGA), 5 bp upstream from the translational start codon.
FIG. 1.
Comparison of the amino acid sequences of AlnA and the OmpA of E. coli. Dashes, sequence gaps black, identity; shaded, similarity.
Expression of alnA in E. coli.
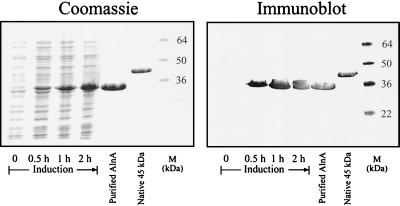
After PCR, the full-length 1.044-kb (minus the signal peptide coding sequence) gene was ligated to pET15b, a high-expression vector containing a T7 promoter and the coding sequence for an N-terminal six-His tag, forming pAT1. After transformation into E. coli, expression of the recombinant protein at 30°C following induction with IPTG was examined. Figure 2 shows that a single protein with the predicted size of 37 kDa was induced. As can be seen from the Coomassie blue-stained gel (Fig. 2), the induced protein is apparent after 0.5 h and accumulates for 2 h. The induced protein reacted with anti-45-kDa protein monoclonal antibodies. An induced 2-liter culture allowed for the purification of 25 mg of the recombinant protein, AlnA-6 His, which was used in the subsequent studies.
FIG. 2.
Expression of AlnA in E. coli. An exponentially growing culture of E. coli (A600 = 0.5) containing pAT1 was induced with IPTG. After 0, 0.5, 1, and 2 h, samples were removed for SDS-PAGE (lanes 1 to 4). Lanes 5 and 6, column-purified AlnA and the native 45-kDa protein, respectively; lane 7, protein standards, prestained for the gel used for immunoblotting.
Emulsifying properties of recombinant bioemulsifier AlnA.
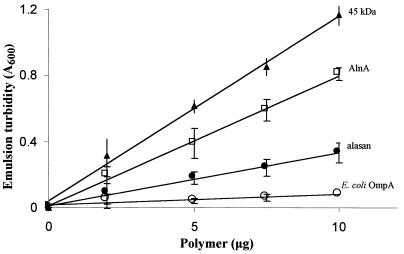
A modification of the standard emulsion assay was used to determine the emulsifying activities of pure recombinant protein AlnA, the native 45-kDa bioemulsifier, alasan, and E. coli OmpA (Fig. 3). Units of emulsifying activity are defined in Materials and Methods. At 10 μg of polymer, the weight ratio of hydrocarbon to emulsifier was 1,600. The specific emulsifying activities of the 45-kDa protein, AlnA, and alasan were 179, 126, and 56 U per mg, respectively. On a molar basis, the specific activity of AlnA was 44% lower than that of the native 45-kDa protein. In all cases, emulsion turbidity was proportional to polymer concentration. E. coli OmpA showed very low emulsifying activity.
FIG. 3.
Emulsifying activity of AlnA. The abilities of recombinant AlnA, E. coli OmpA, alasan, and the native 45-kDa protein to stabilize oil-in-water emulsions were determined as a function of polymer concentration as described in Materials and Methods. The assay tubes contained 0.02 ml of hydrocarbon and the indicated amounts of polymer in a final volume of 1.5 ml.
Solubilization of PHE by AlnA.
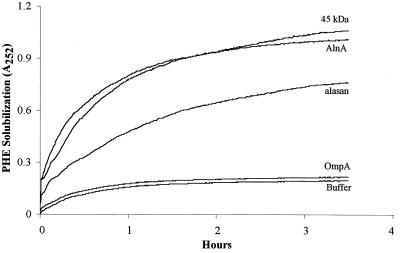
Previously it was demonstrated that the alasan complex greatly enhances the aqueous solubility of polyaromatic hydrocarbons (2). To test if this property was expressed by the isolated 45-kDa alasan protein and recombinant AlnA, the solubility of PHE in quartz cuvettes was measured spectrophotometrically (Fig. 4). Without any mixing, equilibrium was obtained in ca. 3 h. The quantity of PHE solubilized by the polymers was directly proportional to their concentrations (data not presented). In the presence of 40 μg of AlnA or the 45 kDa protein per ml, the amounts of PHE solubilized were similar (A252 of 1.00 to 1.05, compared to 0.19 for the buffer control, or a net increase in A252 of 0.81 to 0.86). Both of these proteins were more efficient in solubilizing PHE than alasan, (equilibrium net A252 = 0.58). E. coli OmpA had little effect on the solubility of PHE. By using the molar extinction coefficient of PHE at 252 nm, the data in Fig. 4 have been converted into micrograms of PHE solubilized per milligram of protein and molecules of PHE solubilized per molecule of protein (Table 1). On a weight basis, AlnA and the 45-kDa protein solubilized 8.1 and 7.4% PHE, respectively, which corresponds to 16.3 and 18.8 molecules of PHE per molecule of protein. Thus, on a molar basis the recombinant AlnA contains 87% of the solubilizing activity of the native 45-kDa protein.
