Abstract
Cell surface mannan is implicated in almost every aspect of pathogenicity of Candida albicans. In Saccharomyces cerevisiae, the Vrg4 protein acts as a master regulator of mannan synthesis through its role in substrate provision. The substrate for mannosylation of proteins and lipids in the Golgi apparatus is GDP-mannose, whose lumenal transport is catalyzed by Vrg4p. This nucleotide sugar is synthesized in the cytoplasm by pathways that are highly conserved in all eukaryotes, but its lumenal transport (and hence Golgi apparatus-specific mannosylation) is a fungus-specific process. To begin to study the role of Golgi mannosylation in C. albicans, we isolated the CaVRG4 gene and analyzed the effects of loss of its function. CaVRG4 encodes a functional homologue of the S. cerevisiae GDP-mannose transporter. CaVrg4p localized to punctate spots within the cytoplasm of C. albicans in a pattern reminiscent of localization of Vrg4p in the Golgi apparatus in S. cerevisiae. Like partial loss of ScVRG4 function, partial loss of CaVRG4 function resulted in mannosylation defects, which in turn led to a number of cell wall-associated phenotypes. While heterozygotes displayed no growth phenotypes, a hemizygous strain, containing a single copy of CaVRG4 under control of the methionine-repressible MET3 promoter, did not grow in the presence of methionine and cysteine, demonstrating that CaVRG4 is essential for viability. Mutant Candida vrg4 strains were defective in hyphal formation but exhibited a constitutive polarized mode of pseudohyphal growth. Because the VRG4 gene is essential for yeast viability but does not have a mammalian homologue, it is a particularly attractive target for development of antifungal therapies.
Candida albicans is the most important human fungal pathogen. It causes infections that range from superficial colonization of oral and vaginal tissues to life-threatening infections in severely immunocompromised hosts. An essential step for the colonization and infection of host tissues is adhesion, which is initiated by the outermost components of the fungal cell wall. Cell wall-associated mannosylated proteins (mannans) on the external layer of the cell wall have been implicated as key determinants that mediate these initial and critical interactions between the fungus and its host (for reviews, see references 8, 10, and 11). The mannose branches on these glycoproteins, attached by N- and O-glycosidic linkages, are the structures recognized during the immune response against the pathogen (for reviews, see references 3, 14, and 15). Therefore, the enzymes that regulate addition of the mannose molecules are the focus of intense research for understanding fungal biology and mechanisms of host defense and as potential antifungal drug targets.
Biogenesis of mannoproteins in Saccharomyces cerevisiae has been well characterized (for a review see reference 40). Following the initial glycosylation steps in the endoplasmic reticulum, yeast mannoproteins are elongated by addition of mannose or mannosylphosphate in the Golgi apparatus. Extension and branching of N-linked oligosaccharides in the Golgi apparatus result in structures that contain up to 200 mannose units per site, while each O-linked chain is extended up to 5 mannose units (16). In addition to protein mannosylation, sphingolipids and glycophosphatidylinositol (GPI) anchors are also modified by addition of mannose in the Golgi apparatus (5, 47). The sugar donor for all of these Golgi apparatus-localized reactions is GDP-mannose, whose site of synthesis is the cytoplasm (for a recent review see reference 6). Utilization of GDP-mannose by the mannosyltransferases requires transport of this compound into the lumen of the Golgi apparatus by a specific nucleotide sugar transporter (1) that is encoded by the VRG4 gene (19). Through its role in substrate provision, the activity of Vrg4p is the first and rate-limiting step in the Golgi apparatus-mediated mannosylation of cell surface glycoproteins in yeast cells. While none of the genes that encode various Golgi protein or lipid mannosyltransferases appear to be essential, deletion of the VRG4 gene is lethal in S. cerevisiae. Therefore, while a defect in any one pathway can be tolerated in this yeast, a complete loss of Golgi mannosylation leads to death.
Much of the machinery that governs oligosaccharide biosynthesis in C. albicans and S. cerevisiae appears to be conserved. The structure of the outer chains of N-linked oligosaccharides on glycoproteins in C. albicans is similar, although not identical, to the structure of the outer chains of N-linked oligosaccharides on glycoproteins in S. cerevisiae (53). Genes related to S. cerevisiae mannosyltransferase genes have been identified as functional homologues (50, 54–56) and by their sequence similarities. As a first step towards studying the global effects of mannan defects on pathogenesis, here we describe identification of the C. albicans VRG4 (CaVRG4) gene and phenotypic characterization of C. albicans vrg4 mutants. Conditional loss of CaVRG4 function leads to lethality, reduction in the amount of cell wall mannan, and profound induction of morphological abnormalities.
MATERIALS AND METHODS
Strains, media, growth conditions, and transformation protocols.
The C. albicans and S. cerevisiae strains used in this study and their genotypes are shown in Table 1. S. cerevisiae strains were routinely grown on YPAD (1% yeast extract, 2% Bacto Peptone [Difco], 50 mg of adenine sulfate per liter, 2% glucose). C. albicans strains were grown on YPAD supplemented with 50 μg of uridine (Uri) per ml [YAPD(+Uri)] (in medium designations a plus sign before a compound indicates that the compound is present in the medium, and a minus sign before a compound indicates that the compound is not present in the medium). C. albicans ANC3 (MET3p-VRG4/vrg4Δ) was maintained on SD(−Ura,−Met,−Cys) (0.67% yeast nitrogen base without amino acids supplemented with 2% glucose and amino acids but lacking methionine [Met], cysteine [Cys], and uracil [Ura]). To repress the MET3 promoter, C. albicans strains were grown in SD or YPAD(+Uri) containing 5 mM methionine and 2 mM cysteine. To induce hyphal formation in solution, strains were inoculated at a concentration of 5 × 106 cells/ml into YPAD(+Uri) supplemented with 10% fetal calf serum and, as indicated below, with methionine and cysteine and were grown at 37°C. To examine morphological defects on solid media, strains were grown on Spider medium (1% nutrient broth, 1% mannitol, 2% potassium phosphate [pH 7.2)]) or YPAD(+Uri) with 4% fetal calf serum, solidified with 2% agar. When necessary (see below), these media were supplemented with 2 mM methionine and 0.5 mM cysteine.
TABLE 1.
C. albicans and S. cerevisiae strains used in this study
| Strain | Parental strain | Genotype | Reference or source |
|---|---|---|---|
| C. albicans strains | |||
| CA14 | SC5314 | ura3Δ::λimm434/ura3Δ::λimm434 | 22 |
| JPC16J | CAI4 | ura3Δ::λimm434 vrg4Δ::hisG-URA3-hisG/ura3Δ::λimm434 VRG4 | This study |
| JPC16J-1 | CAI4 | ura3Δ::λimm434 vrg4Δ::hisG/ura3Δ::λimm434 VRG4 | This study |
| BWP17 | CAI4 | ura3Δ::λimm434 his1::hisG arg4::hisG/ura3Δ::λimm434 his1::hisG arg4::hisG | 58 |
| ANC1 | BWP17 | ura3Δ::λimm434 his1::hisG arg4::hisG vrg4Δ::hisG-URA3-hisG/ura3Δ::λimm434 his1::hisG arg4::hisG VRG4 | This study |
| ANC2 | BWP17 | ura3Δ::λimm434 his1::hisG arg4::hisG vrg4Δ::hisG/ura3Δ::λimm434 his1::hisG arg4::hisG VRG4 | This study |
| ANC3 | BWP17 | ura3Δ::λimm434 his1::hisG arg4::hisG vrg4Δ::URA3-Met3p-/ura3Δ::λimm434 his1::hisG arg4::hisG vrg4Δ::hisG | This study |
| S. cerevisiae strains | |||
| SEY6210 | MATα ura3-52 his3Δ200 trp1Δ901 lys2-801 suc2-Δ9 leu2-3,112 | S. Emr | |
| RSY255 | MATα ura3-52 leu2-211 | R. Sheckman | |
| NDY5 | RSY255 | MATα ura3-52 leu2-211 vrg4-2 | 41 |
| XGY14 | W303-1A | MATα ura3-1 his3-11 trp1-1 ade2-1 leu2-3,112 can1-100 GAL1p-VRG4::HIS3 | 25 |
C. albicans was transformed by the method described by Gietz et al. (26), with the following modifications. For each transformation, 5 ml of cells grown in YPAD(+Uri) (optical density at 600 nm [OD600], 2) was harvested, washed once with 100 mM lithium acetate, and incubated with 5 to 10 μg of transforming DNA and 50 μg of salmon sperm DNA in 40% polyethylene glycol-100 mM lithium acetate in a 360-μl (final volume) mixture. After overnight incubation at 30°C, the cells were spread on selective medium.
Plasmid construction.
All DNA manipulations were carried out by using standard protocols (43). pCaHHB contains the genomic clone of the entire CaVRG4 open reading frame (ORF), as well as 3.3 kb of upstream flanking sequences and 0.6 kb of downstream flanking sequences, cloned in the HindIII/BamHI site of pRS316, a URA3/CEN6 yeast vector (46). pSK−CaVRG4 R/X contains the CaVRG4 ORF, flanked by an EcoRI site upstream of the initiating start codon and an XbaI site downstream of the TAG stop codon. This 1.1-kb fragment was cloned into the EcoRI/XbaI site of pTαO (25) to produce pTiM1, which placed the CaVRG4 ORF under control of the constitutive triose phosphate isomerase (TPI1) promoter for expression in S. cerevisiae. Digestion with XhoI in the URA3 gene in pTiM1 targeted integration of this gene into the S. cerevisiae ura3-52 locus. To produce a hemagglutinin (HA)-tagged allele, the CaVRG4 ORF was amplified by PCR so that it included an EcoRI site upstream of the initiating codon and a 3′ PstI site that replaced the TAG stop codon. The EcoRI/PstI fragment was cloned into pSK−P/X HA3 (37), which resulted in in-frame fusion of CaVRG4 to sequences encoding three copies of the HA epitope to produce pSK−M1-HA3. An EcoRI/XbaI fragment from pSK−M1-HA3 was subcloned into pTαO to produce the integration plasmid pTiM1-HA3, which was digested with XhoI to target integration in S. cerevisiae. To produce green fluorescent protein (GFP)-tagged alleles, the CaVRG4 ORF was amplified by PCR as a BglII/SacI fragment and subcloned into pAW6 (a gift from A. Warenda, State University of New York, Stony Brook). pAW6 encodes the yeast optimized GFP yEGFP (13) at the MluI/BglII sites of YPB1-ADHp. The resulting plasmid, pCaV:GFP, contained the CaVRG4 ORF, tagged at the 3′ end with sequences encoding GFP, under the control of the strong Candida ADH promoter in a Candida URA3 ARS-containing vector.
pTiM1-A316D contains the Cavrg4-A316D allele, which carries a single missense mutation. The GCA codon, encoding Ala316, was changed to GAT (encoding Asp) by site-directed mutagenesis with a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) used according to the manufacturer’s instructions.
