Abstract
Streptomyces griseus NP4, which was derived by UV mutagenesis from strain IFO13350, showed a bald and wrinkled colony morphology in response to glucose. Mutant NP4 formed ectopic septa at intervals along substrate hyphae, and each of the compartments developed into a spore which was indistinguishable from an aerial spore in size, shape, and thickness of the spore wall and in susceptibility to lysozyme and heat. The ectopic spores of NP4 formed in liquid medium differed from “submerged spores” in lysozyme sensitivity. Shotgun cloning experiments with a library of the chromosomal DNA of the parental strain and mutant NP4 as the host gave rise to DNA fragments giving two different phenotypes; one complementing the bald phenotype of the host, and the other causing much severe wrinkled morphology in the host. Subcloning identified a gene (dasR) encoding a transcriptional repressor belonging to the GntR family that was responsible for the reversal of the bald phenotype and a gene (dasA) encoding a lipoprotein probably serving as a substrate-binding protein in an ATP-binding cassette (ABC) transport system that was responsible for the severe wrinkled morphology. These genes were adjacent but divergently encoded. Two genes, named dasB and dasC, encoding a membrane-spanning protein were present downstream of dasA, which suggested that dasRABC comprises a gene cluster for an ABC transporter, probably for sugar import. dasR was transcribed actively during vegetative growth, and dasA was transcribed just after commencement of aerial hypha formation and during sporulation, indicating that both were developmentally regulated. Transcriptional analysis and direct sequencing of dasRA in mutant NP4 suggested a defect of this mutant in the regulatory system to control the expression of these genes. Introduction of multicopies of dasA into the wild-type strain caused ectopic septation in very young substrate hyphae after only 1 day of growth and subsequent sporulation in response to glucose. The ectopic spores of the wild type had a thinner wall than those of mutant NP4, in agreement with the observation that the former was sensitive to lysozyme and heat. Disruption of the chromosomal dasA or dasR in the wild-type strain resulted in growth as substrate mycelium, suggesting an additional role of these genes in aerial mycelium formation. The ectopic septation and sporulation in mutant NP4 and the wild-type strain carrying multicopies of dasA were independent of a microbial hormone, A-factor (2-isocapryloyl-3R-hydroxymethyl-γ-butyrolactone), that acts as a master switch of aerial mycelium formation and secondary metabolism.
The gram-positive, soil-inhabiting, filamentous bacterial genus Streptomyces shows complex morphological differentiation, which makes this genus one of the model prokaryotes for studying multicellular differentiation (7, 9). On agar medium, one or more substrate hyphae formed from a germinating spore branch frequently and grow rapidly by cell wall extension at the hyphal tips. Filamentous development is strong, and septation in the substrate mycelium occurs very rarely. Subsequently aerial hyphae emerge by reuse of material assimilated into the substrate mycelium, such as DNA, proteins, and storage compounds. Many cells in substrate hyphae thus lyse and die (67). When apical growth of aerial hyphae stops, in contrast to substrate mycelium, septa are formed at regular intervals along the hyphae to form many unigenomic compartments within a sheath composed of elongated hollow or grooved elements, finer fibrillar elements, and amorphous material.
The sporulation septa consist of two membrane layers separated by a double layer of cell wall material, which permits the eventual separation of adjacent spores (14, 17, 38, 68). Spore chains usually consist of many tens of spores. The aerial spores thus formed are resistant to heat treatment and lysozyme digestion. Some Streptomyces strains produce spores in submerged culture when critical nutritional and environmental conditions are met (10, 12, 32, 41). Streptomyces griseus B-2682, for example, produces abundant submerged spores in nutrient-depleted media (32). The submerged spores of S. griseus are similar but never identical to aerial spores and are sensitive to lysozyme digestion, probably because of the thinness of the spore wall.
The combination of morphological mutants and gene cloning techniques is useful for the study of morphological differentiation, giving insight into structural and regulatory genes important for the developmental processes. Many bld mutants lacking an aerial mycelium and whi mutants lacking spores have facilitated isolating such genes from Streptomyces coelicolor A3(2) (7, 40). whiG, which encodes an RNA polymerase ς factor resembling the motility ς factor of Bacillus subtilis, is one of those genes and determines the timing of sporulation (8, 39). Interestingly, overexpression of whiG causes the host to form septa even in substrate mycelium, resulting in occasional ectopic sporulation in the substrate hyphae. Ectopic sporulation also results from deletion of a region close to the glkA locus (31), from overexpression of ssgA encoding a small acidic protein (29, 62), and from a mutation in ssfR encoding an iclR-type transcriptional regulator for ssgA (26). Thus, an increase and decrease of expression of the genes involved in regulation of the programmed, ordered developmental processes yields morphologically deprogrammed mutants.
We have isolated an S. griseus mutant, NP4, that shows marked ectopic sporulation in substrate hyphae in response to glucose both on agar and in liquid media. Shotgun cloning experiments with this mutant as the host revealed a gene cluster encoding an ATP-binding cassette (ABC)-type transporter and a transcriptional factor. Some ABC transporters have been adapted to transport specific regulatory molecules and, hence, regulate cell physiology. Examples are the Spo0K system of B. subtilis (47, 50), which mediates the uptake of a regulatory peptide and is required for the initiation of sporulation; the BldK system of S. coelicolor A3(2) (43), which probably imports an oligopeptide and is required for aerial mycelium formation; and the AmfAB system, which is probably involved in transport of some substance and aerial mycelium formation in S. griseus (61) and S. coelicolor A3(2) (34).
In the present study, overexpression of one of the cloned ABC transporter genes in the wild-type S. griseus strain caused ectopic sporulation in the substrate hyphae in response to glucose. The ectopic spores of the wild-type strain harboring multicopies of one of the transporter genes differed in various aspects from those formed by mutant NP4 on glucose-containing medium. The ectopic sporulation in the wild type was A-factor independent, which shows that the ectopic septation and subsequent development into spores occur independently of the A-factor regulatory cascade. A-factor is a representative of γ-butyrolactones serving as microbial hormones in a wide variety of Streptomyces spp., and the chemical structure of A-factor in S. griseus is 2-isocapryloyl-3R-hydroxymethyl-γ-butyrolactone (20–22). It switches on aerial mycelium formation and secondary metabolism at a concentration as low as 10−9 M in S. griseus. An A-factor-deficient mutant S. griseus strain neither forms aerial mycelium nor produces streptomycin.
We thus expected that the S. griseus mutant showing ectopic sporulation would give useful information on morphological development. The purposes of this study were (i) to characterize the S. griseus mutant strain forming ectopic spores by septation in substrate hyphae, (ii) to clone and characterize the genes that altered the phenotype of the mutant strain, (iii) to characterize the ectopic spores formed by the mutant and similar but not identical ectopic spores formed by the wild-type strain carrying multicopies of one of the cloned ABC transporter genes by scanning and transmission electron microscopy, and (iv) to analyze transcription of the ABC transporter gene cluster.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
S. griseus IFO13350 (13) was obtained from the Institute of Fermentation (IFO), Osaka, Japan. S. griseus HH1 is A-factor deficient due to the deletion of afsA (23). S. griseus NP4 was derived from the wild-type strain IFO13350 by UV mutagenesis. S. griseus ΔadpA, which had a deletion in adpA, encoding an A-factor-dependent transcriptional activator (45), and S. griseus ΔadsA, which has a deletion in adsA, encoding an extracytoplasmic function ς factor of RNA polymerase (70), were described previously.
Streptomyces strains were grown in YMPD medium (yeast extract [Difco], 0.2%; meat extract [Wako Pure Chemicals], 0.2%; Bacto-peptone [Difco], 0.4%; NaCl, 0.5%; MgSO4 · 7H2O, 0.2%; glycine, 0.5%; and glucose, 1%). YMPD agar medium contained 2% agar. Phenotypes of S. griseus strains were examined at 28°C on YMP agar and in YMP liquid medium containing various sugars at a concentration of 1% instead of glucose. Minimal medium for S. griseus has been described (42). Thiostrepton (50 μg/ml) and neomycin (20 μg/ml) were added when necessary. R2YE medium (19) was used for regeneration of protoplasts. A-factor production was assayed by the streptomycin cosynthesis method with the A-factor-deficient S. griseus mutant HH1, as described (23).
As Streptomyces plasmids, high-copy-number plasmids pIJ486, containing the kanamycin and thiostrepton resistance genes (66), and pIJ702, containing the melanin production and thiostrepton resistance genes (28), both with copy numbers of 40 to 100 per genome, were used. A low-copy-number plasmid, pKU209, with a copy number of 1 to 2 per genome, containing the ampicillin and thiostrepton resistance genes (27), was also used.
Escherichia coli JM109 and pUC19 (71) for DNA manipulation were purchased from Takara Shuzo. E. coli JM110 containing dam and dcm mutations was used for preparing nonmethylated Streptomyces DNA used for gene disruption. Media and growth conditions for E. coli were those described by Maniatis et al. (35). Ampicillin (50 μg/ml) and kanamycin (20 μg/ml) were used when necessary.
General recombinant DNA studies.
Restriction enzymes, T4 DNA ligase, reverse transcriptase, and other DNA-modifying enzymes were purchased from Takara Shuzo. [α-32P]dCTP (110 TBq/mmol) for DNA labeling with the Takara BcaBest DNA labeling system and [γ-32P]ATP (220 TBq/mmol) for end labeling at 5′ ends with T4 polynucleotide kinase were purchased from Amersham Pharmacia Biotech. DNA was manipulated in Streptomyces (19) and in E. coli (3, 35), as described earlier. Nucleotide sequences were determined by the dideoxy chain termination method (51) with the Termo Sequenase fluorescence-labeled primer cycle sequencing kit (Amersham) or the DNA sequencing kit (ABI Prism) on an automated DNA sequencer. Open reading frames in the nucleotide sequence were predicted by Frame Plot analysis (25).
Shotgun cloning.
