Abstract
DNA arrays were used to investigate the functional role of Rox1 in mediating acclimatization to anaerobic conditions in Saccharomyces cerevisiae. Multiple growth conditions for wild-type and rox1 null strains were used to identify open reading frames with a statistically robust response to this repressor. These results were compared to those obtained for a wild-type strain in response to oxygen availability. Transcripts of nearly one-sixth of the genome were differentially expressed (P < 0.05) with respect to oxygen availability, the majority (>65%) being down-regulated under anoxia. Of the anaerobically induced genes, about one-third (106) contain putative Rox1-binding sites in their promoters and were significantly (P < 0.05) up-regulated in the rox1 null strains under aerobiosis. Additional promoter searches revealed that nearly one-third of the anaerobically induced genes contain an AR1 site(s) for the Upc2 transcription factor, suggesting that Upc2 and Rox1 regulate the majority of anaerobically induced genes in S. cerevisiae. Functional analyses indicate that a large fraction of the anaerobically induced genes are involved in cell stress (∼1/3), cell wall maintenance (∼1/8), carbohydrate metabolism (∼1/10), and lipid metabolism (∼1/12), with both Rox1 and Upc2 predominating in the regulation of this latter group and Upc2 predominating in cell wall maintenance. Mapping the changes in expression of functional regulons onto metabolic pathways has provided novel insight into the role of Rox1 and other trans-acting factors in mediating the physiological response of S. cerevisiae to anaerobic conditions.
Nearly all organisms respond to changes in their environment by differentially transcribing genes that are important for cell fitness and survival. Using cDNA arrays to track these changes can provide insight into the gene networks and control circuitry underlying the adaptive response. In this study, we used such an approach to obtain a more comprehensive view of the response of Saccharomyces cerevisiae to anaerobiosis and determine the role of Rox1, a key regulator of anaerobic gene expression, in mediating this response. S. cerevisiae is one of few yeast species that can grow in the complete absence of oxygen (82), making it an excellent eukaryotic model for investigating the genomic remodeling of gene expression that is elicited by oxygen deprivation.
Until recently, Rox1 was the only known transcriptional regulator of anaerobically induced genes in S. cerevisiae (reviewed in references 45, 49, and 84). It is a sequence-specific DNA-binding protein in the SOX class of HMG proteins (25) that binds to the consensus sequence YYYATTGTTCTC in double-stranded DNA (8, 28). Rox1-dependent gene repression requires Tup1-Ssn6 (Cyc8) (8, 25), which interacts with a number of other regulon-specific DNA-binding factors (reviewed in reference 73). Repression is thought to occur through at least two mechanisms: nucleosome phasing and direct interactions of the Tup1-Ssn6 repression complex with the basal transcriptional machinery. Although Rox1 functions in an O2-independent manner, its expression is oxygen (heme) dependent, activated by the heme-dependent transcription factor Hap1 (46). Thus, as oxygen levels fall to those that limit heme biosynthesis (51), ROX1 is no longer transcribed (84), its protein levels fall (83), and the genes it regulates are derepressed.
Many of the previously identified Rox1-regulated genes encode proteins involved in oxygen-dependent pathways, such as respiration (COX5b and CYC7) (58, 79) and the biosynthesis of heme (HEM13) (46), sterols (HMG2, ERG11, and NCP1) (77, 80), and unsaturated fatty acids (ATF1 and OLE1) (20, 37; reviewed in references 45, 49, and 84). Others may provide essential functions under anaerobiosis (ACC3 and ANB1) (57, 69) or are themselves transcription factors (SUT1) (60, 66). The expression of nearly all of these Rox1-regulated genes increases as oxygen concentrations fall to levels approaching anoxia (49). Their up-regulation may, therefore, serve to maintain or activate flux through essential pathways by increasing key enzyme levels, increasing the efficiency of oxygen utilization (e.g., expression of hypoxic isoforms), and/or providing alternative pathways for metabolism. The importance of maintaining flux through sterol and unsaturated-fatty-acid biosynthetic and/or import pathways is underscored by an absolute nutritional requirement for these components during anaerobic growth (4, 5).