Disruption plasmid pSK−Cavrg4Δ::URA3 was constructed in several steps. First, 200 bp of the 5′ end (as an EcoRI/BamHI fragment) and 200 bp of the 3′ end (as a BamHI/XbaI fragment) flanking the CaVRG4 ORF were amplified by PCR and cloned into pBluescript SK(−), which resulted in a unique BamHI site between the 5′ and 3′ CaVRG4 sequences. The 2.86-kb BglII/BamHI hisG-CaURA3-hisG fragment from p5921 (22) was inserted into this BamHI site, resulting in a plasmid (pSK−Cavrg4Δ::URA3) containing the hisG-CaURA3-hisG region flanked by exactly 200 bp of the 5′ and 3′ ends of CaVRG4. This fragment was released prior to transformation by digestion with SacI and HindIII.
To generate a derivative of CaVRG4 that was under the control of the MET3 promoter, pDisCaVRG4 was constructed. About one-half of the 5′ portion of the CaVRG4 ORF (673 bp), including the first ATG, was amplified by PCR and cloned in the BamHI/PstI site of pCaDIS (9). Linearization of this plasmid at a unique BstEII site within the CaVRG4 sequence targeted this fragment to integrate at the chromosomal locus of CaVRG4, simultaneously truncating the endogenous VRG4 gene and inserting the MET3p-driven CaVRG4 gene.
Isolation of chromosomal DNA and Southern hybridization.
Overnight cultures (4 ml) were harvested, washed with water, and suspended in 0.4 ml of TENTS (2% Triton X-100, 1% sodium dodecyl sulfate [SDS], 100 mM NaCl, 10 mM Tris [pH 8.0], 1 mM EDTA). After addition of 0.4 ml of phenol-chloroform (1:1), cells were broken by vortexing them with glass beads. Nucleic acids in the aqueous phase were ethanol precipitated and digested with 10 μg of RNase per ml, and the remaining chromosomal DNA was ethanol precipitated and dissolved in 100 μl of Tris-EDTA. DNA probes were prepared by the random priming method with a Prime-It Fluor labeling kit (Stratagene). Southern transfer and hybridization were carried out as described previously (17). Detection was carried out with the Illuminator chemiluminescent detection system (Stratagene) used according to the manufacturer’s instructions.
Cloning and sequence analysis of the C. albicans VRG4 gene.
A single 3.5-kb BamHI/HindIII fragment, containing the 5′ end of the C. albicans VRG4 gene and flanking sequences, was detected by hybridization to the S. cerevisiae VRG4 (ScVRG4) gene. To isolate this BamHI/HindIII fragment, a mini-genomic library enriched for the DNA fragment was prepared. A 100-μg portion of genomic C. albicans CAI4 DNA was digested with BamHI and HindIII, and fragments that were between 3.0 and 5.0 kb long were size fractionated and cloned into pBluescript SK(−) (Stratagene). The resulting library was used to transform Escherichia coli for use in a modified bacterial hybridization screening analysis, performed as described previously (17), in which increasingly smaller pools of transformants were screened by Southern blot analysis to identify individual positive colonies. In the initial hybridization procedures we employed the ScVRG4 gene as a probe. A sequence from the Candida sequencing project that resembled the 3′ portion of the ScVRG4 gene allowed the corresponding C. albicans fragment to be amplified by PCR. This fragment was subsequently used as a homologous probe. Positive clones were confirmed by Southern blot analysis. An additional 1.2-kb HindIII/HindIII fragment, containing the 3′ end of the CaVRG4 ORF and flanking DNA, was isolated as described above to allow reconstruction of the entire C. albicans VRG4 gene. Sequence analysis was performed by the dideoxy chain termination method using a Thermo Sequenase cycle sequencing kit (Amersham Pharmacia Biotech). DNA sequence analysis was performed with an automated LI-COR 4000L DNA sequencer.
Disruption of the CaVRG4 locus.
Gene disruption was performed in two steps. The first allele of CaVRG4 was deleted by utilizing the URA blaster protocol and the hisG-CaURA3-hisG module (22). pSK−CaVRG4Δ::URA3 (see above) was linearized with HindIII and SacI and transformed into C. albicans BWP17 (58). Uracil prototrophs were isolated on SD(−Ura) medium. Putative heterozygotes were screened by Southern blotting to confirm disruption of the CaVRG4 locus (e.g., strain ANC1). Forced excision of the URA3 gene by selection on 5′-fluoroorotic acid (5′-FOA) allowed recovery of uracil auxotrophs. A suitable strain (ANC2) was verified by Southern blot analysis and transformed with pDISCaVRG4 (see above) in order to simultaneously delete the remaining VRG4 allele and introduce the MET3p-VRG4 allele. The genotypes of the resulting hemizygous strains (e.g., ANC3) were verified by Southern blot analysis (Table 1).
Complementation analyses.
The CaVRG4 gene was introduced into S. cerevisiae NDY5, which contains the vrg4-2 allele (41), or S. cerevisiae XGY14, which contains the glucose-repressible GAL1p-VRG4 allele (25). CaVRG4 and ScVRG4 were introduced into each of these strains on integrative plasmids pTiM1 and pTαO-ScVRG4 (25), respectively. Complementation of the hygromycin B-sensitive phenotype of NDY5 was assayed by growth on YPAD agar supplemented with 30 μg of hygromycin B per ml (18). Complementation of the lethality associated with the glucose-dependent repression of VRG4 expression in XGY14 was assayed by comparing growth on YPA containing galactose and growth on YPAD (25).
Alcian blue binding assay.
Alcian blue dye binding assays were carried out as described previously (38), with the following modifications. C. albicans strains were grown to the stationary phase in YPAD(+Uri) with or without Met/Cys at 30°C. After washing and resuspension in H2O, each of the cultures was divided into two aliquots. One aliquot was dried to determine the dry weight, while the other was subjected to Alcian blue dye binding. The amounts of dye bound by different C. albicans strains were determined spectrophotometrically (at 600 nm) by measuring the optical density of Alcian blue that remained in the supernatant after the binding reaction. For each sample, values (see Fig. 8B) were normalized to the cell dry weight and expressed as a percentage of the value obtained for parental strain BWP17 that had been grown in YPAD, as follows: DB = (OD600 of dye solution − OD600 of cell supernatant)/cell dry weight; and relative dye binding = (DB of strain/DB of BWP17 grown in YPAD) (100), where DB is dye binding.
FIG. 8.
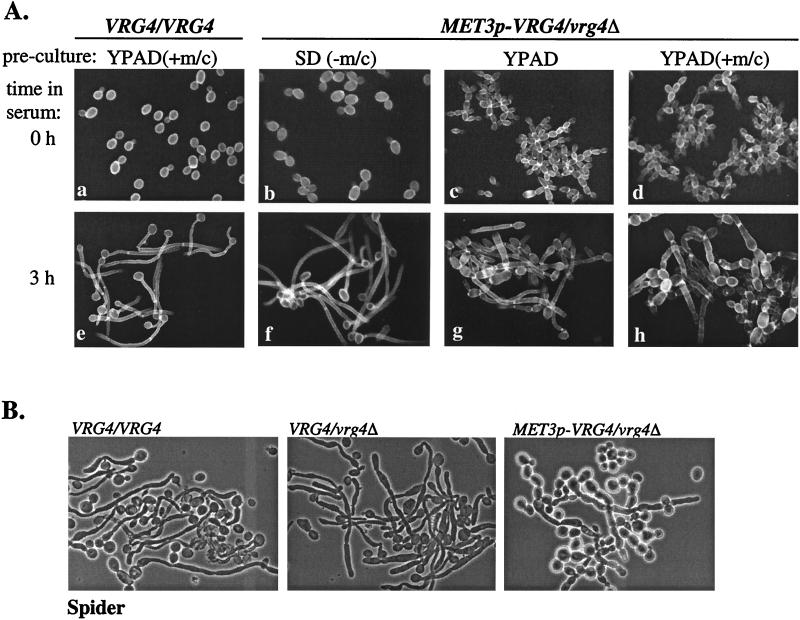
Repression of VRG4 prevents hyphal formation on liquid and solid media. (A) C. albicans parental wild-type strain BWP17 and MET3p-VRG4 hemizygous strain ANC3 were precultured in SD(−Met/Cys), YPAD, or YPAD(+Met/Cys) for 12 h prior to induction of hyphal formation by growth at 37°C in YPAD containing 10% bovine calf serum. At zero time or 3 h after serum induction, aliquots were collected, stained with Calcofluor white, and examined by fluorescence microscopy. (B) Candida strains BWP17, ANC1 (VRG4/vrg4), and ANC3 were spotted on Spider medium plates and incubated for 3 days at 37°C. Individual cells from each colony were examined microscopically.
Immunoprecipitation, Western blot, and immunofluorescence analyses.
HA-tagged CaVrg4p was detected by immunoblot analysis of whole-cell lysates prepared from S. cerevisiae by trichloroacetic acid precipitation as described previously (12), using anti-HA mouse antibodies. Primary antibodies were bound to horseradish peroxidase (HRP)-coupled mouse immunoglobulin G and were detected by the enhanced chemiluminescence system (Amersham Pharmacia Biotech).
For analysis of concanavalin A (ConA)-reactive proteins, proteins in whole-cell lysates were isolated and processed as described above, but the membranes were blocked with 0.2% Tween 20 in phosphate-buffered saline. Glycoproteins were probed with 0.2 μg of HRP-labeled ConA (Sigma Chemical Co., St. Louis, Mo.) in phosphate-buffered saline and were detected by chemiluminescence.