Chromosomal DNA of the wild-type S. griseus IFO13350 was partially digested with Sau3AI and ligated with either BamHI-digested pIJ486 or BglII-digested pIJ702. The ligation mixture was introduced by protoplast transformation into S. griseus NP4 cells that showed a bald phenotype. Thiostrepton-resistant transformants were selected on YMPD medium, and seven sporulating colonies were isolated. A colony showing a severely wrinkled morphology was also isolated. The seven sporulating colonies contained a common region, and one of them (plasmid pNS5), which carried a 6-kb insert on pIJ702, was chosen for further study. The wrinkled colony contained a plasmid, named pES1, which carried a 2.7-kb insert on pIJ486.
Subcloning.
A 1,379-bp HincII fragment containing dasR (see Fig. 4A) was cloned in the HincII site in the multilinker of pUC19. The dasR sequence was excised as a HindIII-BamHI fragment and placed between the HindIII and BamHI sites of pIJ486 to construct pHR3. The dasR sequence, excised as a HincII fragment, was cloned into the EcoRI site of pKU209 (plasmid pLR3) after the EcoRI site had been flush-ended with Klenow fragment. A 1,801-bp EcoRI fragment containing the whole dasA sequence was placed in the EcoRI site of pKU209 to construct pLA1. A 3.2-kb KpnI-SacI fragment containing the truncated orf1, orf2, and orf3 was cloned between the KpnI and SacI sites of pUC19. This fragment was excised as an EcoRI-HindIII fragment and placed between the EcoRI and HindIII sites of pIJ486 to construct pKS3. A 3-kb SacI fragment containing dasB and dasC was flush-ended with Klenow fragment and cloned in the HincII site of pUC19. The dasBC sequence was excised as a BamHI-HindIII fragment and placed in pIJ486 to construct pHBC1. A 1.7-kb NruI fragment containing the dasB sequence was similarly cloned in pIJ486 to construct pHB1.
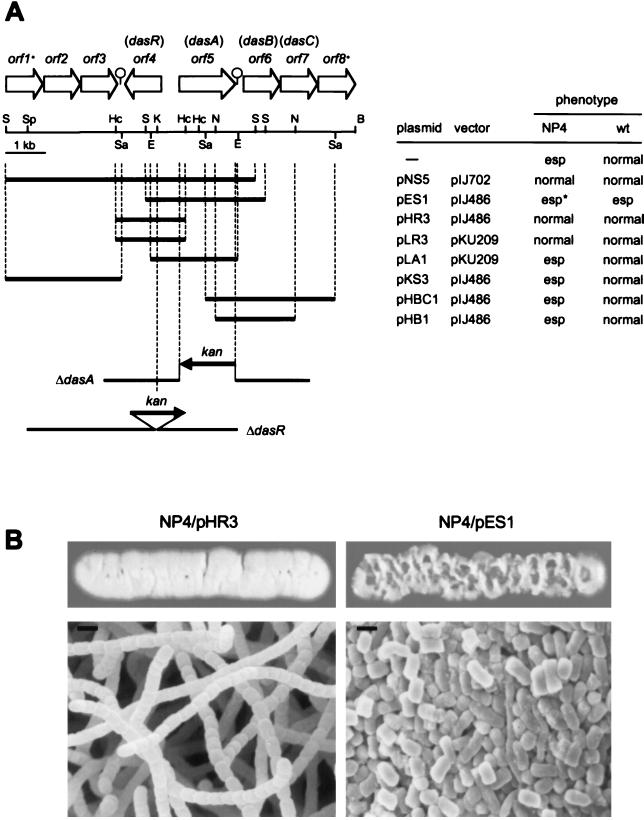
FIG. 4.
Restriction map and subcloning of the cloned fragment. (A) The extents and directions of open reading frames on the cloned fragments are indicated by arrows. orf1* and orf8* are truncated. Plasmid pNS5 was harbored in one of the seven NP4 transformants showing normal development culminating in sporulation. Plasmid pES1 was harbored in the NP4 transformant showing a severe wrinkled morphology (esp*). pES1 caused the wild-type strain S. griseus IFO13350 to form ectopic, substrate spores (esp). An inverted repeat sequence downstream of dasR is TGA (termination codon of orf3)-(N)10-GGGCGGTGGCCCGGGGA-(N:loop)5-TCCCCGGGCCACCGGCCC-(N)46-TCA (termination codon of dasR). An inverted repeat sequence downstream of dasA is TGA (termination codon of dasA)-(N)67-TCCGGGGC-(N:loop)2-GCCCCGGA-(N)22-GTG (start codon of dasB). The insertion of the kanamycin resistance determinant (kan) into dasA and dasR is shown schematically. Abbreviations for restriction enzymes: B, BamHI; E, EcoRI; Hc, HincII: K, KpnI; N, NruI; S, Sau3AI; Sa, SacI; and Sp, SphI. (B) Mutant NP4 containing only dasR on either the high-copy-number or the low-copy-number plasmid developed normally and formed spores after growth at 28°C for 4 days on YMPD agar medium, whereas NP4 containing dasA on pIJ486 showed a more severe wrinkled morphology. Scanning electron micrographs reveals the formation of normal arthrospores by mutant NP4 harboring pHR3 and substrate spores by NP4 harboring pES1.
Gene disruption.
For disruption of the chromosomal dasA gene, the upstream and downstream regions of dasA were amplified by PCR with the following primers. The upstream region of about 2 kb in size (see Fig. 4A) was amplified with 5′-GAAGCTTGCTGAAGAGACAGCGCCGTCA-3′ (the underlining indicates a HindIII site) and 5′-GGATCCGTTAACACGATGAGCTTGCGCTTCACG-3′ (the underlined and italic letters indicate a BamHI and HpaI site, respectively; the boldface letters indicate the start codon of dasA). The downstream region was amplified with 5′-GTTAACGATCAACGCGCTGATCAACAACAAG-3′ (corresponding to the region 6-bp upstream of the stop codon of dasA) and 5′-GGATCCGTCAGGGTGTCCGTGGTGGAAACG-3′. The HindIII-HpaI fragment from the upstream region, the HpaI-BamHI fragment from the downstream region, and pUC19 DNA digested with HindIII plus BamHI were ligated by three-fragment ligation. The HincII-SmaI fragment containing the kanamycin resistance gene (4) was then inserted in the HpaI site of the pUC19 recombinant plasmid. In the resultant plasmid, most of the dasA sequence was replaced by the kanamycin resistance gene. The plasmid was linearized by DraI digestion, alkali denatured with 0.1 M NaOH, and introduced by transformation into S. griseus IFO13350, as described (44). Correct insertion of the kanamycin resistance gene by homologous recombination was checked by Southern hybridization with the 0.6-kb HincII fragment containing the dasA sequence and the 1.3-kb HincII-SmaI fragment containing the kanamycin resistance gene as probes against the EcoRI-digested chromosomal DNA.
For disruption of the chromosomal dasR gene, the kanamycin resistance gene was inserted in the KpnI site within the dasR sequence (see Fig. 4A). The disrupted dasR sequence, together with the 1.7-kb upstream and 3.2-kb downstream regions, was placed in pUC19. The pUC19 plasmid linearized by DraI digestion was similarly introduced in S. griseus IFO13350. Correct disruption of the dasR gene was checked by Southern hybridization with the 1.4-kb HincII fragment containing the dasR sequence and the 1.3-kb HincII-SmaI fragment containing the kanamycin resistance gene as probes against the HincII-digested chromosomal DNA.
S1 nuclease mapping.
Methods for RNA isolation from cells grown on cellophane on the surface of agar medium and S1 nuclease mapping were described by Kelemen et al. (30). Hybridization probes were prepared by PCR with a pair of 32P-labeled and nonlabeled primers. For dasA, 5′-GGTCACGGGTCGTCGCCCTCG-3′ (corresponding to positions −105 to −85, taking the transcriptional start point of dasA as +1, which was determined later) and 5′-GGTCTTCCTTGCGGTCCTTCG-3′ (corresponding to positions +212 to +196) were used. For dasR, 5′-GGTCTTTAATGGTTTAGACCAGTACCG-3′ (corresponding to positions −142 to −115, taking the transcriptional start point of dasR as +1) and 5′-GTCCGTCATGTCGAGGAGATGGC-3′ (corresponding to positions +167 to +144) were used. hrdB, encoding a ς factor of RNA polymerase, was used to check the purity and amount of RNA used, as described previously (70). Protected fragments were analyzed on 6% polyacrylamide sequencing gels by the method of Maxam and Gilbert (37).
RT-PCR.
Transcription of the dasABC region was analyzed by reverse transcription (RT)-PCR with pairs of the following primers (see Fig. 8C): A1 (5′-GTGAAGCGCAAGCTCATCGTGG-3′; the boldface letters indicate the start codon of dasA) and A2 (5′-GTCAGGATTCCTTGTTGATCAGCG-3′; the boldface letters indicate the stop codon of dasA); AB1 (5′-GCCGCTACTGGTACGCCGCGATG-3′, corresponding to Arg-203 to Met-209 of DasA) and AB2 (5′-GTTCCTTGGGGACCTGGGTGAGG-3′, corresponding to Leu-228 to Glu-222 of DasB); and B1 (5′-GTGTCTGCCGCTGATACCAAGGCCG-3′; the boldface letters indicate the start codon of dasB) and BC2 (5′-GCACTATCCCTTCACAGCGCCGGAAG-3′; the boldface letters indicate the stop codon of dasC).