FIG. 4.
The kinetics of solubilization of PHE by AlnA. To quartz cuvettes containing 100 μg of crystalline PHE was added 1.0 ml of buffer containing 40 μg of AlnA, 40 μg of alasan, 40 μg of the 45-kDa protein, and 20 μg of E. coli OmpA. Absorbance was determined every 5 min without mixing.
TABLE 1.
Enhanced solubilization of PHEa
| Addition | Solubility of PHE
|
|||
|---|---|---|---|---|
| μg/mlb | Δμg/mlc | Δ μg/ml/mg of protein | Molecules of PHEd/molecule of protein | |
| Buffer control | 1.17 | |||
| Alasan | 3.54 | 2.37 | 59.3 | 9.9 (29.9)e |
| AlnA | 4.42 | 3.25 | 81.2 | 16.3 |
| 45-kDa protein | 4.15 | 2.98 | 74.5 | 18.8 |
| E. coli OmpA | 1.34 | 0.17 | 8.6 | 1.5 |
Solubility measurements were performed as described for Fig. 4.
Calculated from A252 after equilibrium.
After subtracting solubility in the buffer control.
Calculated from molecular weights of PHE and the added protein. For alasan, it was assumed that the 16-, 31-, and 45-kDa proteins were present in equal molar quantities (31).
Expressed as molecules of PHE per molecule of the 45-kDa protein in the complex, assuming equal molar quantities of the 16-, 31-, and 45-kDa proteins.
DISCUSSION
Bioemulsans are complex mixtures of anionic polysaccharides and proteins (23). This complexity makes it difficult to analyze their chemical structures and the features required for emulsification. Once we were able to demonstrate that the surface-active component of alasan, the bioemulsan of A. radioresistens KA53, is a protein (31), it became possible to apply the techniques of molecular genetics to characterize the gene and gene product responsible for the surface activity of alasan. Evidence has been presented that the alnA gene that was cloned, sequenced, and expressed in E. coli is the A. radioresistens KA53 gene, which codes for the surface-active protein of alasan, i.e., the 45-kDa protein. The protein coded for by alnA reacted with the monoclonal antibodies prepared against the native 45-kDa protein (Fig. 2), and AlnA and the 45-kDa protein contained identical N-terminal, C-terminal, and internal peptide sequences. Furthermore, the recombinant AlnA showed the remarkable properties of stabilizing oil-in-water emulsions and solubilizing PHE. The surprising finding was that the apparent molecular mass of the native 45-kDa protein was significantly different from both the calculated (35.77 kDa) and gel-determined sizes (ca. 36 kDa; Fig. 2) of the recombinant AlnA. It is likely that the native 45-kDa protein contains modifications which increase its apparent size on SDS gels. In this regard, acid hydrolysis of the 45-kDa protein generated reducing sugars, suggesting that it may be a glycoprotein. Apparently, the modifications are not essential for surface activity since recombinant AlnA emulsifies hydrocarbons and solubilizes PHE. The fact that AlnA contained 87 and 56% of the solubilizing and emulsifying activities, respectively, compared to the 45-kDa protein suggests that modifications of the native protein are more important for emulsification than solubilization of hydrocarbons. If the values for emulsification and solubilization of alasan are expressed per mole of the 45-kDa protein in the complex, then alasan has approximately twice the specific activities of the purified native and recombinant proteins. This is most likely due to the synergistic effect of the 31-kDa protein (31).
