Abstract
Sulfur metabolism in gram-positive bacteria is poorly characterized. Information on the molecular mechanisms of regulation of genes involved in sulfur metabolism is limited, and no regulator genes have been identified. Here we describe the regulation of the lactococcal metC-cysK operon, encoding a cystathionine β-lyase (metC) and cysteine synthase (cysK). Its expression was shown to be negatively affected by high concentrations of cysteine, methionine, and glutathione in the culture medium, while sulfur limitation resulted in a high level of expression. Other sulfur sources tested showed no significant effect on metC-cysK gene expression. In addition we found that O-acetyl-l-serine, the substrate of cysteine synthase, was an inducer of the metC-cysK operon. Using a random mutagenesis approach, we identified two genes, cmbR and cmbT, involved in regulation of metC-cysK expression. The cmbT gene is predicted to encode a transport protein, but its precise role in regulation remains unclear. Disruption of cmbT resulted in a two- to threefold reduction of metC-cysK transcription. A 5.7-kb region containing the cmbR gene was cloned and sequenced. The encoded CmbR protein is homologous to the LysR family of regulator proteins and is an activator of the metC-cysK operon. In analogy to CysB from Escherichia coli, we propose that CmbR requires acetylserine to be able to bind the activation sites and subsequently activate transcription of the metC-cysK operon.
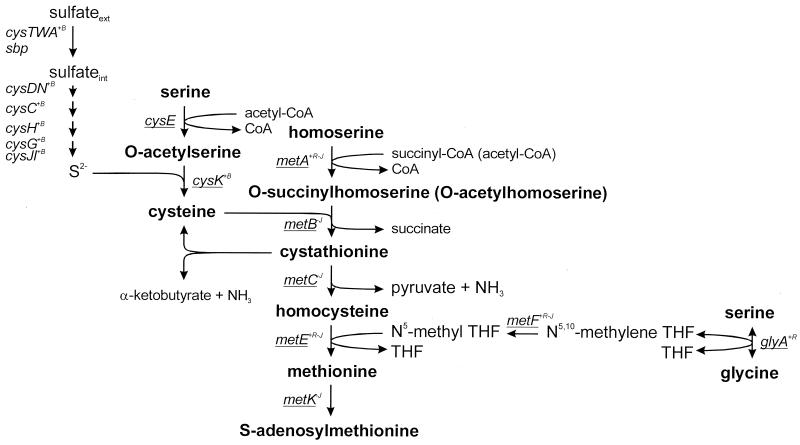
In many bacteria, cysteine biosynthesis is the predominant mechanism by which inorganic sulfur is reduced and incorporated into organic compounds. The pathway involves transport and reduction of inorganic sulfate to sulfide in one branch and the synthesis of O-acetyl-l-serine (OAS) in another. The subsequent reaction of sulfide with OAS results in cysteine synthesis (Fig. 1). For Escherichia coli and Salmonella enterica serovar Typhimurium, at least 22 genes have been identified that are required for the transport and reduction of sulfate and its incorporation into cysteine (21). Full expression of these genes requires a positively regulatory protein encoded by cysB, the presence of the inducer N-acetyl-l-serine (NAS), and sulfur limitation (21, 35).
FIG. 1.
Cysteine and methionine biosynthesis pathways in E. coli and S. enterica serovar Typhimurium and the responsible genes. Activation by CysB (+B) or MetR (+R) and repression by MetJ (−) are indicated in superscript. The genes from this pathway with homologues in the L. lactis IL1403 genome sequence are underlined. The conversion of cystathionine to cysteine is not described for E. coli. CBL from L. lactis is able to catalyze this reaction in vitro (1).
Methionine biosynthesis is closely linked to cysteine biosynthesis (Fig. 1). For E. coli and S. enterica serovar Typhimurium, transcription of all the met genes involved in methionine biosynthesis, except metH, is under negative control of the MetJ repressor, with S-adenosylmethionine, the pathway’s end product, acting as a corepressor (39). Expression of the metE, metA, metF, metH, and glyA genes is also under positive control of the MetR activator (9, 27, 30, 31, 46, 51). Homocysteine may modulate the regulator role of MetR and is required for the metE gene activation (47). Furthermore, vitamin B12 is involved in metE repression, probably by depletion of the coactivator homocysteine (53).
CysB and MetR are members of the LysR family of prokaryotic transcriptional regulatory proteins. Common family features are the size (between 300 and 350 amino acids), the formation of either homodimers or homotetramers, the presence of a helix-turn-helix DNA binding motif in the N-terminal region, and the requirement for a small molecule that acts as a coinducer (41). Both CysB and MetR activate gene expression at one or more loci while negatively regulating the expression of their own genes. The interactions of CysB with responsive promoter regions are well characterized. CysB binds as a tetramer, bending the DNA, and interaction with the inducer results in a conformational change of CysB, allowing it to interact with activation sites of the cysJ, cysK, or cysP promoters (19, 32). Several amino acid residues of CysB involved in DNA binding, response to the inducer, or oligomerization have been identified through mutagenesis (25).
Little is known about the organization and regulation of the sulfur assimilation and methionine biosynthesis pathway in gram-positive bacteria. In Bacillus subtilis, Clostridium acetobutylicum, and Staphylococcus aureus, several genes involved in the biosynthesis of methionine and cysteine are regulated by a global transcription termination control system called the S box regulon (17). Recently Mansilla et al. (29) showed that in B. subtilis, the cysH operon that contains an S box motif in the leader region is not regulated by a transcription terminator control system. The expression of this operon is controlled at the transcription initiation level by a repressor; the expression of the operon is induced by OAS and repressed by cysteine.
For Lactococcus lactis, a lactic acid bacterium commonly used as a starter in the dairy industry, the biosynthesis of methionine and cysteine has been poorly characterized. This microorganism has been described as prototrophic for cysteine and auxotrophic for methionine (8). Nevertheless, it was expected that the genes coding for the enzymes involved in methionine biosynthesis are present, since some autotrophic strains are known (8). This was corroborated by the analysis of the L. lactis IL1403 genome sequence (4). Homologues for all the genes involved in methionine biosynthesis in E. coli (metA, metB, metC, metE, metF, and glyA) are present together with cysE and two cysK homologues from the cysteine biosynthesis pathway (Fig. 1). In contrast, no homologues of the E. coli genes responsible for sulfate uptake and reduction seem to be present, although there is a putative sulfate transporter (yafB) of the SulP family (4).