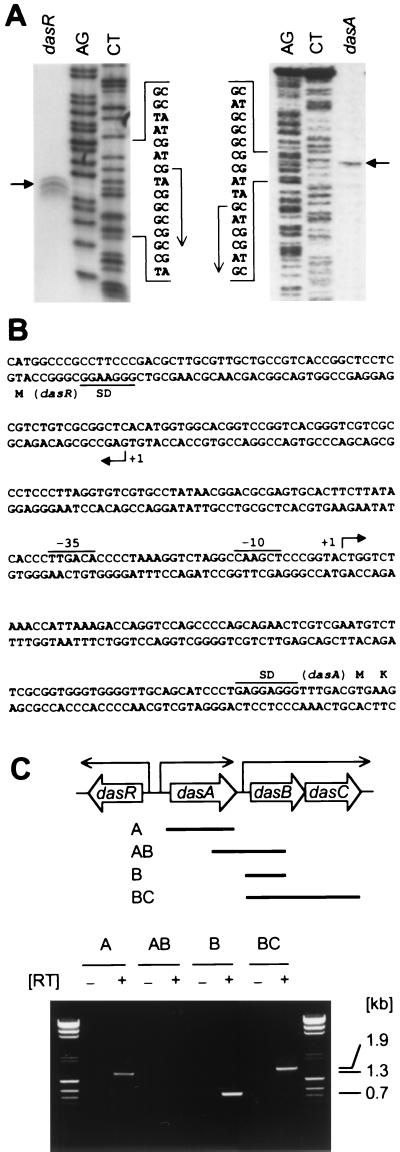
FIG. 8.
Determination of the transcriptional start points of dasA and dasR by S1 nuclease mapping. (A) RNA was prepared from cells grown on YMPD medium at 28°C for 1 day for dasR and 3 days for dasA. The sequence ladders, derived from the same primer, are shown with the A+G and T+C reactions. The deduced transcriptional start points are shown by arrows. (B) The nucleotide sequence covering the promoter regions of dasA and dasR is shown, together with their NH2-terminal amino acid sequences. Probable Shine-Dalgarno (SD) sequences and probable −35 and −10 sequences for dasA are indicated. (C) RT-PCR for determination of transcriptional linkages of dasABC. Pairs of primers were used to amplify the indicated four DNA fragments. No amplification occurred when reverse transcriptase was omitted from the reaction mixture, indicating the absence of DNA in the mRNA samples.
Microscopy.
For light microscopy, S. griseus strains IFO13350 and NP4 were cultured at 28°C in YMPD medium. Cells were harvested at intervals and placed with oil immersion under a phase-contrast light microscope (Zeiss Axiophot 2).
For scanning electron microscopy (59), S. griseus strains were grown on YMPD agar medium at 28°C for 4 days, and agar blocks containing the spores and hyphae were cut. For preparation of the specimens, the agar blocks were fixed with 2% osmium tetroxide for 24 h and then dehydrated by air-drying for 1 h. Each specimen was sputter coated with platinum-gold and examined with a Hitachi S4000 scanning electron microscope.
For transmission electron microscopy, S. griseus strains were grown on YMPD agar medium at 28°C for 2 to 4 days. A colony was scraped and prefixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.3) for 2 to 5 h. The samples were washed three times with 0.05 M potassium phosphate buffer (pH 7.0) and fixed with 1% osmium tetroxide in 0.05 M potassium phosphate buffer for 12 to 14 h. The samples were washed with water three times, prestained with 0.5% uranyl acetate for 2 h, and dehydrated with 70%, 80%, 90%, and absolute ethanol and finally with acetone. The samples were encapsulated in Spurr’s resin, and sections were stained with 3% uranyl acetate and then with lead citrate for 2 h. The sections were placed under a JEOL 2010 electron microscope at 100 kV.
Heat and lysozyme resistance tests.
S. griseus strains were precultured at 28°C for 24 h in YMPD liquid medium. About 1 mg of mycelium was inoculated to a petri dish (diameter, 9 cm) containing YMPD agar medium and further incubated at 28°C. At day 2, when spores were not yet formed, mycelium was transferred to 1 ml of 20 mM TES [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic 0acid] buffer (pH 7.2) and suspended. The mycelium suspension was divided into three parts: one was a control with no treatments, the second was heated at 55°C for 30 min, and the third was incubated at 37°C for 1 h in the presence of 20 μg of egg white lysozyme (Seikagaku Kogyo) per ml. After the treatments, portions of the suspensions were homogenized with a glass homogenizer and spread on YMPD medium, and colonies were counted after incubation at 28°C for 2 days. At day 5, when abundant spores were formed, heat and lysozyme resistance was similarly examined.
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper has been deposited to the DDBJ, EMBL, and GenBank DNA databases under accession no. AB061860.
RESULTS
Properties of S. griseus mutant NP4.
We have long studied the regulation of morphological development and secondary metabolism in streptomycin-producing S. griseus IFO13350, especially in relation to a chemical signaling molecule, A-factor. During these studies, we found mutant NP4, showing a wrinkled and ragged colony shape among UV-mutagenized colonies (Fig. 1A). The defect in aerial mycelium formation of mutant NP4 did not result from A-factor deficiency, since NP4 was found to produce A-factor when assayed by the streptomycin cosynthesis method (23) (data not shown). Mutant NP4 produced little streptomycin when assayed by the bioassay with B. subtilis ATCC 6633 as an indicator. This is probably due to the cessation of development. The morphological defect in mutant NP4 was glucose dependent, because the defect was observed only on YMPD medium containing glucose as the carbon source and not on YMP medium containing maltose, galactose, mannitol, or glycerol instead of glucose (data not shown). Mutant NP4 showed normal morphological development culminating in sporulation on the latter media. As will be described in detail below, the effects of dasA overexpression on the wild-type strain were also glucose dependent.
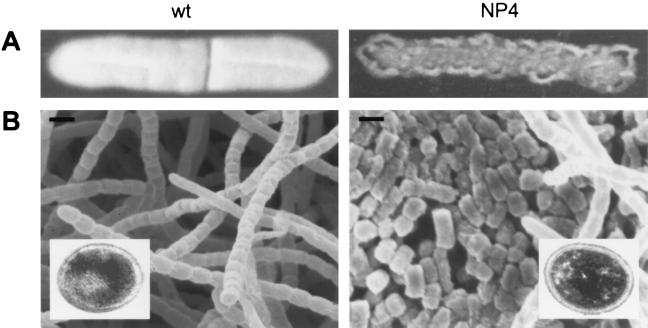
FIG. 1.
Morphology of S. griseus NP4 on solid medium. (A) Mycelium of mutant NP4 and the wild-type (wt) strain S. griseus IFO13350 was spread on YMPD agar medium and grown at 28°C for 4 days. The wild-type strain forms white spores, but mutant NP4 shows a wrinkled and ragged morphology. (B) Scanning electron micrographs of the colony surfaces of the wild-type and NP4 strains grown at 28°C for 4 days. Bars, 1 μm. Transmission electron micrographs of a single spore of both strains are also shown.
Scanning electron microscopy revealed that mutant NP4 grew as substrate mycelium for 4 days, in which septa were subsequently formed at regular intervals (Fig. 1B). Septation in substrate mycelium occurs very rarely in the genus Streptomyces. In the visible field, some aerial hyphae and chains of spores developed normally from the substrate hyphae were also seen. Each compartment separated by the septa in substrate hyphae developed into a spore, which was indistinguishable in size, shape, and thickness of the spore coat from the aerial spore of the parental S. griseus strain IFO13350, as determined by transmission electron microscopy (Fig. 1B). The spores examined were those formed in the substrate hyphae that penetrated the agar. Consistent with the morphology of the ectopic spores from mutant NP4, they were resistant to heat and lysozyme to nearly the same extent as the aerial spores (Fig. 2). The septum is therefore supposed to consist of two separate cross walls, as is observed in sporulation septum formation and arthrospore maturation in S. griseus (14).
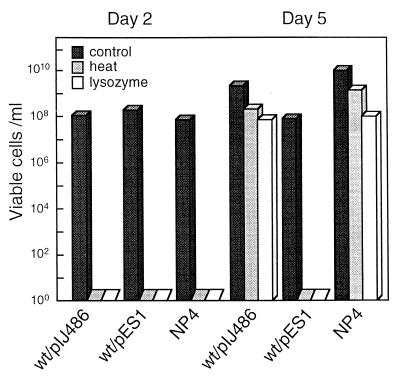
FIG. 2.
Heat and lysozyme resistance of ectopic, substrate spores of S. griseus mutant NP4 and the wild-type strain S. griseus IFO13350 harboring pES1. The three strains were grown on YMPD agar for the indicated days and spread on the same medium after challenges by heat treatment at 55°C for 30 min and by lysozyme treatment at 20 μg/ml and 37°C for 1 h. At day 2, mutant NP4 and the wild-type strain grew as substrate mycelium, as did the wild-type strain harboring pIJ486 as a control, but the wild type carrying pES1 showed a wrinkled colony morphology, indicative of ectopic septation. At day 5, mutant NP4 formed substrate spores, whereas the wild-type strain formed arthrospores.
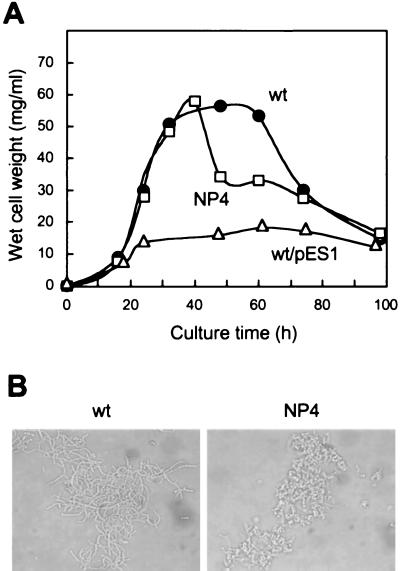
S. griseus IFO13350 rarely forms submerged spores in YMPD liquid medium, although a different S. griseus strain, B-2682, forms abundant submerged spores (32). When mutant NP4 was cultured in YMPD liquid medium, it grew with the same time course as the wild-type strain, but at the beginning of the stationary phase the cell mass suddenly dropped (Fig. 3A). The sudden decrease in the cell mass coincided with the septation in the growing mycelium and formation of apparent “submerged spores” (Fig. 3B). The decrease in cell mass was probably due to the breakdown of the sheath. Submerged spore formation was glucose dependent, as was found for substrate spore formation on solid medium. The spores of mutant NP4 formed in liquid medium were resistant to lysozyme, since the rod-like cells observed 40 h after inoculation gave nearly the same CFU after incubation at 37°C for 1 h in the presence and absence of 20 μg of lysozyme. This is in vivid contrast to the submerged spores of S. griseus B-2682, which are sensitive to lysozyme digestion (32). We assume that the submerged spores of mutant NP4 are formed in a different way from those of several other Streptomyces strains (10, 12, 32, 41), but in the same manner as the substrate spores are formed on solid medium. The growth of NP4 in liquid medium resembles that of “nocardioform organisms,” which produce branching hyphae that sooner or later break up into a bacillary or coccoidal form.