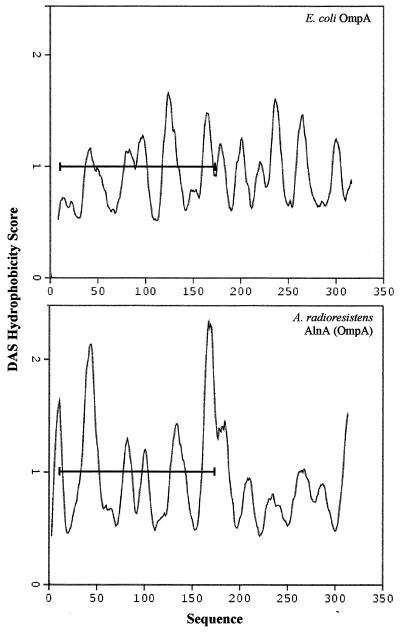
There is significant amino acid sequence similarity (21% identity) between the surface-active AlnA and E. coli OmpA (Fig. 2), yet the latter showed almost no emulsifying or PHE-solubilizing activities. Thus, comparison of the structures of the two proteins might provide some insight into what structural features of AlnA allow it to be a potent emulsifier and solubilizer of hydrophobic compounds. Amino acids 6 to 171 of E. coli OmpA form a β-barrel transmembrane structure, with a hydrophilic interior and hydrophobic exterior (17, 18). Amino acids 6 to 171 of AlnA show a hydrophobicity pattern similar to that of E. coli OmpA (Fig. 5), suggesting that it might also exist as a β-barrel transmembrane structure. The striking difference between E. coli OmpA and AlnA is the high hydrophobicity of the sequences of AlnA on both ends (amino acids 2 to 10 and 168 to 175) of the presumptive β-barrel transmembrane structure. As a working hypothesis, we suggest that these two hydrophobic regions fold over the β-barrel structure, forming hydrophobic regions that are responsible for the remarkable emulsifying and PHE-solubilizing properties of AlnA. X-ray crystallography studies of AlnA are under way.
FIG. 5.
Hydrophobicity patterns of E. coli OmpA and A. radioresistens AlnA (OmpA). The dense alignment surface (DAS) program (5) was used to obtain hydrophobicity scores.
Based on its sequence similarity to other OmpA proteins, AlnA probably functions as an OmpA during exponential phase when it is cell associated (12). When growth ceases as a result of starvation or unbalanced conditions, many Acinetobacter species release bioemulsans into the medium (27). For RAG-1 emulsan, it was shown by immunological (6) and radioactive labeling (26) experiments that the source of the extracellular emulsan was proemulsan which accumulated on the cell surface during the growth phase. It is at present not known whether AlnA exists on the cell surface as a complex with apoalasan and other proteins or if the complex is formed as part of the release mechanism.
The results presented here suggest that AlnA is the OmpA of A. radioresistens. This assumption is supported by the observation that Southern hybridization of enzymatically digested genomic DNA, using probes designed from the sequences of E. coli and Acinetobacter OmpAs, revealed only one gene, corresponding to alnA. The hypothesis is also consistent with the fact that not a single emulsifier-negative mutant was obtained after screening 10,000 randomly inserted transposon strain KA53 mutants (5a). OmpA is an essential protein in E. coli (in the absence of Braun lipoprotein) unless the bacteria are grown in media containing elevated concentrations of magnesium (29).
The ability of AlnA (OmpA of A. radioresistens) to solubilize large quantities of hydrophobic compounds (e.g., 8% PHE by weight) is interesting from both biological and practical points of view. It was recently demonstrated that polyaromatic hydrocarbons were degraded microbiologically more rapidly when they were solubilized by alasan (2). It may be that one of the functions of AlnA is to desorb, solubilize, and bring water-insoluble substrates to the bacteria when they are starved of water-soluble carbon compounds. It has been reported that an OmpA-like protein from an unidentified Acinetobacter species stimulates gastrin and interleukin-8 promoters (15). The mechanism of this promoter-stimulating activity is not known, nor is it known if the surface activity of OmpA is required for the stimulation; however, it would not be surprising to find that a water-soluble protein that binds hydrophobic compounds affects cell physiology.
