Abstract
Polyhydroxyalkanoates (PHAs) are polyoxoesters that are produced by many bacteria and that accumulate as intracellular granules. Phasins (PhaP) are proteins that accumulate during PHA synthesis, bind PHA granules, and promote further PHA synthesis. Interestingly, PhaP accumulation seems to be strictly dependent on PHA synthesis, which is catalyzed by the PhaC PHA synthase. Here we have tested the effect of the Ralstonia eutropha PhaR protein on the regulation of PhaP accumulation. R. eutropha strains with phaR, phaC, and/or phaP deletions were constructed, and PhaP accumulation was measured by immunoblotting. The wild-type strain accumulated PhaP in a manner dependent on PHA production, and the phaC deletion strain accumulated no PhaP, as expected. In contrast, both the phaR and the phaR phaC deletion strains accumulated PhaP to higher levels than did the wild type. This result implies that PhaR is a negative regulator of PhaP accumulation and that PhaR specifically prevents PhaP from accumulating in cells that are not producing PHA. Transfer of the R. eutropha phaR, phaP, and PHA biosynthesis (phaCAB) genes into a heterologous system, Escherichia coli, was sufficient to reconstitute the PhaR/PhaP regulatory system, implying that PhaR both regulates PhaP accumulation and responds to PHA directly. Deletion of phaR caused a decrease in PHA yields, and a phaR phaP deletion strain exhibited a more severe PHA defect than a phaP deletion strain, implying that PhaR promotes PHA production and does this at least partially through a PhaP-independent pathway. Models for regulatory roles of PhaR in regulating PhaP and promoting PHA production are presented.
Polyhydroxyalkanoates (PHAs) are polyoxoesters that are produced by a wide range of bacteria as carbon storage compounds and that accumulate intracellularly as amorphous granules (2, 11, 21). Three classes of proteins, PHA synthase (PhaC), PHA depolymerase (PhaZ), and phasin (PhaP), play important roles in PHA metabolism. PHA synthases catalyze the conversion of R-3-hydroxyalkanoyl coenzyme A molecules to PHAs (4, 11, 16). PHA depolymerases hydrolyze PHAs to yield oligomers and/or monomers of hydroxyalkanoates (5, 7, 22). Phasins accumulate during PHA synthesis, bind PHA granules, and promote further PHA synthesis in a manner that is poorly understood (6, 13, 15, 17, 25, 27). Despite similarities in regulation and function, phasins from different bacterial species are not homologs. This study focuses on the mechanism that couples phasin synthesis with the presence of PHA in cells.
Several recent studies have shed light on phasin regulation. Maehara et al. (13) have identified a Paracoccus denitrificans protein, designated PhaR, that is an excellent candidate for a negative regulator that couples PhaP phasin synthesis to PHA production by preventing PhaP accumulation in the absence of PHA. The P. denitrificans phaR gene was identified based on its proximity to the phaP and phaC genes (13). In a heterologous system, Escherichia coli, PhaP of P. denitrificans accumulates to lower levels in the presence of the phaR gene (13), and PhaR binds the phaP promoter region and blocks phaP expression in vitro (12). One simple model for PhaR function is that PhaR contains a domain that binds DNA and a second domain that binds PHA or a factor associated with PHA. In the absence of PHA, PhaR binds to and represses transcription of phaP. In the presence of PHA, PhaR is titrated from phaP and thus enables transcription of phaP.
Many PHA-producing bacteria encode a PhaR homolog (13), suggesting that PhaR may be important for phasin regulation and PHA biosynthesis. Ralstonia eutropha is an excellent system to study phasin regulation because it is amenable to standard gene replacement techniques (20, 24), it is the only strain for which phasin mutants have been generated (25, 27), and it produces polyhydroxybutyrate (PHB) under a range of cultivation conditions. In R. eutropha the phaR gene (originally designated ORF1 [24]) is located downstream of the PHA biosynthetic operon phaCAB and is not linked to phaP. Here we report that R. eutropha PhaR is a negative regulator of PhaP accumulation and that PhaR prevents PhaP accumulation in the absence of PHB.
MATERIALS AND METHODS
Strains, plasmids, and oligonucleotides.
The strains and plasmids used in this study are listed in Table 1. The oligonucleotides are as follows: dGCGATTTCGCCCGACGCACCCAGCACTAA, H508E; dAGCTTGGATCCGATGCGAGCGCTGCATACC, phaC2; dAGCTTGGATCCGGCGCTCATGTTTTCCTGG, phaC5; dCCGAGGATCCATCGCCGGACAAGGCAGC, phaP2; dCTAGCGAATTCGGATCCGCAATCGCGCATCGTTG, phaP4; dGCCGAGGATCCTTTCGCGGGCCGTCAAGGC, phaR1; dCCCGCCGCTGCCAGTGTCCGGTGCTCTGTCCTTGTGTC, phaR2; dGACACAAGGACAGAGCACCGGACACTGGCAGCGGCGGG, phaR3; dCCGGAGGATCCATGAAGGAACCAACCCGC, phaR4; dGCGCAACCATATGGCCACGACCAAAAAAGGC, phaR5; dCCGGAGGATCCAGTGTCTTACTTCTTGTCCG, phaR6; and dGCCTTGACCGAGCTGGCCGAT, Seq376. Engineered restriction sites are underlined. H508E and phaP2 include mismatches relative to the correct sequences of phaC and phaP, respectively.
TABLE 1.