HA-tagged proteins were immunoprecipitated by incubating a 0.5% Triton X-100 detergent extract with anti-HA mouse antibodies as described previously (24). Proteins in whole-cell lysates and in anti-HA immunoprecipitates were solubilized in Laemmli sample buffer and fractionated by SDS-12% polyacrylamide gel electrophoresis (PAGE). After transfer, proteins on membranes were blotted with anti-HA rabbit polyclonal antibodies (Santa Cruz Biotechnology), and secondary HRP-conjugated anti-rabbit antibodies were detected by chemiluminescence.
Strains harboring CaVRG4-GFP plasmids (pCaV:GFP and pAW6) were grown in SD(−Ura) medium to an OD600 of 2, spotted onto glass slides, and viewed immediately by fluorescence microscopy. Cell were stained with Calcofluor white as described previously (27). Indirect immunofluorescence analysis of yeast cells expressing ScVrg4-HAp or CaVrg4-HAp and data analyses were performed as described previously (19).
In situ acid phosphatase assay.
Native gel electrophoresis and activity staining of acid phosphatase from C. albicans were carried out as described by Schweingruber et al. (45) for the homologous Schizosaccharomyces pombe enzyme, with the following modifications. Cells were grown in YPAD(+Uri) to the log phase (OD600, 5), and acid phosphatase activity was induced by overnight cultivation at 30°C in 20 ml of phosphate-reduced YPAD(+Uri) depleted of phosphate as described previously (59). After the cells were washed once in 62.5 mM Tris-HCl (pH 6.8), they were resuspended in 200 μl of lysis buffer (59) and broken by vortexing with glass beads (three times, 20 s each). The lysate was centrifuged for 10 min, and the resulting supernatant was transferred and centrifuged for 10 min to remove insoluble precipitates. The supernatant was mixed with 0.5 volume of sample buffer (59), and 5 μl was applied to a gel and subjected to 5% native gel electrophoresis at 4°C as described previously (45). After incubation for 10 min in 100 mM sodium acetate (pH 4.0) (SA), the gel was incubated at 37°C in SA containing 0.5 mg of α-naphthyl acid phosphate (Sigma) per ml for 1 h. Color was developed by adding 50 ml of SA (prewarmed to 60°C) containing 0.7 mg of o-dianisidine (tetrazotized; Sigma) per ml and incubating the preparation at 60°C for 10 min. Endoglycosidase H (endo H) (New England Biolabs) was added, but samples were digested in the absence of heat or detergent denaturation. Endo H-digested samples were resolved by 8% polyacrylamide native gel electrophoresis but otherwise were treated as described above.
Nucleotide sequence accession number.
The CaVRG4 gene and protein sequences have been deposited in the GenBank database under accession number AF164627.
RESULTS
Isolation and analysis of the C. albicans VRG4 gene.
Using genomic Southern analysis, we identified and isolated a DNA fragment that cross-hybridized to the S. cerevisiae VRG4 gene (see Materials and Methods). Subsequent cloning and sequence analysis demonstrated that this fragment exhibited sequence homology to the 5′ end of the S. cerevisiae VRG4 ORF. The full-length CaVRG4 gene was reconstructed by isolating a genomic fragment that contained the 3′ end of CaVRG4 and downstream flanking sequences (see Materials and Methods). The sequence of the entire 7.7-kb HindIII fragment was determined. In addition to the CaVRG4 gene, this fragment also contained in its 5′ portion a sequence that is very similar to the S. cerevisiae ADE4 gene, suggesting that the CaVRG4 gene is adjacent to and 3′ with respect to CaADE4.
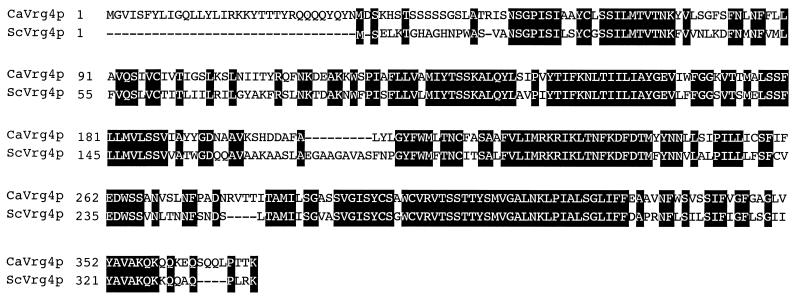
The sequence of the predicted CaVrg4 protein (GenBank accession no. AF164627.) and alignment of this sequence with the S. cerevisiae Vrg4p sequence are shown in Fig. 1. The CaVRG4 ORF encodes a 371-amino-acid polypeptide with a predicted molecular mass of 41.2 kDa; the molecular mass of ScVrg4p is 36.8 kDa. The two proteins are related along their lengths (65% identical and 79% similar) except for an additional 33-amino-acid N-terminal region in CaVrg4p that is not present in ScVrg4p.
FIG. 1.
Alignment of the S. cerevisiae and C. albicans Vrg4 proteins. Identical residues are shaded. The GenBank accession number of ScVRG4 is L33915.
CaVRG4 encodes a functional homologue of S. cerevisiae VRG4.
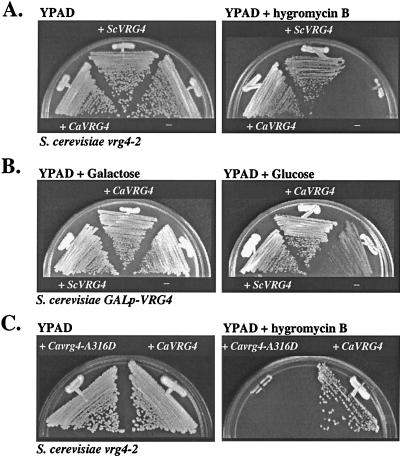
Although deletion of the S. cerevisiae VRG4 gene is lethal, vrg4-2 is a viable allele. Like growth of other S. cerevisiae glycosylation mutants, growth of a vrg4-2 mutant is inhibited by the aminoglycoside hygromycin B (4, 18). As a test for functionality, the CaVRG4 gene was examined to determine its ability to suppress the drug sensitivity of an S. cerevisiae vrg4-2 mutant. Expression of the CaVRG4 gene, driven by the constitutive S. cerevisiae TPI1 promoter, fully restored growth of the vrg4-2 mutant on medium supplemented with hygromycin B (Fig. 2A).
FIG. 2.
Complementation of S. cerevisiae vrg4 mutants by the CaVRG4 gene. (A) Complementation of hygromycin B-sensitive growth of the S. cerevisiae vrg4-2 mutant. Integrative plasmids expressing CaVRG4 (pTiM1) or ScVRG4 (pTαO-VRG4) (25) were introduced into NDY5 (vrg4-2), as described in Materials and Methods. Cells were plated on YPAD with or without 30 μg of hygromycin B per ml and incubated at 30°C for 3 days. (B) Complementation of the lethality associated with loss of VRG4 function, as assayed in the S. cerevisiae GAL1p-VRG4 strain. Integrative plasmids expressing CaVRG4 (pTiM1) or ScVRG4 (pTαO-VRG4) (25) were introduced into S. cerevisiae GAL1p-VRG4 strain XGY14 as described in Materials and Methods. Strains were incubated for 3 days on YPA plates containing galactose or glucose. (C) Integrative plasmids containing wild-type CaVRG4 (pTiM1) or the Cavrg4-A316D allele (pTiM1-A316D) introduced into S. cerevisiae vrg4-2 strain NDY5 and assayed for complementation of the hygromycin B-sensitive growth phenotype as described above.
As a second test for functionality, complementation of the lethality associated with loss of VRG4 function was examined. The CaVRG4 gene was introduced into an S. cerevisiae strain containing VRG4 under the control of the glucose-repressible GAL1 promoter. This strain grows normally in the presence of galactose but does not grow when VRG4 gene expression is repressed in the presence of glucose. Expression of the CaVRG4 gene and expression of the ScVRG4 gene in this strain resulted in comparable growth rates on glucose-containing media (Fig. 2B), further suggesting that the CaVRG4 gene is a functional homologue of its S. cerevisiae counterpart.
ScVrg4p is specific for transport of GDP-mannose, and a region required for binding to GDP-mannose has been mapped to a cytoplasmically oriented region of the protein within the GALNK motif (25). The vrg4-2 allele has a single missense mutation in the GALNK region (A286D) that impairs binding to GDP-mannose (25). As another, more stringent test for the functional redundancy of the C. albicans and S. cerevisiae Vrg4 proteins as GDP-mannose transporters, we examined whether this single amino acid, which is a critical component of the GDP-mannose binding domain, is functionally conserved in the CaVrg4 protein. Site-directed mutagenesis was used to replace the corresponding amino acid, Ala316 of the CaVrg4 protein, with Asp (see Materials and Methods), and the Cavrg4-A316D allele was analyzed for functionality by complementation. When expressed in an S. cerevisiae strain containing the vrg4-2 allele, the Cavrg4-A316D allele did not complement the growth defect on medium supplemented by hygromycin B (Fig. 2C). This mutant gene also did not complement growth of the GAL1-VRG4 strain on glucose-containing media (data not shown). These results demonstrate that mutation of this single amino acid, which was previously shown to define the substrate specificity of GDP-mannose transport by Vrg4p, results in loss of CaVrg4p function and that the same amino acid is important for the functions of both proteins. Together with data which suggest that CaVrg4p resides in the Golgi apparatus (see below), these results imply that the CaVRG4 gene mediates lumenal transport of GDP-mannose into the Golgi apparatus.
CaVrg4 protein resides in the Golgi apparatus.