FIG. 3.
Morphology of S. griseus NP4 in liquid medium. (A) The wild-type strain S. griseus IFO13350 (wt), mutant NP4, and the wild-type strain carrying multicopies of dasA on pES1 were grown at 28°C in YMPD liquid medium, and their wet cell weights were monitored. (B) Phase-contrast photomicrographs of the wild-type and NP4 strains grown at 28°C for 4 days in YMPD medium.
Cloning and nucleotide sequencing of DNA fragments conferring morphological changes on S. griseus mutant NP4.
We constructed a bank of partially Sau3AI digested chromosomal fragments of S. griseus IFO13350 by using multicopy plasmids, pIJ486 and pIJ702, as the cloning vectors. After transformation of mutant NP4 by the library, we found two types of colonies with different morphology. One showed the normal morphological development and formed spores, as does the wild-type strain. The other showed a more severely wrinkled colony morphology than mutant NP4 (Fig. 4). We isolated seven colonies of the former type and one colony of the latter type.
Restriction mapping and partial nucleotide sequencing of the DNA fragments in the transformants showed that they all derived from the same stretch of DNA. The largest DNA fragment (plasmid pNS5) was contained in the colony of the former type, and its nucleotide sequence was determined. The Frame analysis of the nucleotide sequence predicted four complete open reading frames, orf2 to orf5, and two truncated frames, orf1 and orf6. The seven colonies showing wild-type morphology carried orf4 as a common region, and the colony showing the severe wrinkled morphology carried orf5 as a single complete open reading frame.
We also cloned the neighboring regions and determined the whole nucleotide sequence in a total of 8,741 bp. The prediction from a homology search by the computer was as follows. Orf2 (400 amino acids [aa]), hypothetical protein found in Deinococcus radiodurans (DNA database accession no. G75219); Orf3 (341 aa), oxidoreductase in Streptomyces fradiae (accession no. AF147704); Orf4 (258 aa), transcriptional repressor belonging to the GntR family (15); Orf5 (432 aa), sugar-binding protein; and Orf6 (335 aa) and Orf7 (278 aa), integral membrane proteins. Orf1, homologous with a putative sugar transferase (2), and Orf8, homologous with β-glucosidase (16), were truncated.
On the basis of this information, we subcloned the DNA fragment to identify the gene(s) responsible for the phenotypes of the NP4 transformants. As a result, orf4 on either a high-copy-number or a low-copy-number plasmid was sufficient to reverse the wrinkled morphology and orf5 was sufficient to cause mutant NP4 to show the severe wrinkled morphology and to form substrate spores (Fig. 4). Introduction of pKS3 containing orf1 to orf3, pHB1 containing orf6, or pHBC1 containing orf6 and orf7 did not change the morphology of mutant NP4. orf4 was named dasR (deficient in aerial mycelium and spore formation) and orf5 was named dasA, since a mutation or overexpression of these genes affected morphological development even in the wild-type strain, as described below.
Cloned genes are members of the gene cluster for an ABC transporter.
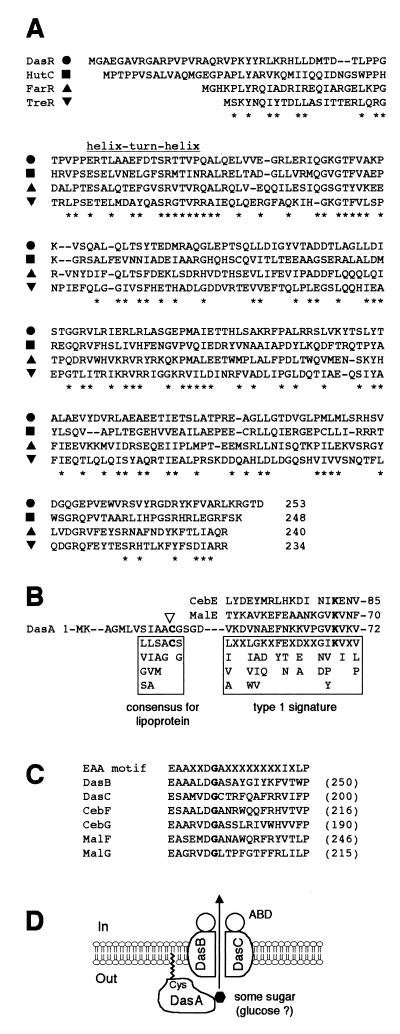
Amino acid alignment of DasR with transcriptional repressors in the GntR family is shown in Fig. 5A. They contain a helix-turn-helix DNA-binding motif at the NH2-terminal portions. DasR shows 23% identity and 46% similarity to the transcriptional repressor TreR for the trehalose operon in Pseudomonas fluorescens (36), 31% identity and 50% similarity to the repressor HutC for the histidine utilization genes in Pseudomonas putida (1), and 32% identity and 49% similarity to the repressor FarR, responsible for the fatty acyl-responsive regulator in E. coli (48).
FIG. 5.
Homologies of DasRABC products with components of the ABC-type transport systems. (A) Amino acid alignment of DasR with the transcriptional repressors in the GntR family. TreR from P. fluorescens (DNA database accession number AAG31030), HutC from P. putida (P22773), and FarR from E. coli (S04645) are shown. Asterisks indicate similar amino acids that are conserved in three of the four proteins. (B) Probable cleavage site of the signal peptide (indicated by a triangle), the signature sequence in SBPs specific for maltooligosaccharides, multiple sugars, α-glycerol phosphate, and iron (60), and amino acid alignment of this region of DasA with MalE of S. coelicolor A3(2) and CebE of S. reticuli are shown. X represents any amino acid. The Lys residue in boldface is the highly conserved amino acid in this family. (C) The consensus sequence conserved in MSDs of ABC transporters (52) and amino acid alignment of this region of DasBC with MalFG and CebFG are shown. X represents any amino acid. The Gly residue in boldface is the highly conserved amino acid in this family. (D) Predicted topology of DasABC as components of an ABC transporter. DasA, a substrate-binding protein, is anchored to the outer surface of the membrane by a lipid attached to Cys-21. DasBC are integrated in the membrane, to each of which an ATP-hydrolyzing subunit (ABD) is associated.
The NH2-terminal portion of DasA shows features typical of a signal peptide (65), i.e., positively charged residues at the NH2 terminus followed by a stretch of hydrophobic residues (Fig. 5B). In addition, the amino acid sequence around Cys-21 resembles the consensus, L(S, A)(A, G)*C(S, G), of the sites (indicated by the asterisk) cleaved by lipoprotein-specific signal peptidases in gram-positive bacteria (58). A more degenerate and longer consensus is (L, V, G, S)(L, I, V, A)(S, A, M)(A, G)*C(S, G), and the sequence including Cys-21 perfectly matches this consensus, suggesting that Cys-21 is at the NH2 terminus of the mature form and is covalently modified by the typical ester-linked and amide-linked acylation of lipoproteins.
The sequence from Val-54 to Val-72 shows similarity to the signature sequence (cluster 1) of binding proteins specific for maltooligosaccharides, multiple sugars, α-glycerol phosphate, and iron (60). These include MalE, essential for import of maltose in S. coelicolor A3(2) (63), and CebE, essential for import of cellobiose and cellotriose in Streptomyces reticuli (53). Therefore, DasA seems to serve as a substrate-binding protein of the components comprising an ABC-type transport system. ABC transporters consist of multisubunit permeases that transport various molecules across the cytoplasmic membrane (6, 11, 18, 55, 60). They consist of two hydrophobic membrane-spanning domains (MSDs) associated with two cytoplasmic ATP-binding domains (ABDs) and a high-affinity extracytoplasmic substrate-binding protein (SBP). Lipoproteins serving as SBPs are anchored in the outer leaflet of the cell membrane by the lipid, usually palmitic acid, attached to the NH2-terminal Cys residue.
The MSD and SBP genes of an ABC transport system are generally found organized together as an operon. The termination codon (TGA) of DasB, which is encoded by the region just downstream of dasA, overlaps the start codon (ATG) of DasC. Both DasB and DasC were predicted to span the membrane six times when their hydrophobicity was analyzed with the PSORT WWW server (http://psort.ims.u-tokyo.ac.jp). Furthermore, they contain the consensus sequence EAAX2DGAX8IXLP, conserved in MSDs of ABC transporters (52) (Fig. 5C). MalFG of S. coelicolor A3(2) and CebFG of S. reticuli also contain a sequence similar to the consensus. Thus, it is probable that DasABC are components of an ABC transport system (Fig. 5D), although no ATP-binding proteins as ABDs are encoded in the vicinity of this operon. DasR seemed to be a regulator for this das operon because of the gene organization and the transcriptional analysis (see below). malEFG and cebEFG are also accompanied by a gene encoding a repressor belonging to the LacI-GalR family, which is adjacent but divergently encoded (53, 64).
Ectopic sporulation of wild-type S. griseus containing multiple copies of dasA.