Several other transcription factors have recently been shown to regulate anaerobically expressed genes in S. cerevisiae. These include the activators Sut1 (66) and Upc2 (Mox4) (1, 2, 21, 22) and the repressors Mox1, Mox2, and Mot3 (44); Mot3 can apparently also activate the expression of some genes (1). In addition, an antagonist interaction between Ord1 and Yap1 has been shown to regulate some anaerobic genes (11). A number of these factors in concert regulate the expression of cell wall-related genes in the PAU gene family, including the DAN/TIR (2) and TIP/SRP subfamilies (31), suggesting a fairly complex regulatory network for this group of genes in contrast to the apparent regulatory simplicity of those repressed by Rox1. The extent to which these factors control the expression of other functional groups of anaerobically induced genes is currently unknown.
In this study, we focused on the gene network controlled by Rox1. Recent microarray studies have identified hundreds of anaerobically expressed genes in S. cerevisiae (76), yet only 11 Rox1-regulated genes have been identified through biochemical and/or genetic means. These results suggest that other regulators control the expression of a large fraction of anaerobically induced genes and/or the number of Rox1-regulated genes is much larger than is currently appreciated. To gain a more comprehensive understanding of the role this factor plays in mediating anaerobic acclimatization, we used cDNA arrays to examine the genome-wide response to both inactivation of ROX1 and oxygen availability. Our results suggest that Rox1 controls nearly one-third of the anaerobically induced genes in yeast, including many involved in lipid, isoprenoid, and sphingolipid metabolism. In addition, nearly one-third of the anaerobically induced genes contain a consensus binding site(s) for Upc2, suggesting that this factor regulates the expression of a much larger set of genes than is currently appreciated. Many of these genes are predictably involved in cell wall-related functions. Finally, mapping the changes in gene expression of functional regulons onto metabolic pathways has provided novel insight into the role that Rox1 and other trans-acting factors play in mediating the physiological response of S. cerevisiae to anaerobiosis.
MATERIALS AND METHODS
Yeast strains and media.
The following yeast strains were used: JM43 (MATα his4-580 trp1-289 leu2-3,112 ura3-52 [ρ+]) (23) and the isogenic strain KKY6, which contains a rox1Δ::LEU2 disruption (this study), and RZ53-6 (MATα trp1-289 leu2-3,112 ura3-52 ade1-100 [ρ+]) (56) and the isogenic strain RZ53-6Δrox1, which contains a rox1Δ::LEU2 disruption (26). Strain KKY6 was obtained by transforming JM43 with a BamHI-XbaI fragment from prox1::LEU2 (58). The gene disruption was confirmed by both Southern blot analysis and Northern blots of known Rox1-regulated genes.
Liquid precultures were grown at 28 to 30°C with shaking (200 rpm) in YPGal (71) or a semisynthetic galactose medium containing Tween 80 (a source of oleic acid), ergosterol, and silicon antifoam (SSG-TEA medium) (14). Amino acids and nucleotides were added, as appropriate, at a concentration of 40 mg/liter. Precultures were kept in mid-log growth phase (<100 Klett units; optical density at 600 nm [OD600] < 0.2) for typically 3 days before the final cultures were inoculated. Batch cultures were grown in a New Brunswick BioFlo III fermentor using either air (aerobiosis) or 2.5% CO2 in O2-free N2 (anaerobiosis) as the sparge gas (flow rate = 1.15 volumes/volume of medium). For anaerobiosis, the gas mixture was passed through an OxyClear O2 absorber capable of reducing trace O2 to 5 ppb, and the cultures were grown in the dark (15). Oxygen concentration was monitored with a polarographic O2 electrode (12 mm) and a model 4300 dissolved oxygen transmitter (both from Ingold Electronics, Wilmington, Mass.). The oxygen electrode was calibrated prior to inoculation of the cultures by equilibration with air (full scale) and oxygen-free N2 (zero) and after harvesting by immersion in zeroing gel (Mettler Toledo). Temperature, pH, and agitation were maintained at 28°C, 5.0, and 300 rpm, respectively (14). For all cultures, the inoculation volume was adjusted so that the cell density upon harvesting, after six doublings in cell mass, was equal to 60 Klett units (OD600 ≈ 0.12). During harvesting, the cells were quick-chilled to ∼4°C by passage through several feet of coiled copper tubing immersed in a salt-water ice bath, pelleted by centrifugation (5,000 × g for 15 min at 4°C), washed once with ice-cold diethyl pyrocarbonate-treated Milli-Q water, flash-frozen in liquid N2, and stored at −80°C.
Respiration measurements.