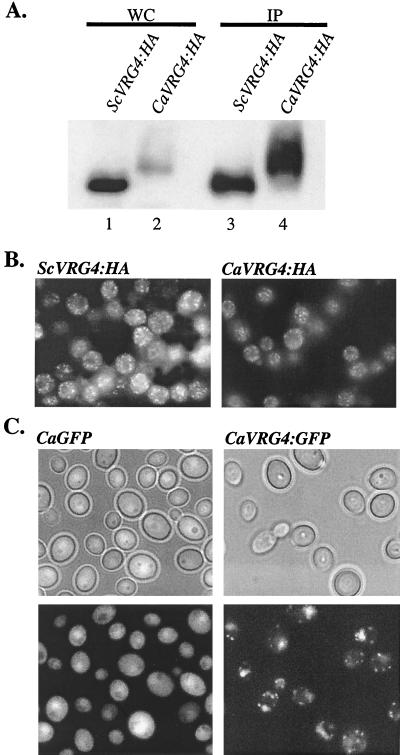
To examine the properties of the CaVrg4 protein, an allele that encodes HA-tagged CaVrg4p was constructed and expressed in S. cerevisiae. The HA-tagged CaVrg4 protein was functional in S. cerevisiae, as the CaVRG4-HA allele could complement an S. cerevisiae vrg4 mutant (data not shown). When examined by immunoblot analysis in whole-cell protein extracts, the level of CaVrg4-HAp that accumulated at steady state was much lower (about 10-fold lower) than the level of ScVrg4-HAp (Fig. 3A, lanes 1 and 2). Detection of CaVrg4p at levels that were comparable to ScVrg4p levels required enrichment by immunoprecipitation (lanes 3 and 4). The reduced level of CaVrg4p was probably not due to differences between the transcription rates of the S. cerevisiae and Candida VRG4-HA genes, since both genes were integrated at the same locus and expressed under the control of the TPI promoter. Since the level of CaVRG4 gene expression fully complemented S. cerevisiae vrg4 mutants, the presence of even very low levels of CaVrg4p at steady state is apparently sufficient for function in S. cerevisiae. CaVrg4p migrated more heterogeneously and at a higher molecular weight than ScVrg4p during SDS-PAGE (Fig. 3A, lanes 1 and 2). The heterogeneity with which CaVrg4p migrated indicates that it may undergo posttranslational modification, although this was not investigated further. The increased molecular weight of CaVrg4p is consistent with its predicted additional N-terminal domain that is not present in S. cerevisiae Vrg4p.
FIG. 3.
Expression and intracellular localization of CaVrg4p. (A) Western blot analysis of CaVrg4-HA and ScVrg4-HA proteins. Whole-cell protein extracts were prepared from S. cerevisiae strains that were grown in YPAD to repress the endogenous GAL1p-VRG4 gene (XGY14) (25) and harbored plasmids expressing either ScVRG4-HA (pTiVRG4-HA) (25) or CaVRG4-HA (pTiM1-HA). Equivalent amounts of proteins were directly applied to SDS-polyacrylamide gels (WC) (lanes 1 and 2) or were first immunoprecipitated with anti-HA mouse antibodies (IP) (lanes 3 and 4), and then the proteins were Western blotted with anti-HA rabbit antibodies and detected by chemiluminescence as described in Materials and Methods. (B) Indirect immunofluorescence of S. cerevisiae strains expressing ScVRG4-HA or CaVRG4-HA. S. cerevisiae strain XGY14 expressing ScVRG4-HA or CaVRG4-HA (as described above) was grown in YPAD to the log phase. Cells were fixed and treated with anti-HA antibodies, followed by fluorescein isothiocyanate-conjugated anti-mouse immunoglobulin G. (C) CaVRG4-GFP (pCaV:GFP) or GFP alone (pAW6) was expressed in C. albicans strain ANC2, and live cells were examined by fluorescence microscopy.
In S. cerevisiae the Vrg4 protein resides in the Golgi compartment, and this localization is critical for function. Mutations in the N-terminal Vrg4p domain that cause mislocalization result in lethality (24). Since CaVrg4p could functionally substitute for S. cerevisiae Vrg4p, we inferred that it was also localized in the Golgi apparatus. To confirm that this was the case, the intracellular locations of CaVrg4-HAp and ScVrg4-HAp in S. cerevisiae were examined by indirect immunofluorescence analysis. CaVrg4-HAp displayed the same fluorescence pattern as ScVrg4-HA albeit with a reduced intensity, residing in punctate spots throughout the cytoplasm that distinguished the Golgi apparatus in S. cerevisiae (Fig. 3B). The decreased intensity of CaVrg4-HAp fluorescence is in agreement with our finding that suggested that the level of expression of this protein in S. cerevisiae was reduced (Fig. 3A). This finding demonstrated that CaVrg4p contains the information for localization to the Golgi apparatus of S. cerevisiae.
Little is known about the structure of the Candida Golgi apparatus or its spatial distribution during morphogenetic switching. To examine CaVrg4p localization in live Candida cells, a fragment encoding the codon-optimized yEGFP (13) was fused in frame to the 3′ end of CaVRG4. The fusion construct was placed under control of the ADH1 promoter in an ARS/URA3-containing plasmid. A plasmid that was identical except that it lacked the CaVRG4 sequences was also constructed. Both plasmids were introduced into a wild-type strain (BWP17) and a heterozygotic vrg4 Candida strain (ANC2), and GFP localization was monitored by fluorescence microscopy. No apparent difference between the morphology or phenotype of cells expressing ADH-driven CaVRG4-GFP and the morphology or phenotype of the control cells was observed. Cells expressing GFP alone exhibited an evenly distributed pattern of fluorescence throughout the cytoplasm, except for the vacuole. In contrast, the CaVrg4-GFP fusion protein localized in punctate spots (Fig. 3C). Its localization pattern was similar to, but in some ways distinct from, that seen in S. cerevisiae. In Candida, there were fewer spots, which tended to be more rod-like than the spots in S. cerevisiae. In agreement with these observations, previous studies in which the workers compared the morphologies of different yeasts by electron microscopy suggested that there was a difference in the appearance of structures hypothesized to be the Golgi apparatus in C. albicans and S. cerevisiae (42). Although preliminary, the different localization patterns of CaVrg4p in S. cerevisiae and C. albicans suggest that the structures or distributions of the Golgi apparatus in these two organisms may be different.
CaVRG4 is an essential gene.
In order to determine the phenotypic consequences of loss of CaVRG4 function, we attempted to construct a homozygous vrg4Δ strain. One allele of CaVRG4 was deleted by using the URA-blaster technique (22). The disruption replaced the entire VRG4 ORF with the CaURA3 gene that was flanked by Salmonella enterica serovar Typhimurium hisG repeats (see Materials and Methods). Uracil prototrophs (e.g., strain ANC1) were plated on 5′-FOA-containing medium. Forced excision of the URA3 gene and deletion of CaVRG4 in 5-FOA-resistant uracil auxotrophs (e.g., strain ANC2) were confirmed by Southern blot analysis (Fig. 4B). To disrupt the second copy of CaVRG4, this procedure was repeated, but no homozygous null mutants were recovered. This procedure was performed with both C. albicans CAI4 and BWP17. Analysis of more than 50 transformants recovered after the second round of transformation indicated that the Cavrg4Δ::hisG-URA3-hisG disruption fragment had preferentially integrated into the previously disrupted allele (data not shown). This biased integration was due either to selective recovery of wild-type cells because of the lethality associated with loss of the CaVRG4 function or to an allele-specific bias in homologous recombination. As VRG4 is an essential gene in S. cerevisiae, the former possibility was the preferred model.
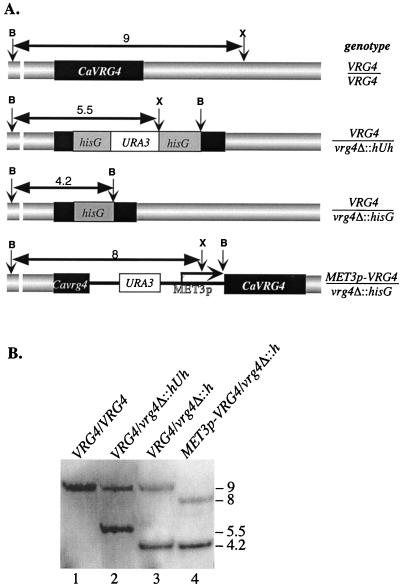
FIG. 4.
Disruption of the CaVRG4 gene. (A) Schematic restriction maps of CaVRG4 and each of the disrupted alleles used to construct the heterozygous VRG4/vrg4Δ and hemizygous MET3p-VRG4/vrg4Δ strains. B and X indicate the positions of BamHI and XbaI restriction sites, respectively, in the CaVRG4 wild-type, vrg4Δ::hisG-URA3-hisG, vrg4Δ::hisG, and MET3p-VRG4 alleles, and the predicted size (in kilobases) of each of the fragments is indicated. (B) Southern blot analysis of C. albicans genomic DNA digested with BamHI and XbaI and isolated from wild-type VRG4/VRG4 (BWP17), heterozygous VRG4/vrg4Δ::hisG-URA3-hisG (ANC1) and VRG4/vrg4Δ::hisG (ANC2), and hemizygous MET3p-VRG4/vrg4Δ::hisG (ANC3) strains. The 3.5-kb BamHI/HindIII fragment that was used as a probe hybridized uniquely to each of the BamHI/XbaI or BamHI/BamHI fragments that are indicated by the arrows in panel A.
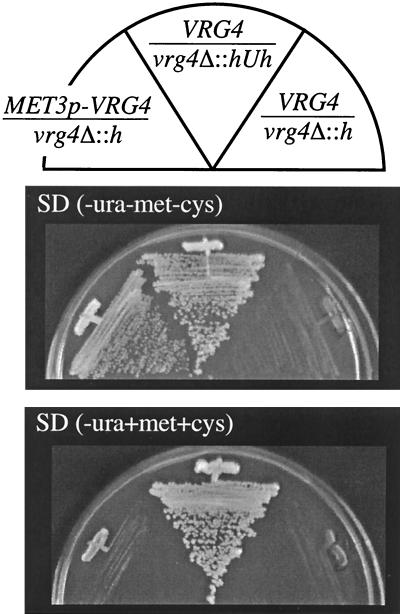
As an alternative strategy to test the null phenotype, we constructed a hemizygous strain that contained a single, conditional VRG4 allele (Fig. 4A). The remaining copy of VRG4 was truncated in a VRG4/vrg4Δ::hisG strain (ANC2), and a methionine-repressible MET3p-driven VRG4 allele was simultaneously introduced (see Materials and Methods) (9). Uracil prototrophic transformants were selected under conditions that allowed CaVRG4 expression (i.e., methionine and cysteine were not present in the growth medium), and the genotypes of the resulting strains (e.g., ANC3) were verified by Southern blot analysis (Fig. 4B). The strains grew on media lacking Met/Cys, although less well than the parental VRG4/VRG4 or VRG4/vrg4Δ strains grew, suggesting that regulated expression by the MET3 promoter did not induce VRG4 at levels or in temporal patterns equivalent to those induced by the VRG4 promoter. The strains did not grow on solid SD medium supplemented with 5 mM methionine and 1 mM cysteine (Fig. 5). This result demonstrated that CaVRG4 is an essential gene.
FIG. 5.