Strains and plasmids used in this studya
| Strain or plasmid | Descriptionb | Reference or source |
|---|---|---|
| Strains | ||
| R. eutropha | ||
| Ae H16 | wt, Gmr, also termed DSM 428 and ATCC 17699 | ATCC 17699 |
| Re1001 | phaP-gfp translational fusion gene replacement strain | 26 |
| Re1034 | ΔphaC strain | 26 |
| Re1052 | ΔphaP strain | 27 |
| Re1099 | ΔphaR strain, derived from Ae H16/pGY95 | This study |
| Re1101 | ΔphaC ΔphaR strain, derived from Re1034/pGY95 | This study |
| Re1102 | ΔphaP ΔphaR strain, derived from Re1052/pGY95 | This study |
| Re1103 | ΔphaR phaP-gfp translational fusion strain, derived from Re1001/pGY95 | This study |
| Re1105 | ΔphaC phaP-gfp translational fusion strain, derived from Re1001/pGY46 | This study |
| Re1106 | ΔphaC ΔphaR phaP-gfp translational fusion strain, derived from Re1103/pGY46 | This study |
| E. coli | ||
| DH5α | Strain for ligation, cloning, and heterologous expression of PHA genes | New England Biolabs |
| Plasmids | ||
| pAeT41 | 5-kb SmaI/EcoRI phaCAB DNA cloned into SmaI/EcoRI sites of pUC18 | 16 |
| pCR2.1-TOPO | High-copy-number plasmid used for cloning, confers Apr and Kmr | Invitrogen |
| pGY1a+ | phaP-gfp translational fusion cloned into pSW213, positive control for GFP expression | 26 |
| pGY4+ | 1.5-kb phaP DNA in BamHI site of pSW213, phaP in same orientation as lac promoter | This study |
| pGY4− | 1.5-kb phaP DNA in BamHI site of pSW213, phaP in opposite orientation as lac promoter | This study |
| pGY46 | ΔphaC gene replacement plasmid, confers Kmr | 26 |
| pGY95 | ΔphaR gene replacement plasmid, confers Kmr | This study |
| pGY104− | 1.35-kb phaR DNA in BglII site of pSW213, phaR in opposite orientation as lac promoter | This study |
| pGY105 | 1.5-kb phaP DNA in BamHI site of pGY104−, phaP oriented divergently from phaR | This study |
| pJQ200mp18Km | Derivative of pJQ200mp 18, Gmr gene disrupted, confers Kmr | 27 |
| pSW213 | Low-copy-number plasmid, confers Tcr | 3 |
| pUC18 | High-copy-number plasmid used for cloning, confers Apr | Invitrogen |
Plasmids constructed in this study that were used only as intermediates for construction of other plasmids are described only in the Materials and Methods section.
Abbreviations: Apr, Gmr, Kmr, and Tcr, ampicillin, gentamicin, kanamycin, and tetracycline resistance, respectively.
Growth media and antibiotics.
R. eutropha strains were cultivated in Luria-Bertani (LB) medium (14), Tryptic Soy Broth-Dextrose Free (TSB) medium (Becton Dickinson Microbiology Systems, Cockeysville, Md.), PHA(med), and PHA(high). PHA(med) and PHA(high) are based on a minimal medium (16) supplemented with fructose (0.5 or 1%, respectively) and ammonium chloride (0.1 or 0.01%, respectively). E. coli strains were cultivated in LB medium. Antibiotics were added to growth media to the following final concentrations: for R. eutropha, gentamicin (10 μg/ml) and kanamycin (270 μg/ml); and for E. coli, ampicillin (100 μg/ml), gentamicin (10 μg/ml), kanamycin (25 μg/ml), and tetracycline (10 μg/ml).
Cultivation conditions.
R. eutropha and E. coli strains were cultivated with aeration at 30 and 37°C, respectively. For preparation of genomic DNA or selection for resistance to antibiotics, R. eutropha strains were cultivated in liquid TSB medium or solid LB agar (1.2%). For R. eutropha immunoblot analyses, strains were cultivated in 2 ml of TSB in test tubes to saturation (24 to 30 h). Aliquots (100 μl) were transferred to 5 ml of TSB in test tubes and were cultivated to saturation (∼16 h). Aliquots of washed cells were transferred to 5 ml of TSB, PHA(med), or PHA(high) to yield cultures with an initial optical density at 600 nm (OD600) of 1.0 (∼109 CFU/ml) and cultivated for 48 h. For R. eutropha PHB production and PhaP quantitation analyses, strains were cultivated as for immunoblot analyses but on a larger scale [growth in 4 ml of TSB in test tubes (36 h), then in 100 ml of TSB in 500-ml flasks (24 h), and finally in 200 ml of LB medium with 2% fructose or PHA(high) in 1-liter baffled flasks (72 h)]. For E. coli immunoblot analyses, strains were cultivated in 2.5 ml of LB(Ap Tc) (that is, LB medium containing ampicillin and tetracycline) in test tubes to saturation (24 h). Aliquots of washed cells were transferred to 5 ml of LB medium plus 2% glucose(Ap Tc) to yield cultures with an initial OD600 of 0.9 and cultivated 24 h. For E. coli PHB production analyses, strains were cultivated as for immunoblot analyses [growth in 5 ml of LB(Ap Tc) in test tubes (16 to 22 h) and then in 100 ml of LB medium plus 2% glucose(Ap Tc) in 500-ml flasks at initial OD600 of 0.15 (72 to 105 h)].
DNA preparation and manipulation.
Standard approaches were used for preparation and manipulation of DNA, for transformation of E. coli, and for the PCR (1). Genomic DNA was prepared from R. eutropha strains as described previously (26). All constructs containing PCR products were confirmed by sequencing at the MIT Biopolymer Lab.
Construction of phaR precise deletion gene replacement plasmid pGY95.
The 347- and 349-bp fragments of R. eutropha DNA corresponding to the regions immediately upstream and downstream of the phaR open reading frame (ORF), respectively, were amplified by PCR with phaR1 and phaR2 or with phaR3 and phaR4. The two PCR products were combined and amplified with phaR1 and phaR4 to yield a 0.7-kb PCR ligation product spanning from upstream to downstream of the phaR ORF, but lacking the phaR ORF, flanked by BamHI sites. The 0.7-kb BamHI fragment of the PCR product was cloned into the BamHI site of pJQ200mp18Km to yield pGY95.
Construction of phaR and phaC gene replacement strains.