Previously we characterized the metC gene encoding a cystathionine β-lyase (CBL) that has both β- and γ-lyase activity (1, 12). In vitro this CBL is able to convert cystathionine to cysteine or homocysteine (Fig. 1). The latter conversion is the penultimate step in methionine biosynthesis. The metC gene forms an operon together with cysK, encoding a cysteine synthase (12). The function of CysK was confirmed by its overproduction, resulting in increased cysteine synthase activity and the construction of a double-crossover cysK knockout in L. lactis MG1363 that resulted in auxotrophy for cysteine (R. van Kranenburg and M. Fernández, unpublished results). The alternative cysK gene seems to be inactive under these conditions. The organization of this metC-cysK gene cluster and its function in sulfur metabolism suggests a putative regulation by cysteine and/or methionine. Dias and Weimer (11) report that cystathionine lyase (CL) activity is influenced by the methionine and cysteine concentrations in the culture medium, confirming this hypothesis.
In this paper we demonstrate that methionine, cysteine, glutathione, and OAS have an effect on metC-cysK gene expression. We identify two genes, cmbT (cysteine and methionine biosynthesis transporter) and cmbR (cysteine and methionine biosynthesis regulator), that are involved in regulation of metC-cysK transcription. CmbR is a LysR-type regulator protein that is essential for metCcysK expression and is the first regulator of sulfur metabolism described for gram-positive bacteria.
MATERIALS AND METHODS
Bacteria, strains, plasmids, and media.
The bacterial strains and plasmids used in this work are listed in Table 1. E. coli was grown in Luria-Bertani broth at 37°C (40). L. lactis cells were routinely grown in M17 broth (Difco Laboratories) supplemented with 0.5% glucose (GM17) or in chemical defined medium (CDM) (26) at 30°C, unless stated otherwise. CDM contains 0.82 mM cysteine (1× cysteine) and 0.84 mM methionine (1× methionine). To test the effect of the addition of various sulfur compounds, the sulfate salts of the CDM were replaced by equimolar amounts of chloride or nitrate salts, cysteine was omitted, the methionine concentration was reduced to 0.1× methionine (0.084 mM), and the medium was supplemented with 1 mM glutathione, 1 mM Na2SO4, 1 mM Na2SO3, 0.5 mM Na2S2O3, or 0.5 mM l-djenkolic acid. The addition of methionine was required for normal growth of L. lactis. The effect of OAS was evaluated by addition of a 1 mM concentration of OAS when the cells reached a turbidity of 0.6 to 0.9 at 600 nm followed by a 2-h incubation. When appropriate, the media contained tetracycline (5 μg ml−1 for E. coli or 2.5 μg ml−1 for L. lactis), ampicillin (100 μg ml−1), chloramphenicol (5 μg ml−1), or erythromycin (5 μg ml−1).
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Relevant characteristic(s)a | Reference |
|---|---|---|
| Strains | ||
| E. coli MC1061 | 6 | |
| L. lactis | ||
| MG1363 | Plasmid-free | 16 |
| MG1363ΔmetC | Eryr derivative of MG1363 with a metC disruption by single crossover | 12 |
| MG1363::pNZ9342 | Eryr derivative of MG1363 carrying a single chromosomal copy of the lacZ gene under the control of PmetC-cysK | This study |
| MG1363::ISS1#1 | Eryr Tetr derivative of MG1363::pNZ9342 with pGh8::ISS1 integrated in cmbR | This study |
| MG1363::ISS1#2 | Eryr Tetr derivative of MG1363::pNZ9342 with pGh8::ISS1 integrated in cmbR | This study |
| MG1363::ISS1#3 | Eryr Tetr derivative of MG1363::pNZ9342 with pGh8::ISS1 integrated in cmbR | This study |
| MG1363cmbR | Eryr derivative of MG1363 with a cmbR disruption by single crossover | This study |
| MG1363cmbT | Eryr derivative of MG1363 with a cmbT disruption by single crossover | This study |
| Plasmids | ||
| pGh8:ISS1 | Tetr, 5.2-kb pWV01 thermosensitive replicon | 28 |
| pNZ124 | Cmr, 2.8-kb pSH71 replicon | 37 |
| pUC19NcoI | Ampr, 2.6-kb pUC19 derivative containing an additional fragment with an NcoI site overlapping an ATG start codon preceded by the L. lactis SK11 prtP ribosome-binding site | This study |
| pNZ9340 | Cmr, 5.2-kb pNZ273 derivative containing the gusA gene fused to PmetC-cysK on a 0.6-kb ScaI-EcoRI fragment | This study |
| pNZ9341 | Eryr, 7.1-kb pIL252 derivative carrying the gusA gene fused to PmetC-cysK on a 0.6-kb ScaI-EcoRI fragment | This study |
| pNZ9342 | Eryr, 7.2-kb pUC19NcoI derivative containing the lacZ gene fused to PmetC-cysK and the Eryr gene from pIL253 | This study |
| pNZ9343 | Tetr, 5.5-kb pGh8:ISS1 derivative carrying a 0.3-kb EcoRI chromosomal DNA fragment | This study |
| pNZ9344 | Tetr, 5.5-kb pGh8:ISS1 derivative carrying a 0.3-kb HindIII chromosomal DNA fragment | This study |
| pNZ9345 | Tetr, 5.7-kb pGh8:ISS1 derivative carrying a 0.5-kb HindIII chromosomal DNA fragment | This study |
| pNZ9346 | Ampr, 6.3-kb pUC18 derivative carrying a 3.6-kb SalI-EcoRI fragment with cmbR | This study |
| pNZ9347 | Ampr, 9.5-kb pUC18 derivative carrying a 6.8-kb SstI-SalI fragment with cmbR | This study |
| pNZ9348 | Eryr, 4.0-kb pUC18Ery derivative carrying a 0.3-kb HindIII-AccI fragment of cmbR gene from pNZ9344 | This study |
| pNZ9349 | Eryr, 4.6-kb pUC18Ery derivative carrying a 0.8-kb PCR fragment of cmbT | This study |
| pNZ9350 | Cmr, 4.6-kb pNZ124 derivative carrying a 1.8-kb EcoRI-PstI fragment from with cmbR | This study |
Ampr, ampicillin resistant; Eryr, erythromycin resistant; Tetr, tetracycline resistant; Cmr, chloramphenicol resistant.
DNA isolation and manipulation.
E. coli MC1061 was used as an intermediate host for cloning (40). Isolation of E. coli plasmid DNA and standard recombinant techniques were performed as described by Sambrook et al. (40). Large-scale isolation of E. coli plasmid DNA for nucleotide sequence analysis was performed using the Jet Star system (Genomed GmbH, Bad Deynhausen, Germany), according to the instructions of the manufacturer. Isolation and transformation of L. lactis plasmid DNA were performed as described previously (10). Southern blots were hybridized with the different probes at 65°C and washed with 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at 65°C before autoradiography.