CaVRG4 is an essential gene. Heterozygous uracil prototrophic and auxotrophic strains ANC1 (VRG4/vrg4Δ::hisG-URA3-hisG) and ANC2 (VRG4/vrg4Δ::hisG) and the hemizygotic MET3p-VRG4 vrg4Δ::hisG strain ANC3 were plated on SD(−Ura) medium in the absence or in the presence of methionine and cysteine.
In contrast to its behavior on solid SD(+Met,+Cys) medium, the hemizygous MET3p-VRG4/vrg4Δ strain continued to grow in liquid media (SD and YPAD) that were supplemented with Met/Cys, although its doubling rate was highly variable compared to that of the parental strain. One factor that contributed to this variability was the density of the inoculum. When cells were inoculated at a very low density (OD600, <0.05), methionine-dependent VRG4 repression was relatively efficient, while cultures inoculated at an OD600 greater than 0.5 reached an OD600 comparable to that of wild-type cell cultures after 12 h (data not shown). Presumably, the uptake or metabolism of methionine in dense cultures depleted the effective methionine and/or cysteine so that the concentration was below the concentration required for efficient repression of growth.
Although not reported previously, concentrations of methionine and cysteine greater than 10 and 5 mM, respectively, also affected the growth and morphology of the VRG4/VRG4 parental strains (CAI4 and its derivative, BWP17). At these concentrations or higher concentrations, methionine and cysteine reduced the growth rates and accelerated hyphal formation, effects that were exacerbated by URA3 prototrophy (data not shown). Therefore, for most of the analyses described below, the effects of MET3p-VRG4 repression were examined in liquid media supplemented with 5 mM methionine and 2 mM cysteine, and overnight cultures grown in SD(−Met,−Cys,−Ura) that were diluted to an OD600 of about 0.2 were used. These conditions had no detectable effect on the growth of the parental strains and reduced, but did not eliminate, the growth of the MET3p-VRG4 strain. Since growth was not eliminated, these growth conditions provided the opportunity to analyze the phenotypes associated with a partial loss of VRG4 function.
Analysis of glycosylation and cell surface defects in Candida vrg4 mutants.
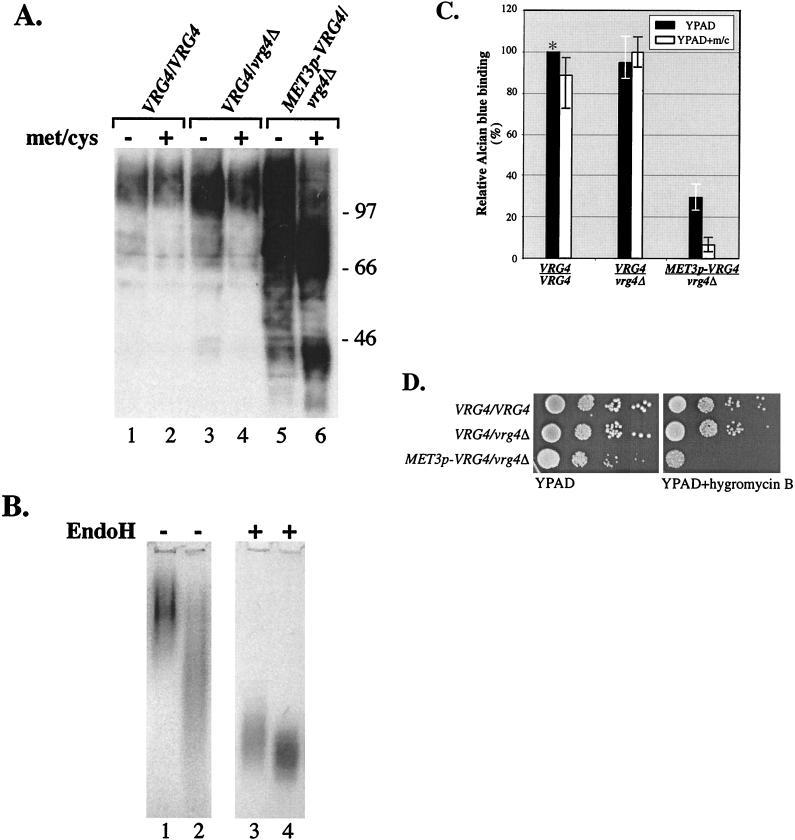
As an indirect measure of mannosylation defects, the electrophoretic mobilities of glycoproteins in wild-type and mutant Candida cell extracts were compared. Protein extracts were prepared from cultures of the parental VRG4/VRG4, heterozygous VRG4/vrg4Δ, and hemizygous MET3p-VRG4 strains grown in YPAD with or without additional Met/Cys. Proteins were fractionated by SDS-PAGE and examined by Western blotting with the mannose-specific lectin HRP-ConA in order to detect changes in the molecular weights of mannoproteins (Fig. 6A). Even when extracts were prepared from cells that had been grown in the absence of additional Met/Cys, the HRP-ConA-reactive proteins in the MET3p-VRG4/vrg4Δ strain (ANC3) had a lower average molecular weight than the HRP-ConA-reactive proteins in either the parental wild-type or heterozygous VRG4/vrg4Δ strain (Fig. 6A, lane 5). This result suggested that YPAD contains sufficient levels of endogenous methionine and/or cysteine to repress MET3p-VRG4 expression. In extracts from mutant Cavrg4 strains grown in the presence of additional Met/Cys, there was a marked decrease in higher-molecular-weight ConA-reactive proteins and a further increase in the lower-molecular-weight ConA-reactive proteins (Fig. 6A, lanes 5 and 6). These results suggest that mannosylation of proteins is severely affected by loss of CaVRG4 function.
FIG. 6.
Loss of CaVRG4 function leads to mannosylation and cell wall-associated defects in C. albicans. (A) Proteins were isolated from wild-type strain BWP17, VRG4/vrg4Δ::hisG strain ANC2, and hemizygotic MET3p-VRG4/vrg4Δ::hisG strain ANC3 grown in YPAD with or without additional methionine and cysteine. After fractionation of equivalent amounts of protein by SDS-8% PAGE, mannoproteins were blotted with ConA-HRP and detected by chemiluminescence, as described in Materials and Methods. (B) Acid phosphatase in wild-type strain BWP17 (lanes 1 and 3) and vrg4 mutant ANC3 (lanes 2 and 4) assayed by native gel electrophoresis and visualized colormetrically (see Materials and Methods). Extracts were not treated or incubated with endo H prior to electrophoresis, as indicated at the top. (C) The strains used in the experiment whose results are shown in panel A were incubated with Alcian blue and subjected to dye binding assays (see Materials and Methods). Relative dye binding activity was calculated by determining the percentage of dye bound compared to the amount bound by the parental strain (BWP17) that had been grown in YPAD (see Materials and Methods). Each bar represents an average based on the results of four experiments. m/c, methionine and cysteine. (D) The strains used in the experiment whose results are shown in panel A were serially diluted and plated on YPAD or YPAD that was supplemented with 150 μg of hygromycin B per ml.
The increased level of lower-molecular-weight glycoproteins described above was consistent with a vrg4-dependent mannosylation defect. To confirm this, we wanted to analyze the effect of the vrg4 mutation on carbohydrate modification of a single glycoprotein. Gel mobility shifts of secreted glycoprotein reporters, such as invertase, chitinase, and acid phosphatase, whose activities and secretion are not dependent on glycosylation, have been important tools for analyzing glycosylation mutants of S. cerevisiae. Therefore, we tried to use this approach to analyze the well-characterized C. albicans acid phosphatase. C. albicans-secreted acid phosphatase is a ∼130-kDa mannoprotein with a pH optimum of 3.6 to 4.5 (39).
To determine if acid phosphatase is undermannosylated in the vrg4 mutant, native gel electrophoresis was used to analyze its mobility. Crude extracts were prepared under nondenaturing conditions from hemizygous MET3p-VRG4 and wild-type strains grown in low-phosphate YPAD to induce acid phosphatase expression. Protein extracts were not treated or incubated with endo H to remove N-linked oligosaccharides. Acid phosphatase was visualized in the gel with a colormetric activity stain (Fig. 6B). Acid phosphatase migrated as a diffuse band whose average molecular weight was lower in vrg4-derived extracts (Fig. 6B, lanes 1 and 2). After removal of N-linked oligosaccharides with endo H, acid phosphatase from both vrg4 mutant and wild-type strains migrated more rapidly, but it was still somewhat diffuse. Consistent with the idea that the remaining heterogeneity might be due to O-linked oligosaccharides whose additions are also affected by loss of VRG4, the mobility of acid phosphatase in vrg4 extracts was slightly greater than the mobility in wild-type extracts (Fig. 6B, lanes 3 and 4). These results demonstrate that the vrg4 mutation affects protein mannosylation.
Cell surface hydrophobicity is considered to be an important virulence determinant of C. albicans. The length of oligomannosides, linked to protein outer chain mannose via phosphodiester bonds (i.e., phosphomannan), has been correlated with cell surface charge, leading to hydrophilic or hydrophobic C. albicans strains (32, 33). In S. cerevisiae, mannosylphosphate gives a net negative charge to cell surface mannoproteins, which in turn allows binding to the dye Alcian blue under weakly acidic conditions (23). To test whether CaVRG4 affects the level of cell surface phosphomannan, we measured the extents of Alcian blue binding to parental wild-type and Cavrg4 mutant strains (see Materials and Methods). As shown in Fig. 6C, the wild-type and vrg4 heterozygotic strains displayed equivalent dye binding affinities, whether they were grown in the presence or in the absence of additional Met/Cys. In contrast, the MET3p-VRG4 strain was markedly defective in dye binding capacity. The MET3p-VRG4 strain exhibited a threefold reduction in dye binding activity when it was grown in YPAD, and the amount of dye bound was less than 10% of the amount of dye bound by the parental wild-type strain when it was grown in YPAD containing Met/Cys. These results suggested that CaVRG4 is required for phosphomannan addition and imply that, as in S. cerevisiae, phosphomannan addition in C. albicans requires a mannosylation step in the Golgi apparatus that utilizes GDP-mannose as a substrate.