Gene replacement was accomplished by adaptation of standard protocols (20, 24). The combinations of starting strains and plasmids used for construction of each strain are indicated in Table 1. Construction of ΔphaR strains was confirmed by PCR using oligonucleotides phaR5 and phaR6, which amplify the phaR ORF (phaR+, 0.6 kb; ΔphaR, no product). Construction of ΔphaC strains was confirmed by PCR with oligonucleotides phaC2 and phaC5 (phaC+, 2.3 kb; ΔphaC, 0.8 kb) and Seq376 and H508E (phaC+, 1.2 kb; ΔphaC, no product).
Plasmid encoding phaP..
Plasmid pGY4+ was constructed as follows. A 1.5-kb fragment of R. eutropha DNA, spanning from 762 bp upstream to 141 bp downstream of the phaP ORF, was amplified by PCR with the oligonucleotides phaP4 and phaP2, such that BamHI sites were introduced at both ends of the fragment. The corresponding 1.5-kb BamHI fragment was cloned into the BamHI site of the multicloning site of pSW213, in the same orientation as the lac promoter present in the plasmid, to yield pGY4+. Plasmid pGY4− contains the same 1.5-kb BamHI fragment cloned into the BamHI site of pSW213 in the opposite orientation.
Plasmid encoding phaR.
Plasmid pGY104− was constructed as follows. A 1.35-kb fragment of R. eutropha DNA, spanning from 347 bp upstream to 349 bp downstream of the phaR ORF, was amplified by PCR with the oligonucleotides phaR1 and phaR4, such that BamHI sites were introduced at both ends of the fragment. This fragment was cloned into the AflII site of pCR2.1-TOPO to yield the plasmid pB3. The 1.35-kb BamHI fragment of pB3 was cloned into the BglII site in the multicloning site of pSW213, such that the phaR ORF is in the opposite orientation as the lac promoter on the plasmid, to yield pGY104−.
Plasmid encoding phaP and phaR.
Plasmid pGY105 was constructed as follows. The 1.5-kb BamHI fragment of pGY4+, containing phaP, was cloned into the BamHI site of pGY104−, such that phaP is in the opposite orientation as phaR on the plasmid (gene order: lac promoter-phaR [reverse orientation]-phaP).
Quantitation of PHB in R. eutropha and E. coli cells.
PHB was quantitated by the sulfuric acid-high-pressure liquid chromatography method of Karr et al. (8) with some modifications (26).
Recombinant PhaP, antibodies, and immunoblot analyses.
Recombinant PhaP was purified as previously described (27) and quantitated based on its extinction coefficient (PhaP ɛ280 = 5,687 M−1 cm−1) as determined by measurements of mass (determined by amino acid analysis) and A280. Preparation and purification of antibodies against PhaP, immunoblotting, and quantitation of PhaP in cells were conducted as described previously (26). Anti-green fluorescent protein (GFP) antibodies were obtained from Clontech (Palo Alto, Calif.).
RESULTS
PhaR negatively regulates PhaP accumulation.
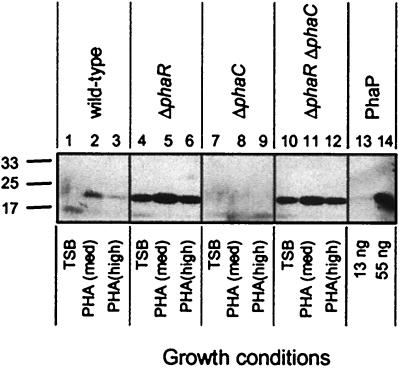
To determine whether R. eutropha PhaR regulates PhaP accumulation, a ΔphaR strain was constructed by precise deletion of the phaR ORF in the wild-type (wt) strain H16. An isogenic ΔphaR ΔphaP strain was also constructed by deletion of the phaR ORF in a ΔphaP strain. Strains were cultivated in TSB, PHA(med), and PHA(high), which are growth media that promote the production of PHB to low, intermediate, or high levels, respectively (27). After 48 h, aliquots of culture, each normalized based on the OD600, were subjected to immunoblot analyses for detection of PhaP. The wt and ΔphaR strains yielded equivalent culture OD600 readings for TSB and PHA(med) (Fig. 1), suggesting similar growth and/or PHA accumulation rates (10, 26). The ΔphaR strain, however, yielded much higher signal for the ∼24-kDa PhaP protein than did the wt strain (Fig. 1, lanes 4 to 6 versus lanes 1 to 3). These observations suggest that PhaR negatively regulates PhaP accumulation across a range of growth conditions. The ΔphaP and ΔphaR ΔphaP strains yielded no signal for PhaP, as expected (data not shown).
FIG. 1.
Immunoblots of PhaP in R. eutropha strains. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and were subjected to immunoblot analysis for detection of PhaP. Cells from R. eutropha cultures were harvested after cultivation for 48 h in TSB, PHA(med), or PHA(high). Bacterial samples correspond to cells from 10 μl of a culture diluted to an OD600 of 0.2. Purified PhaP was included as a control. Molecular mass standards are indicated in kilodaltons. Culture OD600 measurements were as follows: lane 1, 8.9; lane 2, 8.3; lane 3, 9.6; lane 4, 8.6; lane 5, 7.2; lane 6, 5.7; lane 7, 9.1; lane 8, 4.3; lane 9, 1.3; lane 10, 8.9; lane11, 3.8; lane 12, 1.8.
PhaR couples PhaP accumulation to the production of PHB in individual cells.
To test whether PhaR may prevent PhaP accumulation in the absence of PHB, a ΔphaR ΔphaC strain, which is incapable of producing PHB, was constructed, and PhaP accumulation was measured. The ΔphaC and ΔphaR ΔphaC strains exhibited comparable culture OD600 readings under each cultivation condition (Fig. 1). However, while the ΔphaC strain produced no detectable PhaP (Fig. 1, lanes 7 to 9), the ΔphaR ΔphaC strain produced large amounts of PhaP, equivalent to levels produced by the ΔphaR strain (Fig. 1, lanes 10 to 12). Deletion of phaR completely uncouples PhaP accumulation from PHB production in R. eutropha.
PhaR regulates PhaP accumulation at the level of PhaP synthesis.