Acknowledgments
This investigation was supported by the Ministry of Science, Israel, the Pasha Gol Chair for Applied Microbiology, and the Manja and Moris Chair for Biophysics and Biotechnology.
We thank Elisha Orr for antialasan monoclonal antibodies, Juanita L. Merchant for providing us the DNA sequence of Acinetobacter sp. OmpA prior to publication, and Gil Segal for helpful discussions.
REFERENCES
- 1.Banata, I. M. 1995. Biosurfactants production and possible use in microbial enhanced oil recovery and oil pollution remediation. A review. Biosource Technol. 51:1–12 [Google Scholar]
- 2.Barkay, T., S. Navon-Venezia, E. Z. Ron, and E. Rosenberg. 1999. Enhancement of solubilization and biodegradation of polyaromatic hydrocarbons by the bioemulsifier alasan. Appl. Environ. Microbiol. 65:2697–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beher, M., C. A. Schnaitman, and A. P. Pugsley. 1980. Major-modifiable outer membrane protein in gram-negative bacteria: comparison with the OmpA protein of Escherichia coli. J. Bacteriol. 143:906–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belsky, I., D. L. Gutnick, and E. Rosenberg. 1979. Emulsifier of Arthrobacter RAG-1: determination of emulsifier-bound fatty acids. FEBS Lett. 101:175–178 [DOI] [PubMed] [Google Scholar]
- 5.Cserzo, M., E. Wallin, I. Simon, G. von Heijne, and A. Eloffson. 1997. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface. Methods Protein Eng. 10:673–676 [DOI] [PubMed]
- 5a.Dahan, O. 1998. M.Sc. thesis. Tel Aviv University, Ramat Aviv, Israel.
- 6.Goldman, S., Y. Shabtai, C. Rubinowitz, E. Rosenberg, and D. L. Gutnick. 1982. Emulsan in Acinetobacter calcoaceticus RAG-1: distribution of cell-free and cell-associated cross-reacting materials. Appl. Environ. Microbiol. 44:165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaplan, N., and E. Rosenberg. 1982. Exopolysaccharide distribution and bioemulsifier production in Acinetobacter calcoaceticus BD4 and BD413. Appl. Environ. Microbiol. 44:1335–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaplan, N., Z. Zosim, and E. Rosenberg. 1987. Acinetobacter calcoaceticus BD4 emulsan: reconstitution of emulsifying activity with pure polysaccharide and protein. Appl. Environ. Microbiol. 53:440–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klekner, V., and N. Kosaric. 1993. Biosurfactants for cosmetics. Surfactant Sci. Ser. 48:329–372 [Google Scholar]
- 10.Koehler, G., and C. Milstein. 1975. Continuous cultures of fused cells secreting antibodies of predefined specificity. Nature 256:495–497 [DOI] [PubMed] [Google Scholar]
- 11.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 12.Navon-Venezia, S., Z. Zosim, A. Gottlieb, R. Legmann, S. Carmeli, E. Z. Ron, and E. Rosenberg. 1995. Alasan, a new bioemulsifier from Acinetobacter radioresistens. Appl. Environ. Microbiol. 61:3240–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Navon-Venezia, S., E. Banin, E. Z. Ron, and E. Rosenberg. 1998. The bioemulsifier alasan: role of protein in maintaining structure and activity. Appl. Microbiol. Biotechnol. 49:382–384 [Google Scholar]
- 14.Neu, T. R. 1996. Significance of bacterial surface-active compounds in interaction of bacteria with interfaces. Microbiol. Rev. 60:151–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ofori-Darko, E., Y. Zavros, G. Rieder, S. A. Tarlé, M. Van Antwerp, and J. L. Merchant. 2000. An OmpA-like protein from Acinetobacter spp. stimulates gastrin and interlukin-8 promoters. Infect. Immun. 68:3657–3666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel, M. N., and K. P. Gopinathan. 1986. Lysozyme-sensitive bioemulsifier for immiscible organophosphorus pesticides. Appl. Environ. Microbiol. 52:1224–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pautsch, A., and G. E. Schultz. 1998. Structure of the outer membrane protein A transmembrane domain. Nature Struct. Biol. 5:1013–1017 [DOI] [PubMed] [Google Scholar]