S. cerevisiae mutants with defects in protein glycosylation or cell wall synthesis display a number of characteristic phenotypes when they are grown on media supplemented with sodium vanadate, hygromycin B, and Calcofluor white. To determine whether defects in mannan biosynthesis in C. albicans result in similar drug phenotypes, we examined the growth of Cavrg4 strains on media supplemented with these compounds. The MET3p-VRG4 hemizygous strain ANC3 was hypersensitive to hygromycin B and did not grow at all on YPAD plates containing 150 μg of hygromycin B per ml (Fig. 6D). Growth of the parental strains, CAI4 and BWP17, and growth of the VRG4/vrg4Δ heterozygotes were not affected, even at a hygromycin B concentration of more than 200 μg/ml (data not shown). In contrast to the effects of hygromycin B, a defect in CaVRG4 had no effect on the sensitivity of growth to Calcofluor white or resistance to sodium vanadate (data not shown), both of which are phenotypes displayed by Scvrg4 mutants. These results further underscore the differences that exist between S. cerevisiae and C. albicans with regard to cell wall composition and/or drug uptake and efflux mechanisms.
Effect of CaVRG4 on cellular morphology.
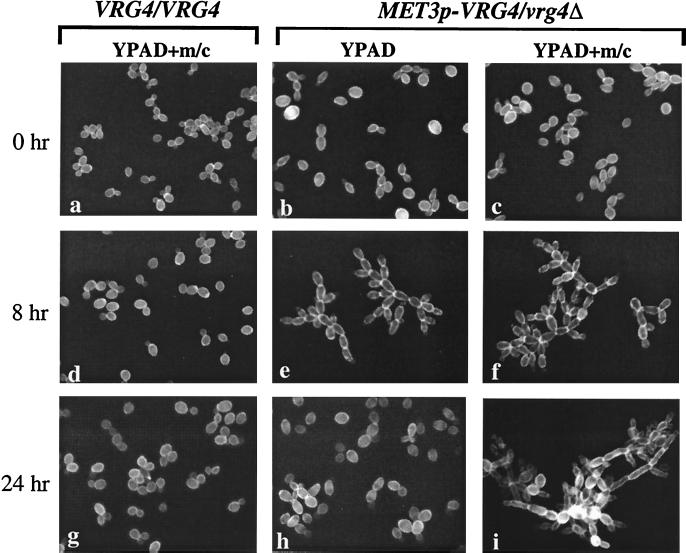
Sustained growth of a hemizygotic MET3p-VRG4 strain in liquid media (either YPAD or SD, supplemented with Met/Cys) eventually led to cultures whose optical densities approached those of cultures of the parental strains. However, the increased optical densities did not wholly reflect increased numbers of cells. Rather, repression of VRG4 expression resulted in a profound change in cell morphology (Fig. 7). Initially, the cells grew as short chains, which aggregated. After 4 to 8 h, long chains of cells attached to or branching from one another were observed, and by 24 h after inoculation most of the cells in each population had assumed a polarized, pseudohyphal mode of growth, and there were obvious constrictions at the sites of septation (Fig. 7). After 24 h in the presence of Met/Cys, many cells became vacuolated and unrefractive, as determined by light microscopy, and there was evidence of considerable cell lysis in the cultures (data not shown). Most of the cells also exhibited irregularities in surface staining by Calcofluor white, suggesting that the thickness of the cell wall was not uniform. The characteristic clumpiness and chain mode of growth occurred even in the absence of additional Met/Cys in YPAD. This was consistent with the results (Fig. 6), which demonstrated that cells grown in YPAD were defective in mannosylation and that the endogenous levels of Met/Cys in rich medium are sufficient for MET3 promoter repression. After 24 h of growth in YPAD (not supplemented with additional Met/Cys) there was no evidence of the elongated, attached cells (Fig. 7) or pseudohyphae that dominated the cultures after 8 h. This observation suggested that continued growth in liquid media might decrease the level of Met/Cys below the level required to repress the MET3 promoter. Together, these results demonstrate that a defect in Golgi mannosylation induces the yeast cells to a transition to pseudohyphal growth and that this transition is reversible.
FIG. 7.
Repression of CaVRG4 promotes pseudohyphal growth. Overnight precultures grown in SD(−Met/Cys) plus 10% YPAD were diluted into YPAD or YPAD containing additional methionine and cysteine (5 and 2.5 mM, respectively). At the times indicated, aliquots were removed, stained with Calcofluor white, and examined by fluorescence microscopy using a UV filter.
Effect of CaVRG4 on hyphal formation.
It is presumed that hyphal formation plays a key role in the pathogenesis of C. albicans. As hyphal formation requires substantial cell wall biosynthesis and/or remodeling, we expected that a defect in mannan biosynthesis would impair hyphal growth. To examine the effect of CaVRG4 on hyphal formation, the morphologies of Cavrg4 strains that were induced to form hyphae under various conditions were examined. Strains were precultured in various liquid media, as indicated in Fig. 8A, and were induced to form hyphae by dilution in YPAD containing 10% calf serum at 37°C. No differences in the rate or appearance of hyphae formed by the MET3p-VRG4 strains were observed when strains were compared to the parental strain and the organisms were precultured in SD(−Met,−Cys) (Fig. 8A, micrograph f). Both the parental strain and the MET3p-VRG4 hemizygous strain initially formed germ tubes (data not shown) and then formed true hyphae that lacked constrictions between septa. This result was surprising since our previous results demonstrated that MET3p-VRG4 expression was partially repressed in YPAD, as shown by the growth of the strain as chains of cells or pseudohyphal cells. No evidence of these growth forms was observed during hyphal induction in YPAD. Since hyphal induction in liquid occurs very rapidly (within 1 to 3 h), one possibility to explain the efficiency of hyphal formation in the MET3p-VRG4 strain was that transcriptional repression of the MET3 promoter phenotypically lagged or was recessive to induction of hyphae. To test this possibility, cells precultured in SD(−Met/Cys) were transferred and first grown in YPAD with or without additional Met/Cys for 12 h prior to hyphal induction. As shown in Fig. 8A, repression of VRG4 expression prior to hyphal induction by growth in YPAD (micrograph g) or in YPAD that contained additional Met/Cys (micrograph h) resulted in cells that failed to form germ tubes (data not shown) or true hyphae. Instead, transfer of these cells into serum-containing media at 37°C resulted in rapid induction of pseudohyphae. Under these conditions, even after prolonged incubation in the presence of serum, no true hyphae were seen in MET3p-VRG4 cell cultures. The mutant strains also did not form hyphae when they were grown at 37°C on mannitol-containing Spider medium plates (Fig. 8B) or on solid YPAD containing 4 or 10% serum (data not shown), whether the media were supplemented or not supplemented with additional Met/Cys. On solid media at 37°C, all strains displayed similar, rough colony morphologies after 2 days of incubation in the presence of Met/Cys or after 5 days of incubation in the absence of Met/Cys, demonstrating that methionine and/or cysteine dramatically accelerated filamentation (data not shown). Despite similar colony morphologies, microscopic examination of cells indicated that the parental and VRG4 heterozygous strains formed true hyphae, while only pseudohyphae or elongated cells were observed for the MET3p-VRG4 strains (Fig. 8B). These results demonstrate that CaVRG4 is required for hyphal formation.
DISCUSSION
In this work we examined the role of mannosylation in C. albicans by analyzing the VRG4 gene. CaVRG4 was cloned on the basis of its structural similarity to the ScVRG4 gene and was shown to be its functional homologue. Like ScVRG4, CaVRG4 is an essential gene, implying that there is no other functionally redundant Candida gene that encodes GDP-mannose transport activity. Construction of a hemizygous Candida strain containing a conditional MET3p-VRG4 allele enabled us to study partial loss of VRG4 function and the requirement of this function for mannosylation. As in S. cerevisiae, the CaVrg4 protein is required for protein hypermannosylation for phosphomannan addition and appears to reside in the Golgi apparatus. All of the data suggest that CaVrg4p is the GDP-mannose transporter that regulates mannosylation in the Golgi apparatus in the same manner that has been demonstrated for S. cerevisiae.
In S. cerevisiae, proper wall construction is essential for maintaining and defining cell shape. Because of the degree of cell wall expansion required to form a hypha, a defect in the synthesis of any wall component is predicted to have severe effects on filamentation in Candida cultures. Compatible with this idea, we found that even a partial loss of CaVRG4 function completely blocked hyphal formation under all conditions tested. Thus, CaVRG4 can be added to the growing list of Candida genes involved in cell wall synthesis that, when mutated, are uniformly impaired to some degree in hyphal formation. These genes include the genes required for synthesis or assembly of glucan (e.g., CaKRE9, PHR1, and PHR2) (21), chitin (e.g., CHS1) (35, 52), and mannan (e.g., MNN9, MNT1, SRB1, PMT1, and PMT6) (7, 50, 55,56,57). While filamentation defects may be a secondary effect due to inactivation of a hyphal regulator in some mutants, the phenotypic similarity of the mutants indicates that the cell wall has an essential role in hyphal formation.
Marked induction of polarized growth was an unexpected phenotype displayed by C. albicans vrg4 mutants. Pseudohyphae or elongated chains of cells that remained attached to one another also suggested that there was polar budding. The elongated cell phenotype was not expected because it is precisely the opposite of what is seen in Scvrg4 mutants and in most S. cerevisiae cell wall mutants, which typically arrest as large, unbudded cells that are defective in polarized growth (34). The relationship between pseudohyphal growth and hyphal growth in C. albicans is unclear. One model to explain induction of these elongated morphological forms in Cavrg4 mutants assumes that pseudohyphae are intermediate forms of hyphae or that the two cell types are induced in response to similar signals. Accordingly, pseudohyphal growth may simply represent a general response to stress. Cells become elongated because mitosis is delayed and the cells continue to grow apically rather than isotropically. Certain Candida cell wall mutants (for instance, mnn9 and chs1 mutants) produce similarly elongated morphological forms (35, 50, 52). On the other hand, other Candida cell wall mutants do not produce an elongated cell phenotype but accumulate as large budded cells that are reminiscent of the cells of S. cerevisiae cell wall mutants (e.g., the gas1-related phr1 mutants [44]). Therefore, the transition to pseudohyphal growth due to loss of CaVRG4 function is probably not a general stress response to cell wall weakening representing a cell wall integrity pathway. In S. cerevisiae, compensatory changes in the composition, assembly, and depolarized localization of cell wall components are activated to ensure the integrity of the cell wall (48), and there is evidence that some aspects of this pathway are similarly activated in C. albicans (30). Further phenotypic analysis of vrg4 and other Candida cell wall mutants is required to determine the basis for the morphological abnormalities that we observed in Cavrg4 mutants, how they relate to a defect in mannosylation, and whether this phenotype is indicative of some type of cell wall-weakening response.