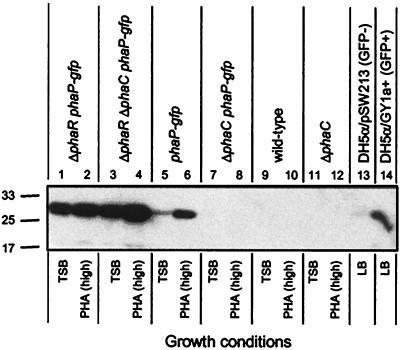
To test whether PhaR may regulate PhaP accumulation at the level of PhaP synthesis, ΔphaR phaP-gfp, ΔphaC phaP-gfp, and ΔphaR ΔphaC phaP-gfp strains were constructed, and GFP expression was measured. The phaP-gfp fusion is designed to reveal regulation of phaP at the level of transcription and/or translational initiation based on expression of GFP (26). Strains were cultivated in TSB and PHA(high) and analyzed for GFP expression by immunoblot. The ΔphaR phaP-gfp and ΔphaR ΔphaC phaP-gfp strains expressed the ∼27-kDa GFP protein to approximately the same extent (Fig. 2, lanes 1 to 4), implying that in the ΔphaR background PhaP is synthesized independently of PHB production and thus that PhaR specifically regulates PhaP synthesis. This interpretation is based on the assumption that if cells express a system specifically to degrade PhaP that such a system would not degrade GFP. Importantly, the phaP-gfp strain, but not the ΔphaC phaP-gfp strain, expressed GFP (Fig. 2, lanes 5 to 8), a finding consistent with our previous observation that in phaR+ strains PhaP synthesis is strictly dependent on PHB production (26). Controls confirmed the specificity of the GFP antibody (Fig. 2, lanes 9 to 14).
FIG. 2.
Immunoblots of GFP in R. eutropha ΔphaR phaP-gfp, ΔphaR ΔphaC phaP-gfp, and control strains. Proteins were separated by SDS-PAGE and then subjected to immunoblot analysis for detection of GFP. Molecular mass standards are indicated in kilodaltons. Cells from R. eutropha cultures were harvested after cultivation for 48 h in TSB or PHA(high) and resuspended in 10 μl at an OD600 of 0.4 for analysis. The E. coli strains DH5α/pGY1a+, which carries the phaP-gfp fusion on a plasmid, and DH5α/pSW213, which carries the corresponding vector lacking the phaP-gfp fusion, were also included as positive and negative controls, respectively (24-h cultivation in LB plus tetracycline, resuspended in 10 μl at an OD600 of 1.0 for analysis). Culture OD600 measurements were as follows: lane 1, 9.0; lane 2, 2.7; lane 3, 9.3; lane 4, 1.5; lane 5, 9.2; lane 6, 4.8; lane 7, 9.7; lane 8, 1.5; lane 9, 8.8; lane 10, 14; lane 11, 9.2; lane 12, 5.0; lane 13, 4.2; lane 14, 4.0.
PhaR directly links PhaP accumulation to the presence of PHB in cells.
We proceeded to test whether PhaR is sufficient to regulate PhaP synthesis in a heterologous system. Plasmids containing phaP (pGY4+), phaR (pGY104−), and phaP and phaR (pGY105), were constructed from the broad-host-range plasmid pSW213. pAeT41, which contains the R. eutropha phaCAB operon and which thus confers upon E. coli the ability to produce PHB, has been constructed previously from ColE1-based pUC18. DH5α strains carrying various combinations of the pSW213- and pUC18-based plasmids, and thus varying in their genetic capacities to produce PhaR, PhaP, and PHB, were generated. Strains were cultivated in LB medium and LB medium plus 2% glucose, and culture OD600 and PhaP immunoblot analyses were conducted. LB medium was used to prevent PHB production in DH5α/pAeT41 strains (PHB ≤ 0.1% cell dry weight [cdw] at 24 h) and LB 2% glucose was used to promote PHB production (PHB ≥ 60% cdw at 24 h).
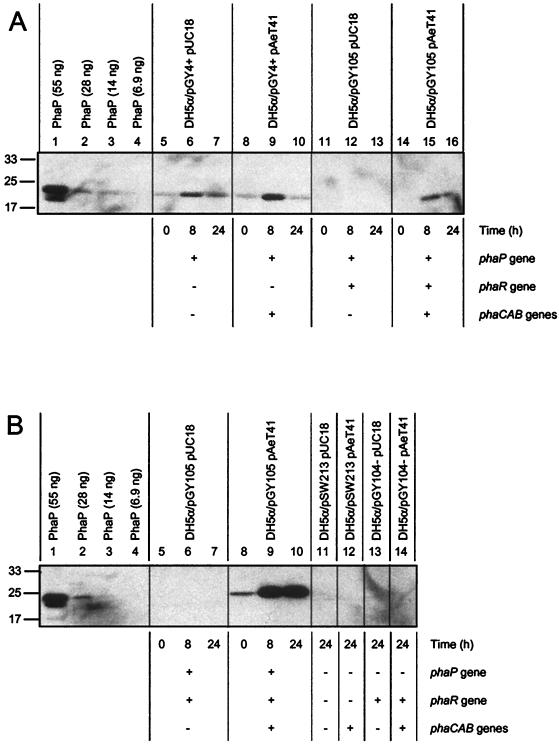
DH5α/pGY4+ pUC18 and DH5α/pGY4+ pAeT41 (analogous to R. eutropha ΔphaR ΔphaC and ΔphaR strains, respectively) expressed PhaP constitutively (Fig. 3A, lanes 5 to 10). DH5α/pGY105 pUC18 (analogous to the R. eutropha ΔphaC strain) did not express PhaP (Fig. 3A, lanes 11 to 13; Fig. 3B, lanes 5 to 7). DH5α/pGY105 pAeT41 (analogous to the R. eutropha wt strain) expressed PhaP in a manner dependent on PHB production (Fig. 3A, lanes 14 to 16; Fig. 3B, lanes 8 to 10). Specifically, PhaP accumulated to low levels during cultivation in LB (Fig. 3B, lane 8, for which the 0-h time point corresponds to 24-h starter culture) and to high levels during cultivation in LB medium plus 2% glucose (Fig. 3B, lanes 9 to 10). As expected, strains that lack phaP (DH5α/pSW213 and DH5α/pGY104− strains) yielded no signal for PhaP (Fig. 3B, lanes 11 to 14). These results demonstrate reconstitution of the PhaR/PhaP regulatory system in a heterologous system and strongly suggest that PhaR regulates PhaP accumulation directly, by negatively regulating PhaP synthesis and coupling PhaP accumulation to PHB production.