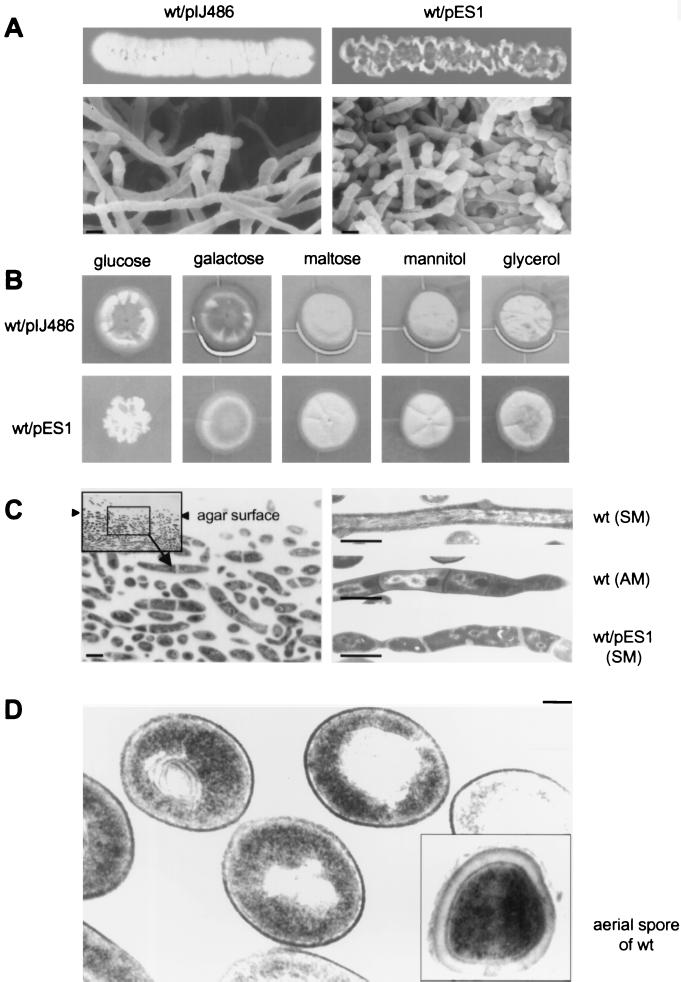
We introduced pES1 and pHR3 into the wild-type strain S. griseus IFO13350 to see the effects of multiple copies of dasA and dasR. No detectable phenotypic changes were observed for S. griseus IFO13350 harboring pHR3. However, the wild-type strain harboring pES1 showed a wrinkled colony morphology on YMPD medium, as did mutant NP4 (Fig. 6A). The effect of dasA on the wild-type strain was observed only on YMPD medium containing glucose and not on YMP medium containing other sugars (Fig. 6B), as was observed for mutant NP4. Vertical sections of the colony showed that the cells far below the agar surface were a mixture of short substrate hyphae with septa and spore-like cells (Fig. 6C). Sporulation of Streptomyces strains rarely occurs in substrate hyphae that have penetrated the agar (8, 67). The ectopic septa in the substrate hyphae and the cell wall of the wild-type strain harboring pES1 are thicker and have a lower electron density than those in the aerial hyphae of the wild-type strain. The envelope and septa of the aerial mycelium of the wild-type strain are electron dense. However, the main wall of an aerial spore of the wild-type strain is apparently thicker than that of the ectopic spore, which is enclosed by a thinner, electron-dense envelope (Fig. 6D).
FIG. 6.
Morphology of the wild-type S. griseus strain IFO13350 containing extra copies of dasA. (A) S. griseus IFO13350 harboring pES1 shows a wrinkled colony morphology on YMPD agar medium, as does mutant NP4, and forms substrate spores, as examined by scanning electron microscopy. Photographs were taken after growth at 28°C for 4 days. Bars, 1 μm. (B) The morphological defect of S. griseus IFO13350 harboring pES1 depends on glucose as a carbon source, because on YMP medium containing other sugars at a concentration of 1% instead of glucose, the wild-type strain harboring pES1 shows normal sporulation. Photographs were taken after growth at 28°C for 5 days. (C) Sporulation in the substrate mycelium of agar-grown S. griseus IFO13350 harboring pES1. Bars, 1 μm. An ultrathin section containing substrate mycelium penetrated into agar after growth at 28°C for 2 days was examined at different magnifications (left panel). A substrate spore chain of the wild type harboring pES1 is shown, together with the substrate (SM) and aerial (AM) mycelium of the wild-type strain as controls (right panel). (D) Ectopic, substrate spores of S. griseus IFO13350 harboring pES1, together with the aerial spore of the wild-type strain as a control, is shown. The samples were prepared after growth at 28°C for 4 days. Bar, 0.1 μm.
Consistent with the apparent difference in the main spore wall, the ectopic spores were susceptible to both heat treatment and lysozyme digestion (Fig. 2). As described above, the ectopic spores of mutant NP4 were almost the same in morphology and susceptibility to heat and lysozyme as the aerial spores. The difference can probably be ascribed to the difference in the texture and thickness of the spore walls. Nevertheless, the two different ectopic spores were found to germinate at nearly the same frequency, as high as more than 90%, when the CFU were measured.
The wild-type strain carrying pES1 appeared to show the wrinkled morphology earlier than mutant NP4. We then examined the courses of the ectopic septation of the wild type carrying pES1 and mutant NP4 in relation to the development of the wild-type strain by scanning and transmission electron microscopy. A lump of mycelium on a petri dish was picked with a toothpick and streaked on YMPD agar medium. The wild-type strain grew as substrate mycelium for 2 days and then as a mixture of aerial and substrate mycelia and at day 4 formed spores. Mutant NP4 grew as substrate mycelium for 4 days and abruptly formed septa in the substrate hyphae. On the other hand, the wild-type carrying pES1 formed septa in the substrate hyphae as early as day 1. Because of the very early septation, the spread of mycelium of the wild type carrying pES1 was poor. The early septation and sporulation of the wild type carrying pES1 also occurred in YMPD liquid medium; no increase in the cell mass, indicative of sporulation, 20 h after inoculation was observed (Fig. 3A). The thin spore wall and sensitivity to lysozyme of the ectopic spores formed by the wild-type strain carrying pES1 likely result from this very early septation.
A-factor independence of ectopic sporulation of S. griseus carrying dasA.
In S. griseus, A-factor serves as a master switch for aerial mycelium formation and secondary metabolism; the development of an A-factor-deficient mutant, S. griseus strain HH1, remains in substrate mycelium. We introduced pES1 into mutant HH1 to see the effect of A-factor on ectopic septation in substrate hyphae caused by overexpression of dasA. Mutant HH1 harboring pES1 apparently spread the mycelium poorly on YMPD agar and showed wrinkled colony morphology. Scanning electron microscopic analysis of the wrinkled colony of mutant HH1 harboring pES1 showed ectopic septation as early as day 1 (data not shown), indicating that the ectopic septation in substrate hyphae and subsequent sporulation were independent of A-factor. Consistent with this idea, ΔadpA and ΔadsA mutants, both containing a mutation in the respective regulatory step essential for normal development in the A-factor regulatory cascade, also showed a wrinkled colony morphology at day 1 on YMPD agar. The poor spread of mycelium of these strains carrying pES1 is presumably due to the very early septation. The septa in the substrate hyphae seem to be formed immediately after the hyphae have elongated.
Aerial mycelium-defective phenotype in dasA and dasR disruptants.
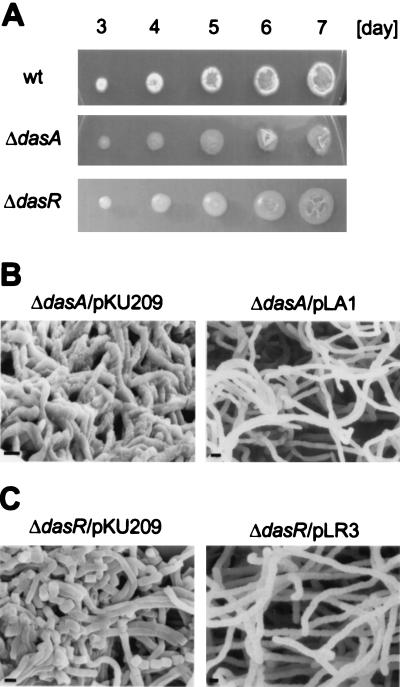
We disrupted the chromosomal dasA gene of S. griseus IFO13350 so that almost the whole dasA coding sequence was replaced by the kanamycin resistance gene. The growth rate of the ΔdasA mutant on YMPD medium was almost the same as on minimal medium containing maltose, mannitol, or glycerol instead of glucose, suggesting that dasRABC are not so actively involved in glucose import, if at all, that loss of the function results in a decrease in growth rate. This is consistent with the observation that dasA is transcribed at the commencement of aerial mycelium formation and during sporulation (see below). However, the ΔdasA strain showed a bald phenotype on YMPD agar medium (Fig. 7A). Introduction of dasA on the low-copy-number plasmid pKU209 (plasmid pLA1) into this mutant reversed the defect, indicating that the inability to form aerial mycelium of the ΔdasA mutant was due solely to the disruption of dasA (Fig. 7B). The aerial mycelium formation and sporulation of this mutant were also greatly reduced on YMP medium containing mannitol, maltose, or glycerol, but sparse spores were still formed.
FIG. 7.
Bald phenotypes of the ΔdasA and ΔdasR mutants derived from S. griseus IFO13350. (A) Colonies of the ΔdasA and ΔdasR mutants were formed with a toothpick on YMPD medium. The growth of the mutants is the same as the wild-type strain, as seen by the diameters of the colonies, but the mutants show a bald phenotype. (B) Scanning electron micrographs of the ΔdasA mutant harboring the vector plasmid pKU209 or pLA1, containing the intact dasA gene. (C) Scanning electron micrographs of the ΔdasR mutant harboring the vector plasmid pKU209 or pLR1, containing the intact dasR gene.