Although CaVRG4 is clearly a functional and structural homologue of the S. cerevisiae VRG4 gene, one difference between these two genes that may be related to differences in their mutant phenotypes is notable. In stark contrast to CaVRG4, the ScVRG4 promoter region contains several consensus recognition elements for the G1 cell cycle transcription factors Swi4/Swi6 (T/CACGAAAA) and MluI (ACGCGT) at positions −125, −245, −267, and −307 relative to the translational start site. As a result, ScVRG4 transcription is strongly cell cycle dependent, and maximal levels of both mRNA and protein occur late in G1, before bud emergence (28; Dean, unpublished results). The cell cycle transcriptional machinery appears to be conserved in S. cerevisiae and C. albicans (2, 31). Therefore, the absence of these conserved cis-acting elements in the CaVRG4 promoter region suggests that regulation of VRG4 in C. albicans may be different from regulation of the gene in S. cerevisiae or that additional, uncharacterized cis and trans elements mediate induction of CaVRG4 transcription late in G1. The transcription of many S. cerevisiae genes whose products regulate cell wall biosynthesis is similarly cell cycle dependent, presumably to meet the high demand for cell wall expansion by an emerging bud (28, 51). It will be interesting to determine if Candida accommodates the need for de novo mannan synthesis during bud emergence or hyphal growth by upregulating CaVrg4p levels or by some other mechanism.
Hypermannosylated cell surface proteins are characteristic of most, if not all, fungal species, although the degrees to which these proteins may be subsequently decorated in the Golgi apparatus by additional types of sugars vary. Unlike S. cerevisiae oligosaccharides, N-linked oligosaccharides of C. albicans contain acid-labile phosphodiester-linked β-1,2-oligomannoside branches, whose lengths have been proposed to contribute to cell surface charge and pathogenicity (33). Although the evidence is indirect, reduced binding to Alcian blue has been strongly correlated with the absence of surface phosphomannan in S. cerevisiae, as shown by studies of MNN6, which encodes the S. cerevisiae Golgi mannosylphosphate transferase (38). As expected, we observed a dramatic reduction in binding to Alcian blue in Cavrg4 mutants, suggesting that there was a reduction in surface phosphomannan and hence in β-1,2-oligomannoside branches. Another factor that has been proposed to contribute to surface hydrophobicity is the presence of sialylated galactosyl glycoconjugates on the surface of C. albicans (49). A search of the Candida genome sequence failed to identify any genes related to the genes that encode CMP-sialic transporters, whose activity is predicted to be required for synthesis of the glycoconjugates. The only gene related to the genes encoding nucleotide sugar transporters that could be identified in the Candida genome was HUT1, whose sequence is most similar to the sequences of the subgroup of genes which encode nucleotide sugar transporters that transport UDP-galactose (29, 36). Although deletion of HUT1 results in little or no detectable change in the phenotype of S. cerevisiae (36), it will be interesting to determine whether the same is true for C. albicans.
We used native gel electrophoresis and in situ activity staining of secreted C. albicans acid phosphatase to directly assess the role of VRG4 in protein mannosylation. Although widely used for analysis of mutants with mutations affecting secretion or glycosylation of S. cerevisiae proteins, these reporter assays have not been utilized for C. albicans glycoprotein analyses. Two predicted acid phosphatase-encoding genes, PHO5 and PHO12, can be identified in the C. albicans genome based on sequence homology. Pho12p has a predicted molecular mass of 53.3 kDa, contains eight recognition sites for addition of N-linked oligosaccharides, and contains clusters of Ser/Thr residues typical of regions modified by sugar chains that are O linked. In contrast, PHO5 encodes a predicted 42.4-kDa protein that contains only one potential site for addition of N-linked chains. While migration on native polyacrylamide gels does not provide an accurate measurement of protein molecular weight, based on the mobility of Pho12p and its susceptibility to endo H, this protein is the best candidate for the low-phosphate induced protein that we detected by the in situ acid phosphatase assay described here. Moreover, multiple protein bands, representing the two different predicted phosphatases, were not readily detected with this assay, suggesting that PHO12 encodes the major inducible secreted acid phosphatase in C. albicans.
CaVRG4 mechanistically affects several parameters that are critical for pathogenesis, including cell growth, viability, hyphal formation, and cell surface mannan, suggesting that VRG4 is required for virulence. Consistent with this assumption, other Candida mutants that are defective in Golgi mannosylation in steps downstream of CaVRG4 (e.g., MNT1 mutants) are avirulent (7). Unfortunately, the morphological phenotype and severe aggregation induced by this mutation posed technical problems that prevented accurate estimation of adhesion to mammalian epithelial cells by Cavrg4 mutants. Studies to directly examine the role of CaVRG4 during pathogenesis in animal models are under way.
Numerous studies have established that cell surface mannoproteins are important for host cell adhesion by fungi, although the relative roles played by the proteins and the carbohydrate portions of cell surface mannoproteins are still not well understood. The VRG4 gene encodes the only GDP-mannose transporter in S. cerevisiae and, as inferred by its essentiality, in C. albicans. Highly related VRG4 homologues can also be identified in S. pombe, Candida glabrata, Aspergillus, and even nonfungal species that contain mannosylated cell surface glyconjugates, such as plants and protozoans. In addition to viability of S. cerevisiae and C. albicans, VRG4 is required for viability of C. glabrata (Nishikawa and Dean, unpublished data) and for virulence of Leishmania (20). Because VRG4 is required for growth and survival of Candida, is conserved in fungi, and is not present in humans, it is likely to be an excellent broad-spectrum target for development of an antifungal compound.
In contrast to our understanding of S. cerevisiae, our understanding of the Candida cell wall is limited. While conservation in S. cerevisiae and C. albicans justifies the validity of using S. cerevisiae as a paradigm to direct studies of the Candida cell wall, the work presented here underscores the differences between these two organisms and the challenges that remain in this field.
Acknowledgments
Part of this work was done while J.B.P. and N.D. were on leave in Amsterdam, The Netherlands. We thank Frans Klis and members of his lab for their hospitality and stimulating discussions. We also thank Amy Warenda and Peter Sudbery for generous gifts of plasmids.
This work was supported by grant GM48467 from the National Institutes of Health (to N.D.)
A.N., J.B.P., and N.D. contributed equally to this work.
REFERENCES
- 1.Abeijon, C., P. Orlean, P. W. Robbins, and C. B. Hirschberg. 1989. Topography of glycosylation in yeast: characterization of GDP mannose transport and lumenal guanosine diphosphatase activities in Golgi-like vesicles. Proc. Natl. Acad. Sci. USA 86:6935–6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andaluz, E., A. Ciudad, J. Rubio Coque, R. A. Calderone, and G. Larriba. 1999. Cell cycle regulation of a DNA ligase-encoding gene (CaLIG4) from Candida albicans. Yeast 15:1199–1210. [DOI] [PubMed] [Google Scholar]
- 3.Ashman, R. B., and J. M. Papadimitriou. 1995. Production and function of cytokines in natural and acquired immunity to Candida albicans infection. Microbiol. Rev. 59:646–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ballou, L., R. A. Hitzeman, M. S. Lewis, and C. E. Ballou. 1991. Vanadate-resistant yeast mutants are defective in protein glycosylation. Proc. Natl. Acad. Sci. USA 88:3209–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beeler, T. J., D. Fu, J. Rivera, E. Monaghan, K. Gable, and T. N. Dunn. 1997. SUR1 (CSG1/BCL21), a gene necessary for growth of Saccharomyces cerevisiae in the presence of high Ca2+ concentrations at 37°C, is required for mannosylation of inositolphosphorylceramide. Mol. Gen. Genet. 255:570–579. [DOI] [PubMed] [Google Scholar]
- 6.Berninsone, P. M., and C. B. Hirschberg. 2000. Nucleotide sugar transporters of the Golgi apparatus. Curr. Opin. Struct. Biol. 10:542–547. [DOI] [PubMed] [Google Scholar]
- 7.Buurman, E. T., C. Westwater, B. Hube, A. J. Brown, F. C. Odds, and N. A. Gow. 1998. Molecular analysis of CaMnt1p, a mannosyl transferase important for adhesion and virulence of Candida albicans. Proc. Natl. Acad. Sci. USA 95:7670–7675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calderone, R. A., and P. C. Braun. 1991. Adherence and receptor relationships of Candida albicans. Microbiol. Rev. 55:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Care, R. S., J. Trevethick, K. M. Binley, and P. E. Sudbery. 1999. The MET3 promoter: a new tool for Candida albicans molecular genetics. Mol. Microbiol. 34:792–798. [DOI] [PubMed] [Google Scholar]
- 10.Cassone, A. 1989. Cell wall of Candida albicans: its functions and its impact on the host. Curr. Top. Med. Mycol. 3:248–314. [DOI] [PubMed] [Google Scholar]
- 11.Chaffin, W. L., J. L. Lopez-Ribot, M. Casanova, D. Gozalbo, and J. P. Martinez. 1998. Cell wall and secreted proteins of Candida albicans: identification, function, and expression. Microbiol. Mol. Biol. Rev. 62:130–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chi, J. H., J. Roos, and N. Dean. 1996. The OST4 gene of Saccharomyces cerevisiae encodes an unusually small protein required for normal levels of oligosaccharyltransferase activity. J. Biol. Chem. 271:3132–3140. [DOI] [PubMed] [Google Scholar]