FIG. 3.
Immunoblots of PhaP in cultures of E. coli strains that carry the R. eutropha phaP, phaR, and/or phaCAB genes. Proteins were separated by SDS-PAGE and then subjected to immunoblot analysis for PhaP. Purified PhaP was included as a control on both gels. Molecular mass standards are indicated in kilodaltons. Cells were harvested after cultivation of strains in LB medium plus 2% glucose(Ap Tc) at various time points (0, 8, and 24 h). (A) Samples correspond to cells from 10 μl of culture diluted to an OD600 of 0.20. Note that lanes 1 to 10 and lanes 11 to 16 correspond to different gels. Culture OD600 measurements were as follows: lane 5, 0.9; lane 6, 4.1; lane 7, 3.3; lane 8, 0.9; lane 9, 11; lane 10, 33; lane 11, 0.9; lane 12, 4.2; lane 13, 4.4; lane 14, 0.9; lane 15, 14; lane 16, 32. (B) Samples correspond to cells from 10 μl of culture diluted to an OD600 of 2.0. Culture OD600 measurements were as follows: lane 11, 4.2; lane 12, 21; lane 13, 4.4; lane 14, 18.
There are two caveats that could potentially affect this interpretation. First, pAeT41 includes the first 132 bp of the phaR ORF in addition to the phaCAB genes and thus could potentially express a truncated but active version of PhaR. However, DH5α/pGY4+ pUC18 and DH5α/pGY4+ pAeT41 accumulated PhaP to approximately the same levels during cultivation in LB (Fig. 3A, lanes 5 and 8, 0-h time point), implying that if such a truncated protein is expressed, it is inactive. Second, pGY4+ contains phaP cloned downstream of, and in the same orientation as, the lac promoter and thus phaP could potentially be transcribed from this promoter. However, two points argue against this possibility. First, transcription from the lac promoter is repressed in DH5α. Second, DH5α strains carrying pGY4+ or pGY4− (a plasmid identical to pGY4+ except for the orientation of the 1.5-kb phaP fragment) expressed PhaP to similar levels from both plasmids (data not shown), implying that in the absence of PhaR the phaP promoter functions constitutively in E. coli.
PhaR promotes PHB production in R. eutropha.
To test whether PhaR affects PHB production, R. eutropha wt and ΔphaR strains were cultivated under two sets of conditions that promote PHB production: PHA(high) and LB medium plus 2% fructose. These growth conditions were chosen to allow comparisons between R. eutropha and E. coli. PHA(high) is a standard growth medium for PHB production in R. eutropha (16, 27), and LB medium plus 2% fructose is analogous to LB medium plus 2% glucose, a standard growth medium for PHB production in E. coli (10, 23). Strains were cultivated in TSB starter cultures, transferred to PHA(high) and LB medium plus 2% fructose treatment cultures, both at an initial OD600 of 1.0 (high titer) and at OD600 of 0.1 (low titer), and cultivated for 3 days. The two inoculation conditions were used to test whether any effect of deletion of phaR might depend on the initial density of cells in cultures. PHB production was measured over a time course. The ΔphaP and ΔphaR ΔphaP strains were also characterized to test the effect of PhaP on PHB production.
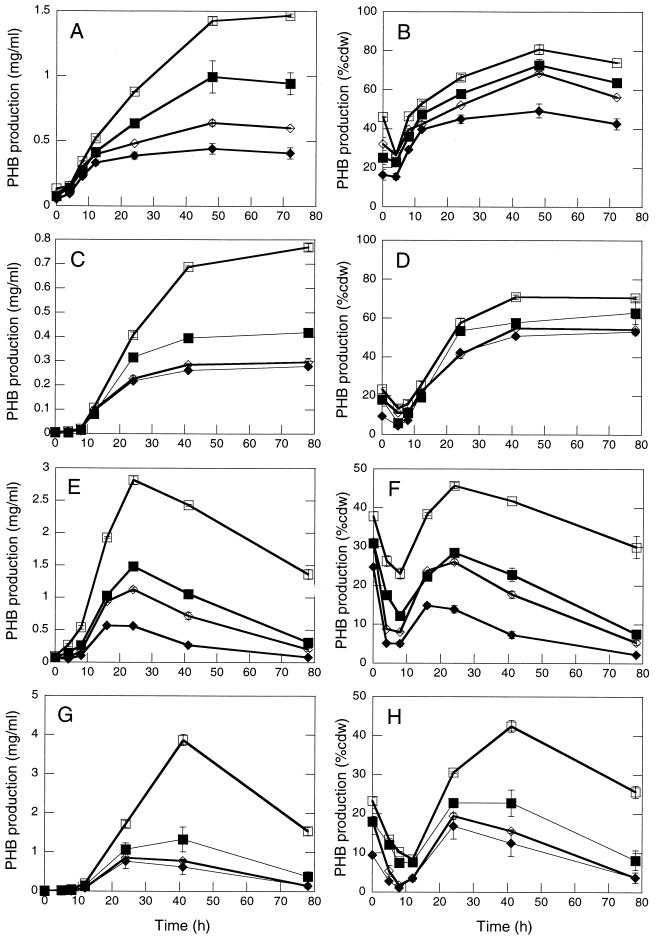
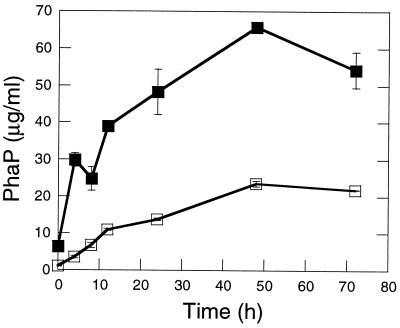
The ΔphaR strain accumulated PHB to ≤70 and ≤55% the level of the wt strain at the peak of PHB accumulation during cultivation in PHA(high) and LB medium plus 2% fructose, respectively (Fig. 4A, C, E, and G). PHB levels rose throughout cultivation in PHA(high) (Fig. 4A to D), whereas PHB levels rose and then fell during cultivation in LB plus 2% fructose (Fig. 4E to H). PHB reached higher levels for high-titer inoculations (except for the wt strain cultivated in LB medium plus 2% fructose) and deletion of phaR had a more dramatic effect on PHB production for low-titer inoculations (Fig. 4A, B, E, and F versus Fig. 4C, D, G, and H). These observations suggest that overexpression of PhaP may have a more dramatic negative effect on PHB production with decreasing titers of inoculation. Deletion of phaR clearly leads to decreased PHB yields, but it is not clear whether PhaR promotes PHB synthesis, prevents PHB degradation, or both. PhaP was present at 2.5- to 8.5-fold-higher levels in ΔphaR cells than in wt cells throughout cultivation [PHA(high)/high-titer inoculation] (Fig. 5), suggesting that PhaP overexpression may contribute to the PHB production defect of the ΔphaR strain. However, the ΔphaR ΔphaP strain exhibited a more severe defect in PHB production than the ΔphaP strain, particularly during cultivation under high-titer conditions (Fig. 4A, B, E, and F), indicating that PhaR also promotes PHB production by a PhaP-independent mechanism.