Construction of strain MG1363::pNZ9342.
To obtain a transcriptional fusion of the metC-cysK promoter (PmetC-cysK) with lacZ, the promoter region, including the 5′ part of the metC gene, was cloned as a 0.6-kb EcoRI-ScaI fragment in the plasmid pUC19NcoI that was digested with NdeI made blunt with Klenow polymerase and digested with EcoRI. The plasmid pUC19NcoI is pUC19 digested with EcoRI and SmaI ligated with a nucleotide linker made with the oligonucleotides 5′-AATTCAGGAGGATATTCCATG-3′ and 5′-CATGGAATATCCTCCTG-3′. The linker contains the upstream sequence of the lactococcal prtP gene, including the ribosome binding site and prtP start codon (boldface), and generates after ligation an NcoI site overlapping the start codon (underlined) that can be used to make translational fusions. The lacZ gene from Streptococcus thermophilus CNRZ 302 was generated by PCR using the primers 5′-CCTTCAAAAAAGGAGAATAATCCATGGACATG-3′ and 5′-ATCGGATCCTAATTTAGTGGTTCAATCATGAAGCTTAATTATAGCTATCTGCTGAGC-3′, introducing an NcoI site overlapping the start codon (underlined) and a BamHI site for cloning (underlined). The PCR fragment was cloned as a 3.0-kb NcoI-BamHI fragment in pUC19NcoI containing PmetC-cysK. The erythromycin resistance gene from pUC18Ery (49) was cloned as a BamHI-PstI fragment in the resulting plasmid, generating the plasmid pNZ9342. This plasmid cannot replicate in L. lactis and was integrated into the chromosome by single-crossover recombination on the PmetC-cysK region. PCR analysis and Southern blotting were used to confirm the site of integration.
Construction of plasmids.
To study the expression of the metC-cysK operon, the gusA gene was cloned under control of PmetC-cysK in a low-copy-number plasmid. First, the PmetC-cysK region was cloned upstream of the promoterless gusA gene as a 0.6-kb EcoRI-ScaI fragment in pNZ273 (37). From the resulting plasmid (pNZ9340), a 2.5-kb BamHI-XhoI DNA fragment containing PmetC-cysK-gusA was cloned into the low-copy-number plasmid pIL252 (44), generating the plasmid pNZ9341.
For the construction of a cmbR disruption strain, an internal fragment of the cmbR gene was isolated as a 0.3-kb HindIII-AccI fragment from the plasmid pNZ9344 in which the HindIII site was made blunt by Klenow polymerase. This fragment was cloned into the plasmid pUC18Ery (49) digested with EcoRI-AccI, with the EcoRI site blunted by Klenow polymerase, generating the plasmid pNZ9348.
To complement the cmbR-knockout strain, the cmbR gene was cloned as an EcoRI-PstI fragment from pNZ9346 into the plasmid pNZ124, resulting in the plasmid pNZ9350.
For the construction of a cmbT disruption strain, an internal fragment of the cmbT gene was generated by PCR using the primers 5′-GGCGCGGAATTCTTGCGATTACGATTCCTCTCTC-3′ and 5′-CGCGGCGGATCCGTAATCGGCTTTCTTGCCCC-3′, introducing EcoRI and BamHI sites (underlined) for cloning. The PCR fragment was cloned as a 0.8-kb EcoRI-BamHI fragment in pUC18Ery (49), generating the plasmid pNZ9349.
Nucleotide sequencing and analysis.
Automatic double-stranded DNA sequence analysis was performed on both strands with an ALFred DNA sequencer (Amersham Pharmacia Biotech). Sequencing reactions were accomplished by using the AutoRead sequencing kit, initiated by using Cy5-labeled universal and reverse primers and continued with synthetic primers in combination with Cy5-13-dATP according to the instructions of the manufacturer (Amersham Pharmacia Biotech). Sequence data were assembled and analyzed using the pc/gene program, version 6.70 (IntelliGenetics). The protein sequences were compared with the swiss-prot all library (6 June 2001) using the Fasta3 WWW service (version 3.3t09 [18 May 2001]) at the European Bioinformatics Institute (36).
RNA isolation and Northern blotting.
Total RNA was isolated from exponentially growing MG1363 cultures by the Macaloid method described by Kuipers et al. (24). For Northern blot analysis, RNA was separated on a 1% formaldehyde agarose gel, blotted, and hybridized as described previously (50). The probes used for hybridization were radiolabeled with [α-32P]dATP by nick translation. The blots were washed with 0.1× SSC at 65°C prior to autoradiography.
Enzymatic assays.
For the different enzymatic assays, the cells were grown in M17 or CDM either with different methionine and cysteine concentrations or with different sulfur sources. CL activity was monitored using cystathionine as a substrate. Enzyme activity was measured in the cell extracts (CFE) by determination of free-thiol group formation with DTNB (5,5′-dithiobis 2-nitrobenzoic acid) as described by Uren (48). Pyridoxal-5′-phosphate was added to the reaction mixture at a final concentration of 20 μM. The data in the figures are from single experiments. The variation of specific activities was less than 10% between different experiments. The relative activities of the different conditions were always alike.
Cysteine synthase activity was measured by the method of Becker et al. (3). The amount of cysteine formed was determined as described by Gaitonde (15).
β-Glucuronidase (GUS) assays were performed with para-nitrophenyl-β-d-glucuronic acid as a substrate, monitoring the optical density at 412 nm as described by Platteeuw et al. (37).
Protein concentrations were determined by a protein assay (Bio-Rad) based on the method of Bradford, with bovine serum albumin as the standard (5).
A qualitative β-galactosidase plate assay was used for blue/white screening of L. lactis colonies by including 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside at a final concentration of 0.5 mM in GM17 plates.
Nucleotide sequence accession number.
The nucleotide sequence reported in this study was submitted to GenBank under the accession no. AF240531.
RESULTS
Regulation of gene expression of the metC-cysK operon.