- 13.Cormack, B. P., G. Bertram, M. Egerton, N. A. Gow, S. Falkow, and A. J. Brown. 1997. Yeast-enhanced green fluorescent protein (yEGFP), a reporter of gene expression in Candida albicans. Microbiology 143:303–311. [DOI] [PubMed] [Google Scholar]
- 14.Cutler, J. E. 1991. Putative virulence factors of Candida albicans. Annu. Rev. Microbiol. 45:187–218. [DOI] [PubMed] [Google Scholar]
- 15.Cutler, J. E., and T. Kanbe. 1993. Antigenic variability of Candida albicans cell surface. Curr. Top. Med. Mycol. 5:27–47. [PubMed] [Google Scholar]
- 16.Dean, N. 1999. Asparagine-linked glycosylation in the yeast Golgi. Biochim. Biophys. Acta 1426:309–322. [DOI] [PubMed] [Google Scholar]
- 17.Dean, N. 1994. Cloning and DNA sequence of a Kluyveromyces lactis ERD1 homologue. Yeast 10:1117–1124. [DOI] [PubMed] [Google Scholar]
- 18.Dean, N. 1995. Yeast glycosylation mutants are sensitive to aminoglycosides. Proc. Natl. Acad. Sci. USA 92:1287–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dean, N., Y. B. Zhang, and J. B. Poster. 1997. The VRG4 gene is required for GDP-mannose transport into the lumen of the Golgi in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 272:31908–31914. [DOI] [PubMed] [Google Scholar]
- 20.Descouteaux, A., Y. Luo, S. J. Turco, and S. M. Beverley. 1995. A specialized pathway affecting virulence glycoconjugates of Leishmania. Science 269:1869–1872. [DOI] [PubMed] [Google Scholar]
- 21.Fonzi, W. A. 1999. PHR1 and PHR2 of Candida albicans encode putative glycosidases required for proper cross-linking of beta-1,3- and beta-1,6-glucans. J. Bacteriol. 181:7070–7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friis, J., and P. Ottolenghi. 1970. The genetically determined binding of alcian blue by a minor fraction of yeast cell walls. C. R. Trav. Lab. Carlsberg 37:327–341. [PubMed] [Google Scholar]
- 24.Gao, X. D., and N. Dean. 2000. Distinct protein domains of the yeast Golgi GDP-mannose transporter mediate oligomer assembly and export from the endoplasmic reticulum. J. Biol. Chem. 275:17718–17727. [DOI] [PubMed] [Google Scholar]
- 25.Gao, X. D., A. Nishikawa, and N. Dean. 2001. Identification of a conserved motif in the yeast Golgi GDP-mannose transporter required for binding to nucleotide sugar. J. Biol. Chem. 276:4424–4432. [DOI] [PubMed] [Google Scholar]
- 26.Gietz, R. D., R. H. Schiestl, A. R. Willems, and R. A. Woods. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355–360. [DOI] [PubMed] [Google Scholar]
- 27.Guthrie, C., and G. R. Fink. 1991. Guide to yeast genetics and molecular biology. Methods Enzymol. 194:3–20. [PubMed] [Google Scholar]
- 28.Igual, J. C., A. L. Johnson, and L. H. Johnston. 1996. Coordinated regulation of gene expression by the cell cycle transcription factor SWI4 and the protein kinase C MAP kinase pathway for yeast cell integrity. EMBO J. 15:5001–5013. [PMC free article] [PubMed] [Google Scholar]
- 29.Kainuma, M., Y. Chiba, M. Takeuchi, and Y. Jigami. 2001. Overexpression of HUT1 gene stimulates in vivo galactosylation by enhancing UDP-galactose transport activity in Saccharomyces cerevisiae. Yeast 18:533–541. [DOI] [PubMed] [Google Scholar]
- 30.Kapteyn, J. C., L. L. Hoyer, J. E. Hecht, W. H. Muller, A. Andel, A. J. Verkleij, M. Makarow, H. Van Den Ende, and F. M. Klis. 2000. The cell wall architecture of Candida albicans wild-type cells and cell wall-defective mutants. Mol. Microbiol. 35:601–611. [DOI] [PubMed] [Google Scholar]
- 31.Loeb, J. D., M. Sepulveda-Becerra, I. Hazan, and H. Liu. 1999. A G1 cyclin is necessary for maintenance of filamentous growth in Candida albicans. Mol. Cell. Biol. 19:4019–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masuoka, J., and K. C. Hazen. 1997. Cell wall protein mannosylation determines Candida albicans cell surface hydrophobicity. Microbiology 143:3015–3021. [DOI] [PubMed] [Google Scholar]
- 33.Masuoka, J., and K. C. Hazen. 1999. Differences in the acid-labile component of Candida albicans mannan from hydrophobic and hydrophilic yeast cells. Glycobiology 9:1281–1286. [DOI] [PubMed] [Google Scholar]
- 34.Mondesert, G., D. J. Clark, and S. I. Reed. 1997. Identification of genes controlling growth polarity in the budding yeast Saccharomyces cerevisiae: a possible role of N-glycosylation and involvement of the exocyst complex. Genetics 147:421–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munro, C. A., K. Winter, A. Buchan, K. Henry, J. M. Becker, A. J. Brown, C. E. Bulawa, and N. A. Gow. 2001. Chs1 of Candida albicans is an essential chitin synthase required for synthesis of the septum and for cell integrity. Mol. Microbiol. 39:1414–1426. [DOI] [PubMed] [Google Scholar]
- 36.Nakanishi, H., K. Nakayama, A. Yokota, H. Tachikawa, N. Takahashi, and Y. Jigami. 2001. Hut1 proteins identified in Saccharomyces cerevisiae and Schizosaccharomyces pombe are functional homologues involved in the protein-folding process at the endoplasmic reticulum. Yeast 18:543–554. [DOI] [PubMed] [Google Scholar]
- 37.Neiman, A. M., V. Mhaiskar, V. Manus, F. Galibert, and N. Dean. 1997. Saccharomyces cerevisiae HOC1, a suppressor of pkc1, encodes a putative glycosyltransferase. Genetics 145:637–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Odani, T., Y. Shimma, X. H. Wang, and Y. Jigami. 1997. Mannosylphosphate transfer to cell wall mannan is regulated by the transcriptional level of the MNN4 gene in Saccharomyces cerevisiae. FEBS Lett. 420:186–190. [DOI] [PubMed] [Google Scholar]
- 39.Odds, F. C., and J. C. Hierholzer. 1973. Purification and properties of a glycoprotein acid phosphatase from Candida albicans. J. Bacteriol. 114:257–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orlean, P. 1997. Biogenesis of yeast cell wall and surface components, vol. 3. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 41.Poster, J. B., and N. Dean. 1996. The yeast VRG4 gene is required for normal Golgi functions and defines a new family of related genes. J. Biol. Chem. 271:3837–3845. [DOI] [PubMed] [Google Scholar]
- 42.Rambourg, A., Y. Clermont, L. Ovtracht, and F. Kepes. 1995. Three-dimensional structure of tubular networks, presumably Golgi in nature, in various yeast strains: a comparative study. Anat. Rec. 243:283–293. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 44.Saporito-Irwin, S. M., C. E. Birse, P. S. Sypherd, and W. A. Fonzi. 1995. PHR1, a pH-regulated gene of Candida albicans, is required for morphogenesis. Mol. Cell. Biol. 15:601–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schweingruber, A. M., F. Schoenholzer, L. Keller, R. Schwaninger, H. Trachsel, and M. E. Schweingruber. 1986. Glycosylation and secretion of acid phosphatase in Schizosaccharomyces pombe. Eur. J. Biochem. 158:133–140. [DOI] [PubMed] [Google Scholar]
- 46.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sipos, G., A. Puoti, and A. Conzelmann. 1995. Biosynthesis of the side chain of yeast glycosylphosphatidylinositol anchors is operated by novel mannosyltransferases located in the endoplasmic reticulum and the Golgi apparatus. J. Biol. Chem. 270:19709–19715. [DOI] [PubMed] [Google Scholar]
- 48.Smits, G. J., H. van den Ende, and F. M. Klis. 2001. Differential regulation of cell wall biogenesis during growth and development in yeast. Microbiology 147:781–794. [DOI] [PubMed] [Google Scholar]
- 49.Soares, R. M., R. M. de A Soares, D. S. Alviano, J. Angluster, C. S. Alviano, and L. R. Travassos. 2000. Identification of sialic acids on the cell surface of Candida albicans. Biochim. Biophys. Acta 1474:262–268. [DOI] [PubMed] [Google Scholar]
- 50.Southard, S. B., C. A. Specht, C. Mishra, J. Chen-Weiner, and P. W. Robbins. 1999. Molecular analysis of the Candida albicans homolog of Saccharomyces cerevisiae MNN9, required for glycosylation of cell wall mannoproteins. J. Bacteriol. 181:7439–7448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spellman, P. T., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9:3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sudoh, M., T. Yamazaki, K. Masubuchi, M. Taniguchi, N. Shimma, M. Arisawa, and H. Yamada-Okabe. 2000. Identification of a novel inhibitor specific to the fungal chitin synthase. Inhibition of chitin synthase 1 arrests the cell growth, but inhibition of chitin synthase 1 and 2 is lethal in the pathogenic fungus Candida albicans. J. Biol. Chem. 275:32901–32905. [DOI] [PubMed] [Google Scholar]
- 53.Suzuki, S. 1997. Immunochemical study on mannans of genus Candida. I. Structural investigation of antigenic factors 1, 4, 5, 6, 8, 9, 11, 13, 13b and 34. Curr. Top. Med. Mycol. 8:57–70. [PubMed] [Google Scholar]
- 54.Thomson, L. M., S. Bates, S. Yamazaki, M. Arisawa, Y. Aoki, and N. A. Gow. 2000. Functional characterization of the Candida albicans MNT1 mannosyltransferase expressed heterologously in Pichia pastoris. J. Biol. Chem. 275:18933–18938. [DOI] [PubMed] [Google Scholar]
- 55.Timpel, C., S. Strahl-Bolsinger, K. Ziegelbauer, and J. F. Ernst. 1998. Multiple functions of Pmt1p-mediated protein O-mannosylation in the fungal pathogen Candida albicans. J. Biol. Chem. 273:20837–20846. [DOI] [PubMed] [Google Scholar]
- 56.Timpel, C., S. Zink, S. Strahl-Bolsinger, K. Schroppel, and J. Ernst. 2000. Morphogenesis, adhesive properties, and antifungal resistance depend on the Pmt6 protein mannosyltransferase in the fungal pathogen Candida albicans. J. Bacteriol. 182:3063–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Warit, S., N. Zhang, A. Short, R. M. Walmsley, S. G. Oliver, and L. I. Stateva. 2000. Glycosylation deficiency phenotypes resulting from depletion of GDP-mannose pyrophosphorylase in two yeast species. Mol. Microbiol. 36:1156–1166. [DOI] [PubMed] [Google Scholar]
- 58.Wilson, R. B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoko-o, T., S. K. Roy, and Y. Jigami. 1998. Differences in in vivo acceptor specificity of two galactosyltransferases, the gmh3+ and gma12+ gene products from Schizosaccharomyces pombe. Eur. J. Biochem. 257:630–637. [DOI] [PubMed] [Google Scholar]