FIG. 4.
Comparison of PHB production for R. eutropha wt, ΔphaR, ΔphaP, and ΔphaR ΔphaP strains. Symbols: open squares, wt strain; closed squares, ΔphaR strain; open diamonds, ΔphaP strain; solid diamonds, ΔphaR ΔphaP strain. Growth media correspond to PHA(high) (A to D) and LB medium plus 2% fructose (E and F). Inoculation conditions correspond to high titer (A, B, E, and F) and low titer (C, D, G, and H). PHB production is expressed as mass PHB per volume culture (A, C, E, and G) and as PHB % cdw (B, D, F, and H). Datum points represent average value for two independent cultures. Error bars indicate the standard deviation.
FIG. 5.
Quantitation of PhaP in R. eutropha wt and ΔphaR strains, as cultivated in PHA(high) by high-titer inoculation and as measured over a time course of 72 h. Symbols: open squares, wt strain; solid squares, ΔphaR strain. All datum points represent the average value for two independent cultures. Error bars indicate the standard deviation. Samples correspond to the cultures of wt and ΔphaR strains in Fig. 4A and B.
PhaP, but not PhaR, promotes PHB production in a heterologous system.
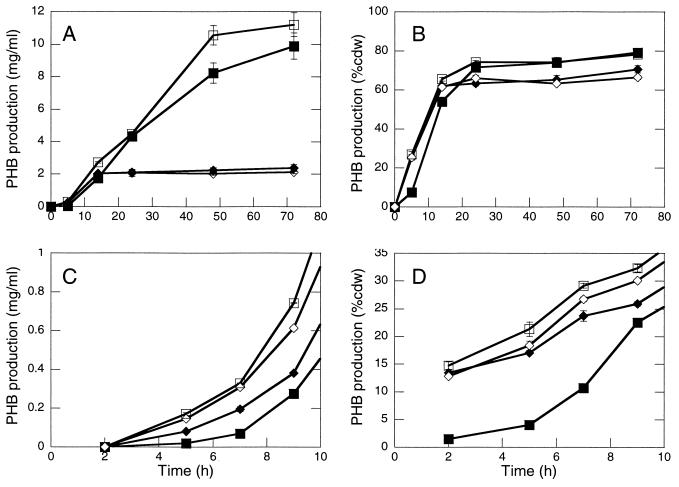
We proceeded to test whether PhaR and PhaP also affect PHB production in E. coli. DH5α/pSW213 pAeT41 and DH5α/pGY104− pAeT41 strains (analogous to R. eutropha ΔphaR ΔphaP and ΔphaP strains, respectively) exhibited similar yields of PHB (Fig. 6A and B), indicating that PhaR does not directly affect PHB production in the absence of PhaP. DH5α/pGY4+ pAeT41 and DH5α/pGY105 pAeT41 (analogous to R. eutropha ΔphaR and wt strains, respectively) exhibited higher yields of PHB than DH5α/pSW213 pAeT41 and DH5α/pGY104− pAeT41, indicating that PhaP promotes PHB production in this system (Fig. 6A and B). DH5α/pGY4+ pAeT41 and DH5α/pGY105 pAeT41 also accumulated PHB to approximately the same levels by 3 days, suggesting that PhaR-mediated regulation of PhaP accumulation does not affect overall PHB production. Interestingly, at 5 h the DH5α/pGY4+ pAeT41 strain, which expresses PhaP constitutively, exhibited low levels of PHB relative to the other three DH5α/pAeT41 strains. Analyses of PHB production by the DH5α/pAeT41 strains at early time points in an independent experiment confirmed this observation (Fig. 6C and D). Apparently, overexpression of PhaP transiently delays or decreases PHB production, implying that PhaR-mediated regulation of PhaP may be important, but only at the onset of PHB synthesis. It is not clear how these results relate to R. eutropha, since R. eutropha cultures were always inoculated with cells that already contained PHB (PHB > 5% cdw).
FIG. 6.