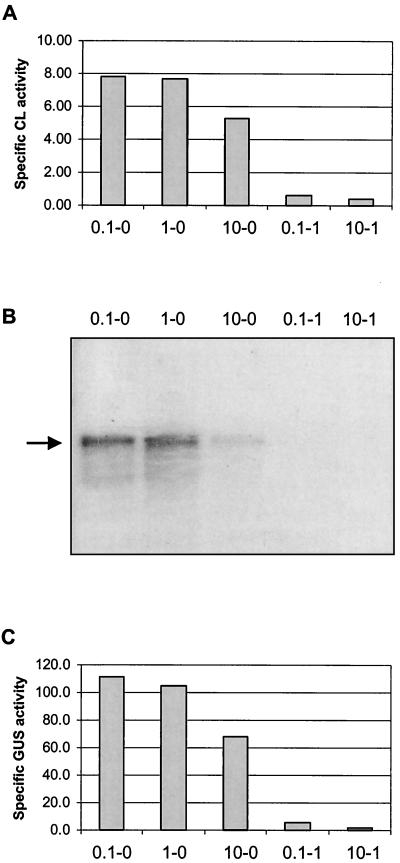
The genes encoding CBL (metC) and cysteine synthase (cysK) are involved in methionine and cysteine biosynthesis pathways and form a single transcriptional unit (12). CL activity in L. lactis has been reported to be affected by the concentrations of cysteine and methionine in the culture medium (11). To test if this effect is caused by regulation of gene expression, both the CL activity and the expression level of the metC-cysK operon were determined using cells grown in CDM supplemented with different concentrations of methionine and cysteine. When the levels of methionine and cysteine were increased, the CL activity decreased (Fig. 2A). The effect of cysteine was more pronounced, as addition of 1× cysteine reduced the activity to that of a metC knockout strain, while addition of 10× methionine resulted in a reduction to two-thirds of the activity of cells grown in CDM supplemented with 0.1× methionine. The cysteine synthase activities were also determined, and these showed the same mode of regulation by methionine and cysteine (data not shown). Northern blot analysis revealed that the effects of cysteine and methionine are at the transcriptional level (Fig. 2B).
FIG. 2.
(A) Specific cystathionine lyase activity in CFE of L. lactis MG1363 cells grown in CDM with various methionine and cysteine concentrations. 0.1-0, 0.1× methionine, no cysteine; 1-0, 1× methionine, no cysteine; 10-0, 10× methionine, no cysteine; 0.1-1, 0.1× methionine, 1× cysteine; 10-1, 10× methionine, 1× cysteine. The specific activities are expressed as nanomoles of mercaptide formed per minute per milligram of protein. (B) Transcriptional analysis of the metC-cysK operon. Total RNA was extracted from L. lactis MG1363 cells grown in CDM with various methionine and cysteine concentrations (see panel A). The blot was hybridized with an internal gene fragment of MG1363 metC. The arrow indicates the position of the metC-cysK transcript. (C) Specific GUS activities in CFE of L. lactis MG1363 harboring pNZ9341 grown in CDM with various methionine and cysteine concentrations (see panel A). The specific activities are expressed as nanomoles of para-nitrophenyl-β-d-glucuronic acid converted per minute per milligram of protein.
To quantify the effects of cysteine and methionine on metCcysK transcription, a transcriptional fusion of PmetC-cysK with the gusA reporter gene was constructed on a low-copy-number plasmid. The resulting plasmid, pNZ9341, was introduced in MG1363, and the GUS activities of cells grown in CDM with different amounts of cysteine and methionine were determined. Addition of 1× cysteine to CDM resulted in a 19-fold reduction of GUS activity compared to that for CDM containing no cysteine (Fig. 2C). Cells grown in CDM supplemented with 10× methionine still retained 61% of the GUS activity of those grown in 0.1× methionine, confirming the results of the Northern blot analysis.
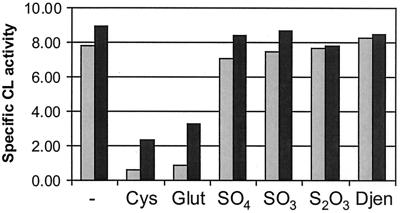
The influence of other sulfur sources on the CL activity was tested (Fig. 3). As described above, sulfur starvation (CDM with 0.1× methionine and no cysteine) resulted in a high level of CL activity. Addition of sulfate, sulfite, thiosulfate, and djenkolate to the medium resulted in a similarly high activity level. In contrast, cysteine and glutathionine almost completely abolished CL activity. Sulfide affected the growth rate of L. lactis dramatically and was therefore not included in the experiments.
FIG. 3.
Effects of sulfur sources and OAS on CL activity in L. lactis. Dark bars represent cells grown in the presence of OAS. Cys, cysteine; Glut, glutathione; SO4, sulfate; SO3, sulfite; S2O3, thiosulfate; Djen, djenkolic acid.
NAS (often provided as OAS) has been described as a coinducer of the cys regulon in E. coli and S. enterica serovar Typhimurium (22). To test if OAS has an effect on the regulation of the metC-cysK operon, CL activity was measured using CFEs from cells grown in a medium with or without addition of OAS in the early log phase (see Materials and Methods). An increase in the CL activity in the presence of OAS was observed for all media tested except for those containing thiosulfate and l-djenkolic acid (Fig. 3). Northern blot analysis of RNA from cells grown in these conditions revealed that the induction by OAS takes place at the transcriptional level (data not shown).
Identification of genes involved in regulation.
To test if the regulation of metC-cysK transcription results from activity of an activator or repressor, a high-copy-number plasmid (pNZ9340) carrying the promoter region was introduced into MG1363. Cells harboring pNZ9340 grown in GM17 showed a fivefold-lower CL activity than MG1363 without the plasmid. This is likely to be the result of a lower level of metC-cysK expression caused by the titration of a putative activator by the multiple copies of the promoter.
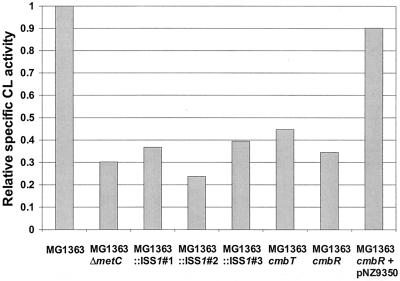
For the identification of the activator gene, a random mutagenesis strategy was followed using the thermosensitive plasmid pGh8:ISS1 (28). Rapid screening of the mutants lacking PmetC-cysK activity was facilitated by the use of the strain MG1363::pNZ9342, in which lacZ is under the control of PmetC-cysK (see Materials and Methods). Random integrants of pGh8:ISS1 in MG1363::pNZ9342 were analyzed on GM17 plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside. From approximately 22,000 colonies, only five showed a white phenotype. Southern blot analysis using lacZ as a probe revealed that in two of those, pGh8:ISS1 was integrated into lacZ, while in the other three, pGh8:ISS1 was inserted in another part of the chromosome. The latter three were designated MG1363::ISS1#1, MG1363::ISS1#2, and MG1363::ISS1#3. The CL activities of these mutants were tested and showed a clear decrease in the CL activity from that of the wild-type strain (Fig. 4).
FIG. 4.
Specific CL activities in CFE of L. lactis strains grown in GM17 relative to that of MG1363. The specific activities are expressed as nanomoles of mercaptide formed per minute per milligram of protein.