Precultures were grown in YPGal medium with shaking (200 rpm) at 28 to 30°C and kept in mid-log growth phase for typically 3 days before inoculation of the final 50-ml aerobic shake cultures. Cultures were harvested in mid-log to late log phase at a cell density of 200 Klett units (OD600 ≈ 0.4) by centrifugation at 5,000 × g for 15 min at 4°C. The cells were washed once with 40 mM potassium phosphate buffer (pH = 7.4), pelleted (14,000 × g for 1 min at 4°C), resuspended in buffer (0.5 g/ml), and kept on ice until use. Whole-cell respiration measurements were made with an Oxygraph (Oroboros, Innsbruck, Austria) using a volume of 2 ml, stir rate of 700 rpm, temperature of 30°C, and sampling rate of 0.5 s (39). Before each experiment, the oxygen sensors were calibrated with air-saturated water and with a high density of respiratory-competent cells in buffer, to exhaust all oxygen (zero). The partial O2 pressure in the chamber was recorded for several minutes under steady-state flux conditions after the sequential addition of air-equilibrated 40 mM potassium phosphate buffer (pH = 7.4) and 1% glucose, followed by ∼2.5 mg of cells, and lastly 1 mM KCN.
Data were analyzed with DatLab software (Oroboros). Oxygen concentration in the chamber (nanomoles of O2 milliliter−1) was calculated from partial O2 pressure measurements and based upon O2 solubility in the respiration medium at 30°C and ambient barometric pressure. Oxygen flux (in picomoles s−1 ml−1) was calculated as the time derivative of the O2 concentration in the chamber. Corrections were made for the consumption of O2 by the sensor and back diffusion of O2 into the chamber and for the exponential time constant of the sensor (39). Duplicate oxygen consumption measurements were made on each of four independent isolates for each yeast strain examined.
Analysis of gene expression.
Total RNA was extracted from frozen cells using a glass-bead phenol-chloroform-isoamyl alcohol method as described elsewhere (50) and treated with RQ1 DNase according to the manufacturer’s protocol (Promega, Madison, Wis.). Ten micrograms of total RNA was then incubated with 2.0 μg of oligo(dT10–20) at 70°C for 10 min, quenched briefly on ice, and reverse transcribed with Superscript II (Gibco BRL, Rockville, Md.) in the presence of [33P]dATP (NEN, Boston, Mass.) according to the ResGen (Huntsville, Ala.) protocol with the modification that the incorporation of [33P]dATP was optimized by increasing the dithiothreitol concentration from 3.3 to 10 mM, increasing the reverse transcriptase concentration from 300 to 350 U, and increasing the [33P]dATP concentration from 1.0 to 1.5 μM. Typical incorporation rates were >40%. After first-strand cDNA synthesis, the probe was purified by passage through a Bio-Spin 6 chromatography column (Bio-Rad, Hercules, Calif.). The purified probe was heat denatured and pipetted into a single hybridization tube (3.5 by 22.5 cm) containing ResGen yeast GeneFilters I, II, and III (comprising 6,218 open reading frames [ORFs]) that had been prehybridized for 6 h at 42°C in 7.5 ml of hybridization solution containing 7.5 μg of poly(dA) (blocking agent). Hybridization was conducted for 16 h at 42°C, followed by two 20-min washes in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-1% sodium dodecyl sulfate at 50°C and one 20-min wash in 0.5× SSC-1% sodium dodecyl sulfate at 42°C. After being washed, the filter arrays were exposed to a phosphorimaging screen for 48 h. The image was scanned at a 50-μm resolution with a Molecular Dynamics STORM 860, and the amount of radioactivity bound to each ORF was quantified using Pathways software (version 2.01; ResGen). Normalization procedures and statistical analyses were performed with SAS (Cary, N.C.) software as described below.
Experimental design and statistical analyses.