Comparison of PHB production for E. coli strains that carry the R. eutropha phaCAB genes and that vary with respect to their capacity to produce PhaP and PhaR. Symbols: open squares, DH5α/pGY105 pAeT41; solid squares, DH5α/pGY4+ pAeT41; open diamonds, DH5α/pGY104− pAeT41; solid diamonds, DH5α/pSW213 pAeT41. The growth medium corresponds to LB medium plus 2% glucose(Ap Tc). Cultures were inoculated at an initial OD600 of 0.15. The data represent two independent cultivation experiments. The datum points represent average values for two independent cultures in a given experiment. The error bars indicate the standard deviation. Results for panels A and B and from panels C and D are from independent experiments. (A) PHB accumulation (milligrams/milliliter of culture). (B) PHB accumulation (% cdw). (C) PHB production (milligrams/milliliter of culture). DH5α/pAeT41 strains accumulated PHB to the following amounts after 105 h of cultivation: pGY105, 8.7 ± 0.13; pGY4+, 8.9 ± 0.47; pGY104−, 4.3 ± 0.071; and pSW213, 4.2 ± 0.25. (D) PHB production (% cdw). DH5α/pAeT41 strains accumulated PHB to the following amounts after 105 h of cultivation: pGY105, 80% ± 0.78%; pGY4+, 81% ± 1.5%; pGY104−, 72% ± 1.4%; and pSW213, 74% ± 0.90%.
DISCUSSION
Our results demonstrate that PhaR is a negative regulator of PhaP synthesis that directly couples PhaP synthesis to PHB production. The simplest model for PhaR regulation of PhaP accumulation is that PhaR binds a regulatory sequence upstream of the phaP ORF and blocks its transcription. The simplest mechanism by which PhaR may couple PhaP synthesis to PHB production is that, upon the onset of PHB synthesis in cells, PhaR is titrated from phaP, possibly by binding to PHB or else by interacting with a factor that is present in R. eutropha and E. coli cells only after these cells have begun to synthesize PHB. PhaP synthesis then occurs to an extent that is determined by the degree of PhaR titration and the availability of nutrients. This model accounts for the observations that in R. eutropha wt cells PhaP synthesis is strictly dependent on PHB synthesis, that in R. eutropha ΔphaR cells PhaP is synthesized constitutively, and that in E. coli cells addition of the phaCAB, phaR, and phaP genes is sufficient to reconstitute the PhaR/PhaP regulatory system.
Our results suggest that PhaR promotes PHB synthesis in R. eutropha by regulating the expression of PhaP and one or more additional proteins, thus functioning by PhaP-dependent and PhaP-independent pathways. For the PhaP-dependent pathway, PhaR represses synthesis of PhaP until after the onset of PHB synthesis, thus preventing diversion of metabolites toward production of excess PhaP and/or preventing PhaP from interfering with the initiation of PHB synthesis. This would account for the observations that E. coli phaCAB+ strains that overexpress PhaP exhibit a defect in PHB synthesis at the onset of PHB synthesis and that R. eutropha phaR mutants exhibit more severe defects in PHB production for low-titer versus high-titer inoculations. For the PhaP-independent pathway, PhaR represses the synthesis of one or more additional proteins that may interfere with PHB production when expressed at inappropriate times or levels or that may promote utilization of PHB under inappropriate conditions. This would account for the observations that ΔphaR ΔphaP mutants exhibit more severe defects in PHB production than ΔphaP mutants and that E. coli phaCAB+ cells exhibit the same final yields of PHB independent of phaR.
Several other groups have reported on PhaR/PhaP and other PHA regulatory systems recently. Maehara et al. (13) transferred P. denitrificans phaR and phaP into E. coli and demonstrated that the presence of phaR dramatically decreased expression of phaP. Maehara et al. (12) also recently reported that PhaR represses phaP expression in vitro. However, in contrast to R. eutropha PhaR/PhaP, P. denitrificans PhaR did not completely block phaP expression in E. coli, even in cells that cannot produce PHA (13). Thus, while P. denitrificans PhaR negatively regulates expression of phaP, it remains to be determined whether it also couples PhaP synthesis to PHB production. Prieto et al. (19) have proposed a model regarding another PHA-related regulatory system, PhaF-mediated regulation of protein expression in P. oleovorans GPo1, that is similar to the R. eutropha PhaR/PhaP system. According to the model, PhaF binds DNA, responds to PHA or factors present during PHA synthesis, and regulates the expression of a granule-associated protein (9, 19). However, PhaF and PhaR are not homologs and, unlike PhaR, PhaF also regulates expression of PHA synthase (19). Thus, the systems are distinct. Povolo and Cadela (18) have demonstrated that deletion of the S. meliloti phaR homolog aniA results in significant changes in carbohydrate polymer production and nitrogenase activity, in addition to a decrease in PHB levels. The observation that many metabolic processes are affected by the aniA mutation is consistent with the idea that PhaR regulates the expression of proteins in addition to phasins.
Comparisons of the properties and yields of a variety of PHAs, as produced by R. eutropha wt, phaRΔ, phaPΔ, and phaRΔ phaPΔ strains and E. coli strains engineered to express PhaR, PhaP, and PHA biosynthetic enzymes, are providing new insights into the role and specificity of phasins in PHA production. A long-range goal is that the PhaR/PhaP regulatory system and PhaP may eventually serve as tools to accomplish the production of novel PHAs in an economically competitive manner.
Acknowledgments
We thank Jiamin Tian, Adam Lawrence, Philip Lessard, Geoff Stamper, and Catherine Drennan for useful discussions.