The ΔdasR mutant also showed a bald phenotype (Fig. 7A), irrespective of carbon source. Scanning electron microscopic analysis of the ΔdasR mutant revealed growth as substrate hyphae and occasional septation in the substrate hyphae (Fig. 7C). Comparison of the substrate hyphae between the ΔdasA and ΔdasR mutants showed that the hyphae of the former were apparently rough and those of the latter were rather smooth, suggesting some difference in wall structure. Consistent with this, the colony surface of the ΔdasA mutant was rough and that of the ΔdasR mutant was smooth and lustrous (Fig. 7A). The rough hyphae was usually observed for the A-factor-deficient mutant HH1. Introduction of dasR on the low-copy-number plasmid pKU209 (plasmid pLR3) into this mutant reversed the defect.
Transcriptional analysis of ABC-type transporter gene cluster.
The transcriptional start points of dasR and dasA in the wild-type strain S. griseus IFO13350 were determined by high-resolution S1 nuclease mapping with RNA prepared from cells that were grown on YMPD agar medium (Fig. 8A). Both genes were transcribed from a single start point. In front of the start point of dasA, a TTGACA sequence, the same as the −35 consensus sequence for many bacteria, including Streptomyces spp. (57), and a CAAGCT sequence, somewhat similar to a typical −10 sequence TATAAT, are present (Fig. 8B). However, no such sequences are present at appropriate positions in front of the transcriptional start point of dasR.
dasBC was hypothesized to be transcribed by a promoter in front of dasB, since an inverted repeat sequence, probably serving as a rho-independent transcriptional terminator, was found downstream of the termination codon of dasA (Fig. 4A). We performed RT-PCR to examine transcription of dasBC by using the four primers indicated in Fig. 8C. A pair of primers for detection of the mRNA stretch corresponding to fragment AB yielded no amplified DNA fragments, and a pair of primers for mRNA corresponding to fragment BC yielded a DNA fragment with the expected size of 1.9 kb. We did not further characterize the transcription of dasBC.
Developmentally regulated expression of dasA and dasR.
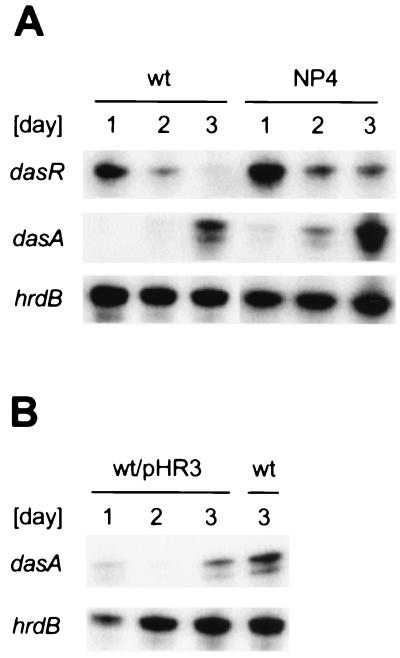
The courses of transcription of dasR and dasA in the wild-type strain and mutant NP4 were examined by low-resolution S1 nuclease mapping with RNAs prepared from cells grown on YMPD agar medium. For preparation of cells in large amounts, cells cultured in liquid medium were collected, concentrated by centrifugation, and spread on cellophane on the surface of the agar medium. hrdB, which encodes ςHrdB and is transcribed throughout growth (56), was used to monitor the quantity and quality of the RNAs used. At day 1 under these conditions, the wild-type strain grew as substrate mycelium. After 2 days, it grew as a mixture of aerial and substrate hyphae, and after 3 days, it grew as a mixture of spores and aerial hyphae. Mutant NP4 began to form septa in substrate hyphae after 3 days. In both the wild-type and mutant strains, dasR was mainly transcribed during vegetative growth, whereas dasA was transcribed just after commitment of aerial mycelium formation and during spore formation (Fig. 9A). The timing of dasA expression contravenes the bald phenotype caused by the dasA deletion. DasA in basal amounts may play a role in the initiation of aerial mycelium formation.
FIG. 9.
Transcription of dasRABC. (A) RNA was prepared from colonies formed on YMPD agar medium. The wild-type S. griseus IFO13350 strain grew as substrate mycelium at day 1, as a mixture of substrate and aerial hyphae at day 2, and as a mixture of aerial hyphae and spores at day 3. (B) Transcription of dasA is repressed in the wild-type strain harboring pHR3.
The gene organization of dasRABC and the homology of DasR with transcriptional repressors belonging to the GntR family suggested that DasR might repress the expression of at least dasA. The above-described course of dasA and dasR expression is in agreement with this idea. In addition, in the wild-type strain harboring multicopies of dasR, transcription of dasA was severely repressed (Fig. 9B), suggesting that DasR represses the promoter of dasA. This is also in agreement with the observation that the ΔdasR mutant, in which dasA is not repressed by DasR, shows occasional septation in substrate hyphae (Fig. 7C).
dasR and dasA were both transcribed more actively in mutant NP4 than in the wild-type strain. It is likely that the active transcription of dasA in NP4 results in the ectopic sporulation, because, as described above, introduction of multicopies of dasA into the wild-type strain caused ectopic sporulation. We determined the nucleotide sequence of the dasR-dasA region, i.e., the SacI-EcoRI fragment covering the whole dasR and dasA genes in mutant NP4 (Fig. 4A), and found no base changes. This excluded the possibility that the mutation(s) in mutant NP4 lay in the coding sequences or promoter regions of dasRA.
DISCUSSION
S. griseus mutant NP4, which was isolated by UV mutagenesis, showed a bald and wrinkled colony morphology because of ectopic septation in substrate hyphae and subsequent spore formation. The ectopic spores were the same as aerial spores in size, thickness of the spore wall, and shape, as determined by transmission and scanning electron microscopy, and in heat and lysozyme susceptibility. Mutant NP4 also formed abundant spores in liquid medium, whereas the parental strain IFO13350 rarely forms submerged spores under these conditions. The wall of the ectopic spores is supposed to be thicker than those of the submerged spores formed by several Streptomyces spp., including S. griseus B-2682 (32), under specific conditions, because the spores of NP4 were resistant to lysozyme. We therefore assume that both on solid and in liquid medium, mutant NP4 forms two separate cross walls in the vegetative hyphae and matures each compartment into a spore indistinguishable from aerial spores in many aspects, as is observed in aerial spore formation in S. griseus (14).
The frequency of septation in substrate mycelium in mutant NP4 is much higher than in S. coelicolor A3(2) strains harboring multicopies of whiG (7, 40) or having a deletion of a region close to the glkA locus (31). These strains form abundant aerial spores and only occasional septa in substrate mycelium and subsequent sporulation. The deprogrammed sporulation of NP4 implies that once septa are formed, even in substrate hyphae, in response to some signal, each compartment is inevitably destined to develop into a spore. In the substrate hyphae of the wild-type S. griseus strain, some signals must block the commitment to septation and subsequent sporulation. A-factor does not release the block, because exogenous addition of an appropriate amount of A-factor to the substrate hyphae of the A-factor-deficient mutant HH1 causes no septum formation but normal formation of aerial spores. An excess amount of DasA appears to release the block, since introduction of pES1 into mutant HH1 results in ectopic septation.
Shotgun cloning of genes on the wild-type chromosome into a mutant is a useful approach for identifying the mutated gene and genes closely related to the mutant phenotype. We at first expected that the mutation(s) responsible for the ectopic sporulation of NP4 was in dasR or dasA, since the ectopic sporulation of mutant NP4 was completely reversed by dasR encoding a transcriptional factor belonging to the GntR family, and dasA made the wrinkled morphology of mutant NP4 more severe. However, nucleotide sequencing of the dasR-dasA region in mutant NP4 revealed no base changes.
On the basis of the observations that introduction of dasA into the wild-type strain caused ectopic sporulation and that the amount of dasA transcript was greater in mutant NP4 than in the wild type, we assume that NP4 has a mutation in the regulatory pathway to control the expression of dasA. Because ectopic sporulation appears to result solely from an increase in the amount of DasA, the regulatory pathway in mutant NP4 seems to lack the ability to repress dasA. The elevated expression of dasR in NP4 suggests that the putative regulatory pathway controls dasR too. The increase in the amount of DasR, which may repress not only dasA but also some other genes involved in programmed development, is a possible explanation for the difference in timing of ectopic septation between the wild-type strain carrying dasA and mutant NP4; an increase in the amount of only DasA in the same background as in the wild type results in early commitment of septation, whereas an increase in the amount of DasR during early growth and the existence thereafter (Fig. 9A) bring forth a different background, resulting in septation at the programmed time.
Is DasRABC involved only in sugar import as an ABC transporter, as predicted by the homology of each of the components? The gene organization dasR-dasA-dasB-dasC and their predicted functions are the same as those for the maltose and cellobiose/cellotriose import systems in Streptomyces spp., malR-malE-malF-malG (63) and cebR-cebE-cebF-cebG (53), respectively. In addition, a gene encoding a glucosidase-like protein is encoded downstream of all three of these gene clusters. Although no ATP-binding proteins as ABDs are encoded in the vicinity of the das operon, the absence of the gene encoding ABD also holds for the malEFG and cebEFG operons. As pointed out by van Wezel et al. (64), MsiK or an MsiK-like ATP-binding protein (24, 53, 54), which is encoded elsewhere on the chromosome and homologous to the ATP-hydrolyzing subunit MalK in the maltose import system in E. coli (5, 11), may serve as a general ATP-hydrolyzing subunit for “orphan” ABC transporters. MalK is an essential component in the E. coli maltose import system, forming MalEFGK2.
The effect of glucose on the ectopic sporulation of mutant NP4 and on the wild type containing multicopies of dasA tempted us to speculate that DasA recognizes and binds to glucose or a glucose derivative and imports it via the Das system. However, dasA is developmentally regulated, and its transcription is enhanced just after commitment of aerial mycelium formation and during spore formation. This means that DasA is not produced until the glucose in the medium is almost consumed. Conceivably, DasA binds a certain sugar compound other than glucose which is needed for septation in aerial hyphae at the programmed point. The bald phenotype of the ΔdasA mutant suggests an additional role of DasA in aerial mycelium formation, although we have no plausible explanation for it. In addition, the bald phenotype, but accompanied with occasional septation in substrate hyphae, of the ΔdasR mutant suggests that the concentration of DasA controlled by dasR is critical for aerial mycelium formation.