For the Rox1 analyses, two different parental strains (JM43 and RZ53-6) and isogenic rox1 null strains (KKY6 [JM43Δrox1] and RZ53-6Δrox1) were each grown in two media (SSG-TEA and YPGal). For each strain and growth condition examined, three independent growth and array analyses were performed (n = 24). For the aerobic-versus-anaerobic comparisons, a total of 12 (8 aerobic and 4 anaerobic) independent growth and array analyses were performed with strain JM43 grown in SSG-TEA medium only. Whenever possible, an entire set of replicates, comprising all strains and growth conditions, were run simultaneously with the same reagents (e.g., isotope lot, reverse transcriptase, hybridization solution, phosphorimaging screen, etc.). All analyses were performed using GeneFilters from the same lot (I, 990927D; II, 990929D; III, 000725E) to avoid interlot printing variability. GeneFilter III contains the remaining ORFs from chromosome XVI that could not be printed on the other two filters, bringing the total to 6,218 ORFs (current annotations recognize 6,150 of these). To facilitate comparisons, the raw hybridization intensity of each ORF on a single array was normalized using the following formula: normalized intensity (NormInt) = (raw intensity − background intensity)/(mean raw intensity of all ORFs − background intensity). This normalization procedure sets the mean NormInt values of all ORFs on a single array to a value of 1. Control spots (1.25 ng of genomic DNA) were excluded from the analyses because of substantially higher spotting variability (K. E. Kwast, unpublished observation).
Statistical analyses for the Rox1 comparisons were performed as three-factor analyses of variance (ANOVA) using the SAS MIXED procedure with the factors Rox1 (wild type or rox1), medium (YPGal or SSG-TEA), and strain (JM43 or RZ53-6). NormInt values, rather than log-transformed intensities, were used, as explained in Results and Discussion. Before the ANOVA were done, outliers whose deviations from the mean in each combination of strain, medium, and genetic background were outside the 99.6% confidence interval were identified and eliminated, based on a pooled standard deviation for each ORF. The NormInt data were then sorted by ORF, and likelihood ratio tests were used to determine the appropriate analysis model (equal variances or unequal variances) using a chi-square value of 0.01. Separate F tests were performed for each group to identify ORFs whose NormInt values were significantly different among the rox1 and wild-type strains. Given that the goal was to identify ORFs whose mRNA abundance was significantly different among the rox1 null and wild-type strains, restrictions based upon the absolute difference in expression levels between these strains were not considered for this step of the analysis. Similar statistical procedures were used for analyzing the aerobic and anaerobic comparisons. Additional post hoc analyses included searches of the promoter region of ORFs for the Rox1-binding site and other cis regulatory sequences using GeneSpring (Silicon Genetics, San Carlos, Calif.) and on-line databases (Yeast Protein Database [YPD] [http://www.proteome.com/databases/YPD] and Munich Information Center for Protein Sequences [MIPS] [http://mips.gsf.de/proj/yeast]).
RESULTS AND DISCUSSION
To identify Rox1-regulated genes, multiple parental and isogenic rox1 null strains were grown in two different galactose media. These conditions were chosen to restrict the analyses to ORFs exhibiting a robust response to the rox1 disruption, independent of slight variations in the genetic background or the growth conditions used. Strains JM43 and RZ53-6 have divergent genetic backgrounds (23, 56), and both have been used previously for the analyses of Rox1-regulated genes (8, 25, 27, 48, 79). SSG-TEA medium was chosen to facilitate comparisons of aerobic and anaerobic cultures and contains nutrients (ergosterol and Tween 80, a source of oleic acid) essential for long-term anaerobic growth (4, 5). These nutritional supplements, however, are known to repress the transcription of several Rox1-regulated genes; for example, unsaturated fatty acids can repress the expression of OLE1 and ATF1 (20, 37). Thus, a comparable galactose medium lacking these supplements (YPGal) was also used. To facilitate statistical analyses, at least three independent growth and cDNA array analyses were performed for each strain and growth condition examined. Data were pooled across strains and media to determine if the rox1 mutation had a significant effect on transcript levels of each ORF. Such pooling of the data, however, compresses the largest differences in transcript abundance observed between the wild-type and rox1 strains for any given pairwise combination of strain and medium. We therefore focused on the statistical differences in transcript abundance, not the absolute difference between treatments per se. Moreover, focusing on all ORFs whose transcript levels are statistically different among treatments, as opposed to those exhibiting differences by a predetermined amount (e.g., a threefold change), is appropriate when the response to a chronic condition is analyzed, because the sustained response is often much smaller than the initial transient response to perturbation (for example, see references 38, 40, and 47). The data presented below, as well as results from studies examining the sensitivity, reproducibility, and specificity of the arrays, are available at http://titan.biotech.uiuc.edu/rox1/rox1.html.