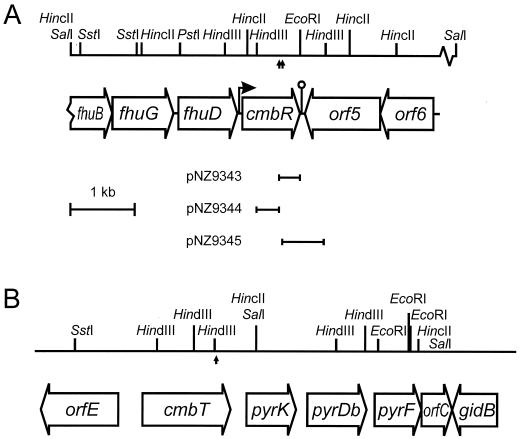
The chromosomal DNA regions flanking the pGh8:ISS1 insertion site were cloned using the unique EcoRI and HindIII sites adjacent to the ISS1 element as described by Maguin et al. (28). From strain MG1363::ISS1#1, both 0.3-kb flanking regions were cloned, while from MG1363::ISS1#2, only the 0.6-kb HindIII flanking region could be cloned (Fig. 5A). Nucleotide sequence analysis revealed that in the two mutants the integration took place in the same gene, which was designated cmbR (cysteine and methionine biosynthesis regulator). From strain MG1363::ISS1#3 a 1.9-kb HindIII flanking region was cloned, and nucleotide sequence analysis showed that pGh8:ISS1 had integrated into a 1,362-bp open reading frame (ORF) putatively encoding an efflux protein with homology to members of the 2.A.1.3.X drug/H+ antiporter-2 subfamily of the major facilitator superfamily (38) (Fig. 5B). This gene, located upstream of the pyrKDbF operon and previously reported as orfA (2), was designated cmbT (cysteine and methionine biosynthesis transporter). The pyrKDbF operon has a promoter upstream of pyrK (2), ruling out possible polar effects of the integration in cmbT.
FIG. 5.
Physical and genetic map of fragments of the L. lactis MG1363 chromosome containing cmbR (A) or cmbT (B) (2). For HindIII, only sites relevant for cloning are listed (A). The upward arrows indicate the sites of insertion of ISS1 in MG1363::ISS1#1, MG1363::ISS1#2, and MG1363::ISS1#3. The flanking regions cloned from MG1363::ISS1#1 (pNZ9343 and pNZ9344) and MG1363::ISS1#2 (pNZ9345) are indicated.
Cloning of the cmbR gene.
The 0.3-kb internal cmbR fragment from pNZ9344 was used as a probe to clone the complete MG1363 cmbR gene on a 3.6-kb SalI-EcoRI chromosomal DNA fragment (pNZ9346) and an overlapping 6.8-kb SstI-SalI fragment (pNZ9347). Sequence analysis of 5.7 kb of the cloned fragments revealed the presence of six ORFs (Fig. 5). The cmbR gene encodes a putative protein of 301 amino acids. It is preceded by a reasonable consensus ribosome binding site for L. lactis (AAGGA) (7) and a putative −35 and −10 region. An inverted repeat sequence, which could form a hairpin structure (ΔG = −18.1 kcal mol−1) and thus function as a terminator, was identified four nucleotides downstream of the stop codon. Northern blot analysis revealed that the cmbR gene is expressed as a 1-kb monocistronic transcript (data not shown).
The predicted CmbR protein is homologous to LysR-type regulators. It differs by only two amino acids from FhuR of L. lactis IL1403 (4), and it shares strongest homology with the gene product of spy0824 from Streptococcus pyogenes (52% identity) (13) and CpsY from Streptococcus agalactiae (30% identity) (20) and moderate homology with CysB from E. coli (24% identity) (34). The N-terminal region containing the DNA-binding domain is the most conserved.
The three ORFs upstream of cmbR encode proteins homologous to FhuB (31% identity), FhuG (38% identity), and FhuD (33% identity) of Bacillus subtilis, involved in ferrichrome transport (42). The ORF downstream of cmbR codes for a putative transmembrane protein with homology (51% identity with YtbD of B. subtilis) to members of the major facilitator superfamily (classified as 2.A.1.X.X) (38). ORF6 encodes a protein that is homologous (59% identity with YvgN from B. subtilis) to (putative) dehydrogenases and oxidoreductases.
cmbT is involved in regulation.
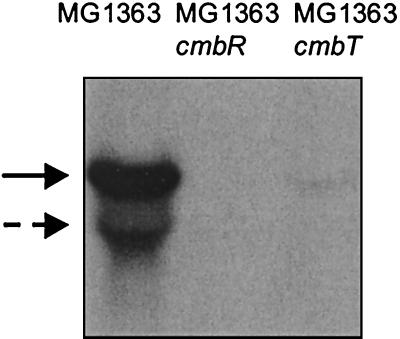
A directed knockout strain of cmbT was constructed to confirm its role in regulation. A 0.8-kb internal gene fragment was generated by PCR and cloned in pUC18Ery, generating the plasmid pNZ9349. This plasmid, which is unable to replicate in L. lactis, was introduced into strain MG1363. Candidate single-crossover mutants in the cmbT locus were isolated as erythromycin-resistant colonies. Plasmid integration was confirmed by Southern blot and PCR analyses. One colony was used for further analysis and designated MG1363cmbT. The CL activity in this strain was assayed and compared to that of MG1363. For MG1363cmbT, the decrease in CL activity was comparable to that of MG1363::ISS1#3 (Fig. 4). Furthermore, MG1363cmbT is autotrophic for cysteine, as it is able to grow on CDM without cysteine, indicating that cysK expression, although reduced, is sufficient to sustain growth. Northern blot analysis showed that disruption of cmbT has an effect on the transcription of the metC-cysK operon (Fig. 6). These results confirm that cmbT is involved in regulation of metC-cysK transcription.
FIG. 6.
Northern blot hybridization of RNA isolated from MG1363 (lane 1), MG1363cmbR (lane 2), and MG1363cmbT (lane 3), hybridized with an internal gene fragment of MG1363 metC. The cells were grown in GM17. The solid arrow indicates the position of the metCcysK transcript, and the dashed arrow indicates that of metC alone.
cmbR is essential for regulation.