In considering the function of DasA, we would like to point out the multiple functions of some substrate-binding proteins of the ABC transporters. For example, ChvE is a multifunctional glucose/galactose-binding protein which participates in the uptake of specific monosaccharides, chemotaxis to these sugars, and virulence gene induction in Agrobacterium. For induction of the virulence genes to form crown gall tumors, monosaccharide-bound ChvE interacts with the periplasmic region of VirA, a sensor kinase in the VirA-VirG two-component signal transduction system (46, 69). For chemotaxis, ChvE is supposed to interact with chemotaxis receptors such as Tar and Trg (33). The maltose-binding protein MalE of E. coli, the oligopeptide-binding protein OppA of E. coli, and the galactose-binding protein MglB of Salmonella enterica serovar Typhimurium are other examples that function as a chaperone for protein folding and protection from stress in the periplasm, in addition to their function in import and chemotaxis (49). These examples, together with the developmentally regulated expression of dasA and involvement in septum formation of DasA, present a possibility that substrate-bound or free DasA interacts with other regulatory proteins in the membrane, thus commencing a regulatory pathway for morphological development.
The ectopic septation and subsequent sporulation of S. griseus triggered solely by overexpression of dasA was independent of A-factor, because the A-factor-deficient mutant HH1 harboring pES1 showed ectopic sporulation. This is consistent with the observation that the wild-type strain harboring pES1 formed ectopic septa at day 1, when the concentration of A-factor is still low (Fig. 3A and 6). In S. griseus, A-factor at a critical concentration triggers aerial mycelium formation and streptomycin biosynthesis by binding a repressor-type receptor protein, ArpA, and dissociating it from the promoter region of adpA, encoding a transcriptional activator (45). A-factor is produced in a growth-dependent manner (20–22).
One of the targets of AdpA is adsA, encoding an extracytoplasmic function sigma factor, ςAdsA, essential for the initiation of aerial mycelium formation (70). AdpA and AdsA supposedly activate many structural genes required for aerial mycelium formation. A-factor thus determines the timing of programmed and ordered development by acting as a master switch for turning on many genes at several hierarchic regulatory steps. The ectopic spores which are triggered by an excess amount of DasA and independently of A-factor germinate at the same frequency as aerial spores, although they are sensitive to lysozyme and heat because of a thinner spore wall. The difference in lysozyme and heat resistance of the aerial spores and ectopic spores of the wild-type harboring multicopies of dasA implies that some of the gene products necessary for the architecture of aerial spores are absent in the maturation of the ectopic spores.
Comparison of transcription of genes necessary for normal morphological and physiological development in the wild-type, A-factor-controlling background and in the dasA-overexpressing background will reveal the difference in the genetic network between the programmed septation in aerial hyphae and ectopic septation in substrate hyphae triggered by an excess of DasA. It will also be useful to study their counterparts in different Streptomyces spp. S. coelicolor A3(2) contains a very similar gene cluster, open reading frames CAB94616 to -94619, in cosmid SC7E4 (www.sanger.ac.uk/Projects/S_coelicolor/), each gene of which shows 33 to 91% identity to the corresponding gene in the dasRABC cluster.
Acknowledgments
This work was supported by the Asahi Glass Foundation, by the Research for the Future Program of the Japan Society for the Promotion of Science, and by the Bio Design Program of the Ministry of Agriculture, Forestry, and Fisheries of Japan (BDP-01-VI-2-2).
REFERENCES
- 1.Allison, S. L., and A. T. Phillips. 1990. Nucleotide sequence of the gene encoding the repressor for the histidine utilization genes of Pseudomonas putida. J. Bacteriol. 172:5470–5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amado, M., R. Almeida, T. Schwientek, and H. Clausen. 1999. Identification and characterization of large galactosyltransferase gene families: galactosyltransferases for all functions. Biochim. Biophys. Acta 1473:35–53. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingstone, D. O. Moore, J. S. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 4.Beck, E., G. Ludwig, A. Auerswald, B. Reiss, and H. Schaller. 1982. Nucleotide sequence and exact localisation of the neomycin phosphotransferase gene from transposon Tn5. Gene 19:327–336. [DOI] [PubMed] [Google Scholar]
- 5.Boos, W., and H. Schuman. 1998. Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol. Mol. Biol. Rev. 62:204–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braibant, M., P. Gilot, and J. Content. 2000. The ATP binding cassette (ABC) transport systems of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 24:449–467. [DOI] [PubMed] [Google Scholar]
- 7.Chater, K. F. 1993. Genetics of differentiation in Streptomyces. Annu. Rev. Microbiol. 47:685–713. [DOI] [PubMed] [Google Scholar]
- 8.Chater, K. F., C. J. Bruton, K. A. Plaskitt, M. J. Buttner, C. Méndez, and J. D. Helmann. 1989. The developmental fate of S. coelicolor hyphae depends upon a gene product homologous with the motility ς factor of B. subtilis. Cell 59:133–143. [DOI] [PubMed] [Google Scholar]
- 9.Chater, K. F., and R. Losick. 1997. Mycelial life style of Streptomyces coelicolor A3(2) and its relatives, p. 149–182. In J. A. Shapiro and M. Dworkin (ed.), Bacteria as multicellular organisms. Oxford University Press, New York, N.Y.
- 10.Daza, A., J. F. Martín, A. Dominguez, and J. A. Gil. 1989. Sporulation of several species of Streptomyces in submerged cultures after nutritional downshift. J. Gen. Microbiol. 135:2483–2491. [DOI] [PubMed] [Google Scholar]
- 11.Ehrmann, M., R. Ehrle, E. Hofmann, W. Boos, and A. Schlösser. 1998. The ABC maltose transporter. Mol. Microbiol. 29:685–694. [DOI] [PubMed] [Google Scholar]
- 12.Glazebrook, M. A., J. L. Doull, C. Stuttard, and L. C. Vining. 1990. Sporulation of Streptomyces venezuelae in submerged cultures. J. Gen. Microbiol. 136:581–588. [DOI] [PubMed] [Google Scholar]
- 13.Hara, O., S. Horinouchi, T. Uozumi, and T. Beppu. 1983. Genetic analysis of A-factor synthesis in Streptomyces coelicolor A3(2) and Streptomyces griseus. J. Gen. Microbiol. 129:2939–2944. [DOI] [PubMed] [Google Scholar]
- 14.Hardisson, C., and M. B. Manzanal. 1976. Ultrastructural studies of sporulation in Streptomyces. J. Bacteriol. 127:1443–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haydon, D. J., and J. R. Guest. 1991. A new family of bacterial regulatory proteins. FEMS Microbiol. Lett. 146:3–12. [DOI] [PubMed] [Google Scholar]
- 16.Henrissat, B., and A. Bairoch. 1993. New families of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 293:781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henssen, A., E. Weise, G. Vobis, and B. Renner. 1981. Ultrastructure of sporogenesis in actinomycetes forming spores in chains, p. 137–146. In K. P. Schaal and G. Pulverer (ed.), Actinomycetes. Gustav Fisher Verlag, Stuttgart, Germany.
- 18.Higgins, C. F. 1992. ABC transporters: from microorganisms to man. Annu. Rev. Cell Biol. 8:67–113. [DOI] [PubMed] [Google Scholar]
- 19.Hopwood, D. A., M. J. Bibb, K. F. Chater, T. Kieser, C. J. Bruton, H. M. Kieser, D. J. Lydiate, C. P. Smith, J. M. Ward, and H. Schrempf. 1985. Genetic manipulation of Streptomyces: a laboratory manual. The John Innes Foundation, Norwich, United Kingdom.
- 20.Horinouchi, S. 1999. γ-Butyrolactones that control secondary metabolism and cell differentiation in Streptomyces, p.193–207. In G. M. Dunny and S. C. Winans (ed.), Cell-cell signaling in bacteria. American Society for Microbiology, Washington, D.C.