Three points pertinent to the statistical analyses are worthy of discussion. First, negative NormInt values were allowed in the analyses given that in all cases their values were not statistically different (P > 0.05) from zero. Moreover, setting their value to zero would artificially decrease the inherent variability observed. Less than 1% of the 221,400 total observations had negative intensity values, and a total of 6 of 6150 ORFs examined had mean intensity values for a given treatment that were negative after pooling of data. It should also be noted that the degree of repression of several known Rox1-regulated genes in aerobically grown wild-type yeasts is such that their transcript abundance would be expected to be equivalent to zero (48, 49) but much higher under the derepressing conditions present in aerobically grown rox1 strains or anaerobically grown wild-type strains.
Second, non-log-transformed hybridization intensity values were used for the analyses. This is a departure from the more common use of the log(intensity) as the dependent variable when differences based upon transcript ratios between treatments are analyzed. However, inspection of the 24 observations within each ORF (for the Rox1 comparison) showed that for the majority of ORFs (>70%), the variances for the non-log-transformed data were similar whereas those of the log-transformed data were not. The use of non-log-transformed intensities, when appropriate, also eliminates problems associated with zero or slightly negative intensity values and the common adjustments that result in biasing variances associated with low-abundance transcripts.
Third, outliers whose deviations from the mean in each combination of strain, medium, and genetic background were outside the 99.6% confidence interval based upon a pooled standard deviation were eliminated. The intensity value of the vast majority of these outliers was greater than the mean of all observations for a given ORF. Visual inspection of the arrays revealed that several of these observations contained small regions of apparently aberrant radioactivity that overlapped or masked the actual hybridization signal, providing further justification for their removal. Although this was a rare event, with the elimination of <0.4% of the total observations, it is a challenge inherent with using a single cDNA population for probing an array that is often overlooked and requires either visual inspection or statistical methods for elimination as used here.
Analyses of differences in transcript abundance among the rox1 and wild-type strains.
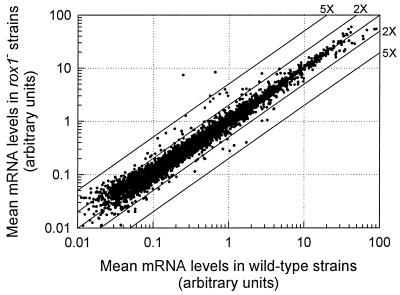
An examination of mean hybridization intensities of the 6,150 ORFs, pooled across media and genetic backgrounds, reveals that the majority of genes were transcribed at similar levels in the rox1 null and wild-type strains, with few genes exhibiting more than a fivefold difference in expression (Fig. 1). The correlation coefficient for this comparison (0.96) is high, with higher variances being associated with high-abundance transcripts as expected. Because the data are pooled across different media and genetic backgrounds, the absolute differences in expression are predictably compressed compared to that observed within a given strain and growth condition. Statistical analyses revealed a surprisingly large number of ORFs with significant differences (P < 0.05) in transcript abundance among the rox1 null and wild-type strains, 595 with significantly higher levels in the rox1 strains and 285 with significantly lower levels. To differentiate between genes that are directly controlled by Rox1 and those that are up-regulated due to changes in metabolic and/or genetic signals elicited by the aerobic derepression of Rox1-regulated genes, additional sequence and expression criteria were used as described below.
FIG. 1.
Mean transcript levels of 6,150 yeast ORFs from wild-type and rox1 null strains. Batch cultures of strains JM43, KKY6 (JM43Δrox1), RZ53-6, and RZ53-6Δrox1 were aerobically grown in SSG-TEA and YPGal media. [33P]dATP-labeled cDNA probes were reverse transcribed from 10 μg of total RNA and hybridized to GeneFilter arrays. Each point represents the mean mRNA abundance for the rox1 null strains compared to that from the wild-type strains (n = 12 for each strain). The diagonal lines indicate ratios of the rox1 and wild-type transcript levels.
Presumably, genes that are significantly up-regulated in the rox1 strains include those that are directly regulated by Rox1 as well as a potentially large group whose expression levels increase in response to the aerobic derepression of Rox1-regulated genes (secondarily affected genes). To identify genes that are directly regulated by Rox1, we searched the promoter region (−10 to −800 bp) for the Rox1-binding site and examined their response to oxygen availability (described below). A promoter search of the 595 genes up-regulated in the rox1 strains revealed several overrepresented (P < 0.05) sequences, which consisted of small runs (e.g., TCATTGTT [P = 0.01] and CTATTGT [P = 0.007]) of the Rox1 consensus sequence (YYYATTGTTCTC). We then eliminated genes whose promoters did not contain inferred permutations of this sequence (NNHWTTGTNNNN) based upon sites identified in all known Rox1-regulated genes (28). Although the inclusion of inferred permutations of the Rox1-binding site is not restrictive and eliminated few genes, the majority of previously identified Rox1-regulated genes would be eliminated using restrictions based upon the entire consensus sequence (YYYATTGTTCTC), and several (ERG11, NCP1, and CYC7) would be eliminated using the core Rox1-binding sequence (YATTGTT) alone (28).