With an approach similar to that for cmbT, a directed knockout strain of cmbR was constructed to confirm its role in regulation. For this purpose, a 0.3-kb HindIII-AccI fragment of the plasmid pNZ9344 was cloned into pUC18Ery, generating the plasmid pNZ9348, which was introduced into the strain MG1363. Plasmid integration of the erythromycin-resistant single-crossover mutants was confirmed by Southern blot and PCR analyses. One colony was selected for further analyses and designated MG1363cmbR. The CL activity in this strain was assayed and compared to that of MG1363. A dramatic decrease in the activity was observed for MG1363cmbR, similar to those of MG1363::ISS1#1 and MG1363::ISS1#2 (Fig. 4). The cysteine synthase activity was also affected, since the knockout strain is auxotrophic for cysteine. Northern blot analysis showed that expression of the metCcysK operon is completely abolished in MG1363cmbR (Fig. 6). Complementation of MG1363cmbR was achieved by introducing the plasmid pNZ9350 carrying the complete cmbR gene under the control of its own promoter. The complementation strain recovered the ability to grow in a medium without cysteine, and the CL activity was comparable to that in the wild-type strain (Fig. 4). These results prove that cmbR is involved in activation of the metC-cysK gene expression.
To test if OAS is an inducer of CmbR, we measured CL activities of MG1363, MG1363cmbT, and MG1363cmbR grown in GM17 medium with and without OAS. CL activity increased 4.5-fold for MG1363 upon addition of OAS and 4-fold for MG1363cmbT. In contrast, MG1363cmbR showed equal low CL activities when grown in medium with or without addition of OAS, indicating that OAS requires intact CmbR to activate CL activity.
DISCUSSION
We studied the regulation of the expression of the metC-cysK operon, encoding CBL and cysteine synthase (12). Previously Dias and Weimer (11) showed that the CL activity in L. lactis is influenced by the concentrations of methionine and cysteine in the culture medium. CBL is expected to be responsible for the majority of this CL activity, since disruption of metC results in a major reduction of CL activity (12). Furthermore, the organization of metC and cysK, involved in methionine and cysteine biosynthesis, respectively, in a single operon suggests a coordinated regulation of at least parts of both biosynthetic pathways. Our results demonstrate that cysteine and, to a lesser extent, methionine affect both CL and cysteine synthase activities and that this is caused by regulation of expression of the metC-cysK operon (Fig. 2). The effect of addition of other sulfur sources to medium containing no cysteine and a small amount of methionine was also evaluated. Addition of glutathione had an effect similar to addition of cysteine and reduced CL activity and metC-cysK expression. In contrast, addition of sulfate, sulfite, thiosulfate, or djenkolic acid had no clear effect on CL activity (Fig. 3). For E. coli, maximal expression of the cys genes is seen during growth with limiting sulfur sources, such as glutathione or djenkolic acid, while sulfate, sulfite, and thiosulfate partially reduce expression of the cys genes, and sulfide, cysteine, and cystine fully repress them (21, 23). For B. subtilis, expression of the cysH operon, encoding part of the pathway for transport and reduction of inorganic sulfate to sulfide, is repressed by cysteine, methionine, and thiosulfate and induced by glutathione (29). Our results demonstrate that for L. lactis, sulfur limitation results in maximum expression of the metC-cysK operon, similar to the cys regulon in E. coli and S. enterica serovar Typhimurium and the cysH operon in B. subtilis, but that the response to sulfate, sulfite, thiosulfate, and glutathione is different. An important difference with E. coli or B. subtilis is that L. lactis seems to lack the genes required for the sulfate reduction pathway (4). Although it contains a putative sulfate transporter (4), it is not likely to be able to use sulfate or sulfite as sulfur sources.
The effect of NAS as a coinducer for the cys regulon is well established with E. coli and S. enterica serovar Typhimurium (22). In these organisms, sulfide and thiosulfate counteract the effect of NAS on transcription. Recently, Mansilla et al. (29) showed that OAS also has a positive effect on transcription of the cysH operon in B. subtilis but that this is independent of sulfur starvation and insensitive to cysteine repression. In L. lactis, OAS induces expression of metC-cysK for all sulfur sources tested except for thiosulfate and djenkolic acid (Fig. 3). The positive effect of OAS is dependent on the sulfur source and is highest under sulfur starvation, resembling more the regulation of the cys regulon in E. coli than that of the cysH operon in B. subtilis.
CmbR is essential for expression of the metCcysK operon. It is a member of the family of LysR-type regulators that includes CysB and MetR, and it activates metCcysK transcription. Since addition of OAS also activates transcription, we assume that CmbR, like CysB, requires NAS as an inducer to allow binding to the activation site. This is corroborated by the lack of effect of addition of OAS on CL activity of MG1363cmbR. The activator binding sequences described for the cys or met promoters in E. coli and S. enterica serovar Typhimurium (18, 52) were not found in the metCcysK promoter. Nevertheless, the PmetC-cysK region contains three inverted repeat sequences and two direct repeats that are located upstream of the −35 region. A role of these repeat sequences in regulation has yet to be established. In our current studies we focus on the interaction of CmbR with the responsive promoter elements and try to define the CmbR binding sites.
The role of CmbT remains unclear. MG1363cmbT still responds to OAS, and we believe that the effect of CmbT is probably indirect, since it is predicted to be a transport protein. In that case the knockout strain would be unable to transport certain compounds, which has a negative effect on the activation of metC-cysK expression. These may be compounds that have an effect on the intracellular level of acetylserine, thereby affecting CmbR-dependent activation of metC-cysK transcription.
Knowledge of regulation of sulfur metabolism in gram-positive organisms is limited. The S box regulon described for B. subtilis, C. acetobutylicum, and S. aureus consists of putative transcriptional units, including metC but not cysK, containing a highly conserved motif in the leader region (17). For one of the members of the S box regulon, the methionine-regulated yitJ gene predicted to be involved in methionine biosynthesis in B. subtilis, it has been demonstrated that the mode of regulation is determined by the leader region rather than by the promoter activity (17). In contrast, regulation of the cysH operon, which is also part of the S box regulon, is independent of the leader region but depends on the promoter (29). A model for regulation of expression of this operon includes the activity of a putative negative regulator CysR, which prevents transcription in the absence of OAS (29). Although the lactococcal metC-cysK operon is not part of an S box regulon, there is an alternative combined regulation of the methionine and cysteine pathways in L. lactis, since this operon is dependent on CmbR, which is the first regulator of sulfur metabolism identified in gram-positive bacteria.