- 21.Horinouchi, S., and T. Beppu. 1992. Autoregulatory factors and communication in actinomycetes. Annu. Rev. Microbiol. 46:377–398. [DOI] [PubMed] [Google Scholar]
- 22.Horinouchi, S., and T. Beppu. 1994. A-factor as a microbial hormone that controls cellular differentiation and secondary metabolism in Streptomyces griseus. Mol. Microbiol. 12:859–864. [DOI] [PubMed] [Google Scholar]
- 23.Horinouchi, S., Y. Kumada, and T. Beppu. 1984. Unstable genetic determinant of A-factor biosynthesis in streptomycin-producing organisms: cloning and characterization. J. Bacteriol. 158:481–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hurtubise, Y., F. Shareck, D. Kluepfel, and R. Morosoli. 1995. A cellulase/xylanase-negative mutant of Streptomyces lividans 1326 defective in cellobiose and xylobiose uptake is mutated in a gene encoding a protein homologous to ATP-binding proteins. Mol. Microbiol. 17:367–377. [DOI] [PubMed] [Google Scholar]
- 25.Ishikawa, J., and K. Hotta. 1999. Frame Plot: a new implementation of the frame analysis for predicting protein-coding regions in bacterial DNA with a high G+C content. FEMS Microbiol. Lett. 174:251–253. [DOI] [PubMed] [Google Scholar]
- 26.Jiang, H., and K. Kendrick. 2000. Characterization of ssfR and ssgA, two genes involved in sporulation of Streptomyces griseus. J. Bacteriol. 182:5521–5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kakinuma, S., Y. Takada, H. Ikeda, H. Tanaka, and S. Omura. 1991. Cloning of large DNA fragments, which hybridize with actinorhodin biosynthesis genes, from kalafungin and nanaomycin A methyl ester producers and identification of genes for kalafungin biosynthesis of the kalafungin producer. J. Antibiot. 44:995–1005. [DOI] [PubMed] [Google Scholar]
- 28.Katz, E., C. J. Thompson, and D. A. Hopwood. 1983. Cloning and expression of the tyrosinase gene from Streptomyces antibioticus in Streptomyces lividans. J. Gen. Microbiol. 129:2703–2714. [DOI] [PubMed] [Google Scholar]
- 29.Kawamoto, S., and J. C. Ensign. 1995. Cloning and characterization of a gene involved in regulation of sporulation and cell division of Streptomyces griseus. Actinomycetologica 9:136–151. [Google Scholar]
- 30.Kelemen, G. H., P. Brian, K. Flärdh, L. Chamberlin, K. F. Chater, and M. J. Buttner. 1998. Developmental regulation of transcription of whiE, a locus specifying the polyketide spore pigment in Streptomyces coelicolor A3(2). J. Bacteriol. 180:2515–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelemen, G. H., K. A. Plaskitt, C. G. Lewis, K. C. Findlay, and M. J. Buttner. 1995. Deletion of DNA lying close to the glkA locus induces ectopic sporulation in Streptomyces coelicolor A3(2). Mol. Microbiol. 17:221–230. [DOI] [PubMed] [Google Scholar]
- 32.Kendrick, K. E., and J. C. Ensign. 1983. Sporulation of Streptomyces griseus in submerged culture. J. Bacteriol. 155:357–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levit, M. N., Y. Liu, and J. B. Stock. 1998. Stimulus response coupling in bacterial chemotaxis: receptor dimers in signalling arrays. Mol. Microbiol. 30:459–466. [DOI] [PubMed] [Google Scholar]
- 34.Ma, H., and K. Kendal. 1994. Cloning and analysis of a gene cluster from Streptomyces coelicolor that causes accelerated aerial mycelium formation in Streptomyces lividans. J. Bacteriol. 176:6401–6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N. Y.
- 36.Matthijs, S., N. Koedam, P. Cornelis, and H. de Greve. 2000. The trehalose operon of Pseudomonas fluorescens ATCC17400. Res. Microbiol. 151:845–851. [DOI] [PubMed] [Google Scholar]
- 37.Maxam, A. M., and W. Gilbert. 1980. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 65:499–560. [DOI] [PubMed] [Google Scholar]
- 38.McVittie, A. 1974. Ultrastructural studies on sporulation in wild-type and white colony mutants of Streptomyces coelicolor. J. Gen. Microbiol. 81:291–302. [DOI] [PubMed] [Google Scholar]
- 39.Mendez, C., and K. F. Chater. 1987. Cloning of whiG, a gene critical for sporulation of Streptomyces coelicolor A3(2). J. Bacteriol. 169:5715–5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merrick, M. J. 1976. A morphological and genetic mapping study of bald colony mutants of Streptomyces coelicolor. J. Gen. Microbiol. 96:299–315. [DOI] [PubMed] [Google Scholar]
- 41.Miguélez, E. M., B. Rueda, C. Hardisson, and M. B. Manzanal. 1997. Colony development in Streptomyces carpinensis: a streptomycete with substrate mycelium spores. FEMS Microbiol. Lett. 157:103–107. [Google Scholar]
- 42.Neumann, T., W. Piepersberg, and J. Distler. 1996. Decision phase regulation of streptomycin production in Streptomyces griseus. Microbiology 142:1953–1963. [Google Scholar]
- 43.Nodwell, J. R., K. McGovern, and R. Losick. 1996. An oligopeptide permease responsible for the import of an extracellular signal governing aerial mycelium formation in Streptomyces coelicolor. Mol. Microbiol. 22:881–893. [DOI] [PubMed] [Google Scholar]
- 44.Oh, S. H., and K. F. Chater. 1997. Denaturation of circular or linear DNA facilitates targeted integrative transformation of Streptomyces coelicolor A3(2): possible relevance to other organisms. J. Bacteriol. 179:122–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohnishi, Y., S. Kameyama, H. Onaka, and S. Horinouchi. 1999. The A-factor regulatory cascade leading to streptomycin biosynthesis in Streptomyces griseus: identification of a target gene of the A-factor receptor. Mol. Microbiol. 34:102–111. [DOI] [PubMed] [Google Scholar]
- 46.Peng, W.-T., Y.-W. Lee, and E. W. Nester. 1998. The phenolic recognition profiles of the Agrobacterium tumefaciens VirA protein are broadened by a high level of the sugar binding protein ChvE. J. Bacteriol. 180:5632–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perego, M., C. F. Higgins, S. R. Pearce, M. P. Gallagher, and J. A. Hoch. 1991. The oligopeptide transport system of Bacillus subtilis plays a role in the initiation of sporulation. Mol. Microbiol. 5:173–185. [DOI] [PubMed] [Google Scholar]
- 48.Quail, M. A., C. E. Dempsey, and J. R. Guest. 1994. Identification of a fatty acyl responsive regulator (FarR) in Escherichia coli. FEBS Lett. 356:183–187. [DOI] [PubMed] [Google Scholar]
- 49.Richarme, G., and T. D. Caldas. 1997. Chaperone properties of the bacterial periplasmic substrate-binding proteins. J. Biol. Chem. 272:15607–15612. [DOI] [PubMed] [Google Scholar]
- 50.Rudner, D. Z., J. R. Ladeaux, K. Breton, and A. D. Grossman. 1991. The spo0K locus of Bacillus subtilis is homologous to the oligopermease locus and is required for sporulation and competence. J. Bacteriol. 173:1388–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saurin, W., W. Köster, and E. Dassa. 1994. Bacterial binding protein-dependent permeases: characterization of distinctive signatures for functionally related integral cytoplasmic membrane proteins. Mol. Microbiol. 12:993–1004. [DOI] [PubMed] [Google Scholar]
- 53.Schlösser, A., J. Jantos, K. Hackmann, and H. Schrempf. 1999. Characterization of the binding protein-dependent cellobiose and cellotriose transport system of the cellulose degrader Streptomyces reticuli. Appl. Environ. Microbiol. 65:2636–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schlösser, A., T. Kampers, and H. Schrempf. 1997. The Streptomyces ATP-binding components MsiK assists in cellobiose and maltose transport. J. Bacteriol. 179:2092–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schneider, E., and S. Hunke. 1998. ATP-binding-cassette (ABC) transport systems: functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol. Rev. 22:1–20. [DOI] [PubMed] [Google Scholar]
- 56.Shinkawa, H., Y. Hatada, M. Okada, H. Kinashi, and O. Nimi. 1995. Nucleotide sequence of a principal sigma factor gene (hrdB) of Streptomyces griseus. J. Biochem. 118:494–499. [DOI] [PubMed] [Google Scholar]
- 57.Strohl, W. R. 1992. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 20:961–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sutcliffe, I. C., and R. R. B. Russell. 1995. Lipoproteins of gram-positive bacteria. J. Bacteriol. 177:1123–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takamatsu, S., H. Kunoh, and H. Ishizaki. 1976. Scanning electron microscopy observations on the perithecia of several powdery mildew fungi. I. Erysiphe and Sphaerotheca. Trans. Mycol. Soc. Jpn. 17:409–417. [Google Scholar]
- 60.Tam, R., and M. H. Saier, Jr. 1993. Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol. Rev. 57:320–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ueda, K., K. Miyake, S. Horinouchi, and T. Beppu. 1993. A gene cluster involved in aerial mycelium formation in Streptomyces griseus encodes proteins similar to the response regulators of two-component regulatory systems and membrane translocators. J. Bacteriol. 175:2006–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Wezel, G. P., J. van der Meulen, S. Kawamoto, R. G. M. Luiten, H. K. Koerten, and B. Kraal. 2000. ssgA is essential for sporulation of Streptomyces coelicolor A3(2) and affects hyphal development by stimulating septum formation. J. Bacteriol. 182:5653–5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Wezel, G. P., J. White, M. J. Bibb, and P. W. Postma. 1997. The malEFG gene cluster of Streptomyces coelicolor A3(2): characterization, distribution and transcriptional analysis. Mol. Gen. Genet. 254:604–608. [DOI] [PubMed] [Google Scholar]
- 64.van Wezel, G. P., J. White, P. Young, P. W. Postma, and M. J. Bibb. 1997. Substrate induction and glucose repression of maltose utilization by Streptomyces coelicolor A3(2) is controlled by malR, a member of the lacI-galR family of regulatory genes. Mol. Microbiol. 23:537–549. [DOI] [PubMed] [Google Scholar]
- 65.von Heijne, G. 1985. Signal sequence: the limits of variation. J. Mol. Biol. 184:99–105. [DOI] [PubMed] [Google Scholar]
- 66.Ward, J. M., G. R. Janssen, T. Kieser, M. J. Bibb, M. J. Buttner, and M. J. Bibb. 1986. Construction and characterization of a series of multicopy plasmid vectors for Streptomyces using the aminoglycoside phosphotransferase gene from Tn5 as indicator. Mol. Gen. Genet. 203:468–478. [DOI] [PubMed] [Google Scholar]
- 67.Wildermuth, H. 1970. Development and organization of the aerial mycelium in Streptomyces coelicolor. J. Gen. Microbiol. 60:43–50. [DOI] [PubMed] [Google Scholar]
- 68.Wildermuth, H., and D. A. Hopwood. 1970. Septation during sporulation in Streptomyces coelicolor. J. Gen. Microbiol. 60:51–59. [DOI] [PubMed] [Google Scholar]
- 69.Winans, S. C. 1992. Two-way chemical signaling in Agrobacterium-plant interactions. Microbiol. Rev. 56:12–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yamazaki, H., Y. Ohnishi, and S. Horinouchi. 2000. An A-factor-dependent extracytoplasmic function sigma factor (ςAdsA) that is essential for morphological development in Streptomyces griseus. J. Bacteriol. 182:4596–4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119. [DOI] [PubMed] [Google Scholar]