G.M.Y. is a DOE-Energy Biosciences Research Fellow of the Life Sciences Research Foundation. This work was supported by NIH grant GM49171 to J.S. and A.J.S.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Barnard, G. N., and J. K. M. Sanders. 1989. The poly-β-hydroxybutyrate granule in vivo. A new insight based on NMR spectroscopy of whole cells. J. Biol. Chem. 264:3286–3291. [PubMed] [Google Scholar]
- 3.Chen, C. Y., and S. C. Winans. 1991. Controlled expression of the transcriptional activator gene virG in Agrobacterium tumefaciens by using the Escherichia coli lac promoter. J. Bacteriol. 173:1139–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerngross, T. U., K. D. Snell, O. P. Peoples, A. J. Sinskey, E. Csuhai, S. Masamune, and J. Stubbe. 1994. Overexpression and purification of the soluble polyhydroxyalkanoate synthase from Alcaligenes eutrophus: evidence for a required posttranslational modification for catalytic activity. Biochemistry 33:9311–9320. [DOI] [PubMed] [Google Scholar]
- 5.Handrick, R., S. Reinhardt, and D. Jendrossek. 2000. Mobilization of poly(3-hydroxybutyrate) in Ralstonia eutropha. J. Bacteriol. 182:5916–5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanley, S. Z., D. J. Pappin, D. Rahman, A. J. White, K. M. Elborough, and A. R. Slabas. 1999. Re-evaluation of the primary structure of Ralstonia eutropha phasin and implications for polyhydroxyalkanoic acid granule binding. FEBS Lett. 447:99–105. [DOI] [PubMed] [Google Scholar]
- 7.Jendrossek, D., A. Schirmer, and H. G. Schlegel. 1996. Biodegradation of polyhydroxyalkanoic acids. Appl. Microbiol. Biotechnol. 46:451–463. [DOI] [PubMed] [Google Scholar]
- 8.Karr, D. B., J. K. Waters, and D. W. Emerich. 1983. Analysis of poly-β-hydroxybutyrate in Rhizobium japonicum bacteroids by ion-exclusion high-pressure liquid-chromatography UV detection. Appl. Environ. Microbiol. 46:1339–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kessler, B., and B. Witholt. 2001. Factors involved in the regulatory network of polyhydroxyalkanoate metabolism. J. Biotechnol. 86:97–104. [DOI] [PubMed] [Google Scholar]
- 10.Lee, S. Y., K. M. Lee, H. N. Chang, and A. Steinbüchel. 1994. Comparison of recombinant Escherichia coli strains for synthesis and accumulation of poly-(3-hydroxybutyric acid) and morphological changes. Biotechnol. Bioeng. 44:1337–1347. [DOI] [PubMed] [Google Scholar]
- 11.Madison, L. L., and G. W. Huisman. 1999. Metabolic engineering of poly(3-hydroxyalkanoates): from DNA to plastic. Microbiol. Mol. Biol. Rev. 63:21–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maehara, A., Y. Doi, T. Nishiyama, Y. Takagi, S. Ueda, H. Nakano, and T. Yamane. 2001. PhaR, a protein of unknown function conserved among short-chain-length polyhydroxyalkanoic acids producing bacteria, is a DNA-binding protein and represses Paracoccus denitrificans phaP expression in vitro. FEMS Microbiol. Lett. 200:9–15. [DOI] [PubMed] [Google Scholar]
- 13.Maehara, A., S. Ueda, H. Nakano, and T. Yamane. 1999. Analyses of a polyhydroxyalkanoic acid granule-associated 16-kilodalton protein and its putative regulator in the pha locus of Paracoccus denitrificans. J. Bacteriol. 181:2914–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 15.McCool, G. J., and M. C. Cannon. 1999. Polyhydroxyalkanoate inclusion body-associated proteins and coding region in Bacillus megaterium. J. Bacteriol. 181:585–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peoples, O. P., and A. J. Sinskey. 1989. Poly-β-hydroxybutyrate (PHB) biosynthesis in Alcaligenes eutrophus H16. Identification and characterization of the PHB polymerase gene (phbC). J. Biol. Chem. 264:15298–15303. [PubMed] [Google Scholar]
- 17.Pieper-Fürst, U., M. H. Madkour, F. Mayer, and A. Steinbüchel. 1995. Identification of the region of a 14-kilodalton protein of Rhodococcus ruber that is responsible for the binding of this phasin to polyhydroxyalkanoic acid granules. J. Bacteriol. 177:2513–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Povolo, S., and S. Cadella. 2000. A critical role for aniA in energy-carbon flux and symbiotic nitrogen fixation in Sinorhizobium meliloti. Arch. Microbiol. 174:42–49. [DOI] [PubMed] [Google Scholar]
- 19.Prieto, M. A., B. Bühler, K. Jung, B. Witholt, and B. Kessler. 1999. PhaF, a polyhydroxyalkanoate-granule-associated protein of Pseudomonas oleovorans GPo1 involved in the regulatory expression system for pha genes. J. Bacteriol. 181:858–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15–21. [DOI] [PubMed] [Google Scholar]
- 21.Rehm, B. H., and A. Steinbüchel. 1999. Biochemical and genetic analysis of PHA synthases and other proteins required for PHA synthesis. Int. J. Biol. Macromol. 25:3–19. [DOI] [PubMed] [Google Scholar]
- 22.Saegusa, H., M. Shiraki, C. Kanai, and T. Saito. 2001. Cloning of an intracellular poly[-d-(−)3-hydroxybutyrate] depolymerase gene from Ralstonia eutropha H16 and characterization of the gene product. J. Bacteriol. 183:94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sim, S. J., K. D. Snell, S. A. Hogan, J. Stubbe, C. Rha, and A. J. Sinskey. 1997. PHA synthase activity controls the molecular weight and polydispersity of polyhydroxybutyrate in vivo. Nat. Biotechnol. 15:63–67. [DOI] [PubMed] [Google Scholar]
- 24.Slater, S., K. L. Houmiel, M. Tran, T. A. Mitsky, N. B. Taylor, S. R. Padgette, and K. J. Gruys. 1998. Multiple beta-ketothiolases mediate poly(beta-hydroxyalkanoate) copolymer synthesis in Ralstonia eutropha. J. Bacteriol. 180:1979–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wieczorek, R., A. Pries, A. Steinbüchel, and F. Mayer. 1995. Analysis of a 24-kilodalton protein associated with the polyhydroxyalkanoic acid granules in Alcaligenes eutrophus. J. Bacteriol. 177:2425–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.York, G. M., B. H. Junker, J. Stubbe, and A. J. Sinskey. 2001. Accumulation of the PhaP phasin of Ralstonia eutropha is dependent on production of polyhydroxybutyrate in cells. J. Bacteriol. 183:4217–4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.York, G. M., J. Stubbe, and A. J. Sinskey. 2001. New insight into the role of the PhaP phasin of Ralstonia eutropha in promoting the synthesis of polyhydroxybutyrate. J. Bacteriol. 183:2394–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]