To test if CmbR is a regulator for other genes involved in sulfur metabolism, Northern blot analyses were performed with probes of the MG1363 metA gene (encoding an O-acetylhomoserine sulfhydrylase), metB gene (encoding a serine acetyltransferase; annotated as cysD for IL1403), cysE gene (encoding a serine acetyltransferase), glyA gene (encoding a serine hydroxymethyltransferase), and the fhuD gene of the adjacent fhu gene cluster. Our preliminary results indicate that transcription of metB, cysE, and glyA is not influenced by the concentrations of methionine and cysteine in the growth media nor is it dependent on cmbR. Although barely detectable, the level of transcription of metA, which in strain IL1403 forms an operon with metB encoding cystathionine γ-synthase (4), was lower for cells grown on CDM with high concentrations of cysteine and methionine than for those grown on CDM with a low concentration of methionine and no cysteine. Disruption of cmbR resulted in a similar reduction of metA transcription, which was equally low for cells grown on CDM containing low and high concentrations of methionine and cysteine. These preliminary results indicate that CmbR may be a regulator of the metAB operon. In addition, the results with fhuD as a probe indicate that CmbR is also involved in regulation of the fhu genes. In the L. lactis IL1403 genome sequence, CmbR is annotated as FhuR, the regulator of the fhu operon (4). The fhu genes are predicted to be involved in ferrichrome uptake, providing Fe3+. Both Fe3+ and S2− are required for Fe-S cluster synthesis, and in E. coli CBL and cysteine synthase are able to generate sulfide, enabling Fe-S cluster synthesis (14). Since lactococcal CBL is known to be able to use cysteine as a substrate, generating sulfide (1), it is tempting to speculate that the metC-cysK gene cluster may also have a role in Fe-S cluster synthesis, suggesting that CmbR is a regulator for this process. Fe-S clusters have been found in several lactococcal enzymes, including the transcription factor FlpA (43), the anaerobic ribonucleotide reductase activase (45), and the B form of dihydroorotate dehydrogenase formed by PyrDB and PyrK (33). Interestingly, the genes for the latter are genetically linked to cmbT, which is the other regulatory gene found in this study.
In conclusion, we have demonstrated that expression of the metC-cysK operon is activated by sulfur limitation. CmbT is involved in regulation, but its precise role remains obscure. CmbR is the activator that is essential for activation of metCcysK transcription and may act as a regulator of metAB and the fhu genes.
Acknowledgments
We thank Jeroen Hugenholtz and Igor Mierau for critically reading the manuscript.
María Fernández was supported by a scholarship of the Ministerio de Educación y Cultura of Spain. This work was partially supported by EU research grant FAIR CT97-3173.
REFERENCES
- 1.Alting, A. C., W. J. M. Engels, S. van Schalkwijk, and F. A. Exterkate. 1995. Purification and characterization of cystathionine β-lyase from Lactococcus lactis subsp. cremoris B78 and its possible role in flavor development in cheese. Appl. Environ. Microbiol. 61:4037–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen, P. S., J. Martinussen, and K. Hammer. 1996. Sequence analysis and identification of the pyrKDbF operon from Lactococcus lactis including a novel gene, pyrK, involved in pyrimidine biosynthesis. J. Bacteriol. 178:5005–5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker, M. A., N. M. Kredich, and G. M. Tomkins. 1969. The purification and characterization of O-acetylserine sulfhydrylase-A from Salmonella typhimurium. J. Biol. Chem. 244:2418–2427. [PubMed] [Google Scholar]
- 4.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254. [DOI] [PubMed] [Google Scholar]
- 6.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179–207. [DOI] [PubMed] [Google Scholar]
- 7.Chiaruttini, C., and M. Milet. 1993. Gene organization, primary structure and RNA processing analysis of a ribosomal RNA operon in Lactococcus lactis. J. Mol. Biol. 230:57–76. [DOI] [PubMed] [Google Scholar]
- 8.Chopin, A. 1993. Organization and regulation of genes for amino acid biosynthesis in lactic acid bacteria. FEMS Microbiol. Rev. 12:21–37. [DOI] [PubMed] [Google Scholar]
- 9.Cowan, J. M., M. L. Urbanowski, M. Talmi, and G. V. Stauffer. 1993. Regulation of the Salmonella typhimurium metF gene by the MetR protein. J. Bacteriol. 175:5862–5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Vos, W. M., P. Vos, H. de Haard, and I. Boerrigter. 1989. Cloning and expression of the Lactococcus lactis subsp. cremoris SK11 gene encoding an extracellular serine proteinase. Gene 85:169–176. [DOI] [PubMed] [Google Scholar]
- 11.Dias, B., and B. Weimer. 1998. Conversion of methionine to thiols by lactococci, lactobacilli, and brevibacteria. Appl. Environ. Microbiol. 64:3320–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandez, M., W. van Doesburg, G. A. Rutten, J. D. Marugg, A. C. Alting, R. van Kranenburg, and O. P. Kuipers. 2000. Molecular and functional analyses of the metC gene of Lactococcus lactis, encoding cystathionine β-lyase. Appl. Environ. Microbiol. 66:42–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658–4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flint, D. H., J. F. Tuminello, and T. J. Miller. 1996. Studies on the synthesis of the Fe-S cluster of dihydroxy-acid dehydratase in Escherichia coli crude extract. J. Biol. Chem. 271:16053–16067. [DOI] [PubMed] [Google Scholar]
- 15.Gaitonde, M. K. 1967. A spectrophotometric method for the direct determination of cysteine in the presence of other naturally occurring amino acids. Biochem. J. 104:627–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grundy, F. J., and T. M. Henkin. 1998. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol. Microbiol. 30:737–749. [DOI] [PubMed] [Google Scholar]
- 18.Hryniewicz, M. M., and N. M. Kredich. 1995. Hydroxyl radical footprints and half-site arrangements of binding sites for the CysB transcriptional activator of Salmonella typhimurium. J. Bacteriol. 177:2343–2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hryniewicz, M. M., and N. M. Kredich. 1994. Stoichiometry of binding of CysB to the cysJIH, cysK, and cysP promoter regions of Salmonella typhimurium. J. Bacteriol. 176:3673–3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koskiniemi, S., M. Sellin, and M. Norgren. 1998. Identification of two genes, cpsX and cpsY, with putative regulatory function on capsule expression in group B streptococci. FEMS Immunol. Med. Microbiol. 21:159–168. [DOI] [PubMed] [Google Scholar]
- 21.Kredich, N. M. 1996. Biosynthesis of cysteine, p. 514–527. In F. C. Neidhart, R. Curtiss, J. L. Ingrahan, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington D.C.