- 18.Pautsch, A., and G. E. Schultz. 2000. High-resolution structure of the OmpA membrane domain. J. Mol. Biol. 298:273–282 [DOI] [PubMed] [Google Scholar]
- 19.Robinson, K., M. Gosh, and Z. Shi. 1996. Mineralization enhancement of non-aqueous phase and soil-bound PCB using biosurfactant. Water Sci. Technol. 34:303–309 [Google Scholar]
- 20.Ron, E., and E. Rosenberg. 2001. Natural roles of biosurfactants. Environ. Microbiol. 3:229–236 [DOI] [PubMed] [Google Scholar]
- 21.Rosenberg, E., and E. Z. Ron. 1997. Bioemulsans: microbial polymeric emulsifiers. Curr. Opin. Biotechnol. 8:313–316 [DOI] [PubMed] [Google Scholar]
- 22.Rosenberg, E., and E. Z. Ron. 1998. Surface active polymers from the genus Acinetobacter, p.281–289. In D. L. Kaplan (ed.), Biopolymers from renewable resources. Springer-Verlag KG, Berlin, Germany
- 23.Rosenberg, E., and E. Z. Ron. 1999. High- and low-molecular-mass microbial surfactants. Appl. Microbiol. Biotechnol. 52:154–162 [DOI] [PubMed] [Google Scholar]
- 24.Rosenberg, E., A. Zuckerberg, C. Rubinovitz, and D. L. Gutnick. 1979. Emulsifier of Arthrobacter RAG-1: isolation and emulsifying properties. Appl. Environ. Microbiol. 37:402–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenberg, E., C. Rubinovitz, A. Gottlieb, S. Rosenhak, and E. Z. Ron. 1988. Production of biodispersan by Acinetobacter calcoaceticus A2. Appl. Environ. Microbiol. 54:317–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rubinovitz, C., D. L. Gutnick, and E. Rosenberg. 1982. Emulsan production by Acinetobacter calcoaceticus in the presence of chloramphenicol. J. Bacteriol. 152:126–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sar, N., and E. Rosenberg. 1983. Emulsifier production by Acinetobacter calcoaceticus strains. Curr. Microbiol. 9:309–314 [Google Scholar]
- 28.Shepherd, R., J. Rockey, I. W. Sutherland, and S. Roller. 1995. Novel bioemulsifiers from microorganisms for use in foods. J. Biotechnol. 40:207–217 [DOI] [PubMed] [Google Scholar]
- 29.Sonntag, I., H. Schwarz, Y. Hirota, and U. Henning. 1978. Cell envelope and shape of Escherichia coli: multiple mutants missing the outer membrane lipoprotein and other major outer membrane proteins. J. Bacteriol. 136:280–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor, W. H., and E. Juni. 1961. Pathways for biosynthesis of a bacterial capsular polysaccharide. I. Characterization of the organism and polysaccharide. J. Bacteriol. 81:688–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toren, A., S. Navon-Venezia, E. Z. Ron, and E. Rosenberg. 2001. Emulsifying activity of purified alasan proteins from Acinetobacter radioresistens KA53. Appl. Environ. Microbiol. 67:1102–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Volkering, F., A. Breure, and W. Rulkens. 1997. Microbiological aspects of surfactant use for biological soil remediation. Biodegradation 8:401–417 [DOI] [PubMed] [Google Scholar]
- 33.Westphal, O., and K. Jann. 1965. Bacterial lipopolysaccharides: extraction with phenol-water and further applications of the procedure, p.83–91. In R. L. Whistler (ed.), Carbohydrate chemistry. Academic Press Inc., New York, N.Y
- 34.Zosim, Z., G. Fleminger, D. L. Gutnick, and E. Rosenberg. 1989. Effect of protein on the emulsifying activity of emulsan. J. Dispers. Sci. Technol. 10:307–317. [Google Scholar]