- 22.Kredich, N. M. 1992. The molecular basis for positive regulation of cys promoters in Salmonella typhimurium and Escherichia coli. Mol. Microbiol. 6:2747–2753. [DOI] [PubMed] [Google Scholar]
- 23.Kredich, N. M. 1971. Regulation of L-cysteine biosynthesis in Salmonella typhimurium. J. Biol. Chem. 246:3474–3484. [PubMed] [Google Scholar]
- 24.Kuipers, O. P., M. M. Beerthuyzen, R. J. Siezen, and W. M. De Vos. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216:281–291. [DOI] [PubMed] [Google Scholar]
- 25.Lochowska, A., R. Iwanicka-Nowicka, D. Plochocka, and M. M. Hryniewicz. 2000. Functional dissection of the LysR-type CysB transcriptional regulator: regions important for DNA binding, inducer response, oligomerization and positive control. J. Biol. Chem. 276:2098–2107. [DOI] [PubMed] [Google Scholar]
- 26.Looijesteijn, P. J., and J. Hugenholtz. 1999. Uncoupling of growth and exopolysaccharide production by Lactococcus lactis subsp. cremoris NIZO B40 and optimisation of its synthesis. J. Biosci. Bioeng. 88:178–182. [DOI] [PubMed] [Google Scholar]
- 27.Lorenz, E., and G. V. Stauffer. 1996. MetR-mediated repression of the glyA gene in Escherichia coli. FEMS Microbiol. Lett. 144:229–233. [DOI] [PubMed] [Google Scholar]
- 28.Maguin, E., H. Prevost, S. D. Ehrlich, and A. Gruss. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178:931–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mansilla, M. C., D. Albanesi, and D. de Mendoza. 2000. Transcriptional control of the sulfur-regulated cysH operon, containing genes involved in l-cysteine biosynthesis in Bacillus subtilis. J. Bacteriol. 182:5885–5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mares, R., M. L. Urbanowski, and G. V. Stauffer. 1992. Regulation of the Salmonella typhimurium metA gene by the metR protein and homocysteine. J. Bacteriol. 174:390–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maxon, M. E., B. Redfield, X. Y. Cai, R. Shoeman, K. Fujita, W. Fisher, G. Stauffer, H. Weissbach, and N. Brot. 1989. Regulation of methionine synthesis in Escherichia coli: effect of the MetR protein on the expression of the metE and metR genes. Proc. Natl. Acad. Sci. USA 86:85–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monroe, R. S., J. Ostrowski, M. M. Hryniewicz, and N. M. Kredich. 1990. In vitro interactions of CysB protein with the cysK and cysJIH promoter regions of Salmonella typhimurium. J. Bacteriol. 172:6919–6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nielsen, F. S., P. S. Andersen, and K. F. Jensen. 1996. The B form of dihydroorotate dehydrogenase from Lactococcus lactis consists of two different subunits, encoded by the pyrDb and pyrK genes, and contains FMN, FAD, and [FeS] redox centers. J. Biol. Chem. 271:29359–29365. [DOI] [PubMed] [Google Scholar]
- 34.Ostrowski, J., G. Jagura-Burdzy, and N. M. Kredich. 1987. DNA sequences of the cysB regions of Salmonella typhimurium and Escherichia coli. J. Biol. Chem. 262:5999–6005. [PubMed] [Google Scholar]
- 35.Ostrowski, J., and N. M. Kredich. 1989. Molecular characterization of the cysJIH promoters of Salmonella typhimurium and Escherichia coli: regulation by CysB protein and N-acetyl-l-serine. J. Bacteriol. 171:130–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Platteeuw, C., G. Simons, and W. M. de Vos. 1993. Use of the Escherichia coli β-glucuronidase (gusA) gene as a reporter for analyzing of promoters in lactic acid bacteria. Appl. Environ. Microbiol. 60:587–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saier, M. H., Jr. 2000. Families of transmembrane sugar transport proteins. Mol. Microbiol. 35:699–710. [DOI] [PubMed] [Google Scholar]
- 39.Saint-Girons, I., C. Parsot, M. M. Zakin, O. Barzu, and G. N. Cohen. 1988. Methionine biosynthesis in Enterobacteriaceae: biochemical, regulatory, and evolutionary aspects. Crit. Rev. Biochem. 23:S1–S42. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 41.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597–626. [DOI] [PubMed] [Google Scholar]
- 42.Schneider, R., and K. Hantke. 1993. Iron-hydroxamate uptake systems in Bacillus subtilis: identification of a lipoprotein as part of a binding protein-dependent transport system. Mol. Microbiol. 8:111–121. [DOI] [PubMed] [Google Scholar]
- 43.Scott, C., J. R. Guest, and J. Green. 2000. Characterization of the Lactococcus lactis transcription factor FlpA and demonstration of an in vitro switch. Mol. Microbiol. 35:1383–1393. [DOI] [PubMed] [Google Scholar]
- 44.Simon, D., and A. Chopin. 1988. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie 70:559–566. [DOI] [PubMed] [Google Scholar]
- 45.Torrents, E., G. Buist, A. Liu, R. Eliasson, J. Kok, I. Gibert, A. Graslund, and P. Reichard. 2000. The anaerobic (class III) ribonucleotide reductase from Lactococcus lactis. J. Biol. Chem. 275:2463–2471. [DOI] [PubMed] [Google Scholar]
- 46.Urbanowski, M. L., and G. V. Stauffer. 1989. Genetic and biochemical analysis of the MetR activator-binding site in the metE metR control region of Salmonella typhimurium. J. Bacteriol. 171:5620–5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Urbanowski, M. L., and G. V. Stauffer. 1987. Regulation of the metR gene of Salmonella typhimurium. J. Bacteriol. 169:5841–5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uren, J. R. 1987. Cystathionine β-lyase from Escherichia coli. Methods Enzymol. 143:483–486. [DOI] [PubMed] [Google Scholar]
- 49.van Kranenburg, R., J. D. Marugg, I. I. van Swam, N. J. Willem, and W. M. de Vos. 1997. Molecular characterization of the plasmid-encoded eps gene cluster essential for exopolysaccharide biosynthesis in Lactococcus lactis. Mol. Microbiol. 24:387–397. [DOI] [PubMed] [Google Scholar]
- 50.van Rooijen, R. J., and W. M. de Vos. 1990. Molecular cloning, transcriptional analysis, and nucleotide sequence of lacR, a gene encoding the repressor of the lactose phosphotransferase system of Lactococcus lactis. J. Biol. Chem. 265:18499–18503. [PubMed] [Google Scholar]
- 51.Weissbach, H., and N. Brot. 1991. Regulation of methionine synthesis in Escherichia coli. Mol. Microbiol. 5:1593–1597. [DOI] [PubMed] [Google Scholar]
- 52.Wu, W. F., M. L. Urbanowski, and G. V. Stauffer. 1995. Characterization of a second MetR-binding site in the metE metR regulatory region of Salmonella typhimurium. J. Bacteriol. 177:1834–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu, W. F., M. L. Urbanowski, and G. V. Stauffer. 1992. Role of the MetR regulatory system in vitamin B12-mediated repression of the Salmonella typhimurium metE gene. J. Bacteriol. 174:4833–4837 [DOI] [PMC free article] [PubMed] [Google Scholar]