To verify the results obtained, we focused on genes previously reported to be regulated by Rox1 (Table 1). Global transcript levels (pooled across media and genetic backgrounds) of AAC3, HEM13, ERG11, NCP1, COX5b, and ANB1 were significantly (P ≤ 4.6 × 10−5 [4.6e−5]) higher in the rox1 strains, with transcript ratios (rox1/wild type) ranging from 1.6 to 13. Nearly all of these genes exhibited a robust response to the rox1 mutation in all pairwise combinations of strain and medium examined (Table 1). Global transcript levels of five of the previously identified Rox1-regulated genes (CYC7, OLE1, SUT1, HMG2, and ATF1) did not, however, show a significant (P ≥ 0.069) rox1 effect. For several of these genes, there are conflicting reports regarding the effects of Rox1 on their expression and/or additional regulatory elements that are know to exert considerable control over expression levels, as described below.
TABLE 1.
Comparison of aerobic transcript levels of previously identified Rox1-regulated genes in rox1 null and wild-type strains by cDNA arraya
| ORF | Gene | Protein | Globalb
|
JM43
|
RZ53-6
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SSG-TEA
|
YPGal
|
SSG-TEA
|
YPGal
|
|||||||||
| P | Ratio | P | Ratio | P | Ratio | P | Ratio | P | Ratio | |||
| YBR085W | AAC3 | ADP/ATP translocase | 4.6e−5 | 1.6 | 2.5e−2 | 1.9 | 1.1e−2 | 1.4 | 6.3e−2 | 1.5 | 6.5e−2 | 1.5 |
| YDR044W | HEM13 | Coproporphyrinogen III oxidase | 7.8e−21 | 13 | 5.1e−9 | 19 | 1.1e−6 | 14 | 1.4e−4 | 8.9 | 7.9e−4 | 3.8 |
| YEL039C | CYC7 | Iso-2-cytochrome c | 0.41 | 1.2 | 0.50 | 1.3 | 0.81 | 0.9 | 0.54 | 1.3 | 0.31 | 1.3 |
| YGL055W | OLE1 | Δ-9 fatty acid desaturase | 0.34 | 1.2 | 0.18 | 0.4 | 0.17 | 1.5 | 0.21 | 1.6 | 3.3e−2 | 2.0 |
| YGL162W | SUT1 | Transcription factor involved in sterol uptake | 0.56 | 0.8 | 0.98 | 1.0 | 0.25 | 0.1 | 0.21 | 1.6 | 0.91 | 1.1 |
| YHR007C | ERG11 | Cytochrome P450 lanosterol 14α-demethylase | 9.8e−7 | 2.9 | 5.2e−4 | 3.1 | 2.8e−3 | 2.4 | 3.6e−4 | 2.5 | 1.9e−3 | 3.5 |
| YHR042W | NCP1 | NADPH cytochrome P450 reductase | 5.5e−7 | 2.1 | 6.5e−2 | 1.6 | 1.0e−3 | 2.1 | 2.3e−2 | 2.4 | 3.4e−4 | 2.3 |
| YIL111W | COX5b | Cytochrome c oxidase subunit Vb | 4.5e−8 | 4.2 | 9.8e−4 | 5.0 | 2.1e−3 | 3.5 | 5.2e−2 | 4.7 | 1.2e−2 | 3.5 |
| YJR047C | ANB1 | Translation initiation factor eIF-5b | 1.9e−7 | 4.1 | 4.9e−3 | 4.2 | 2.3e−4 | 4.1 | 1.7e−2 | 5.3 | 0.18 | 2.2 |
| YLR450W | HMG2 | 3-Hydroxy-3-methylglutaryl CoA reductase | 0.43 | 1.2 | 0.15 | 1.5 | 1.2e−2 | 2.3 | 0.52 | 1.3 | 0.41 | 0.4 |
| YOR377W | ATF1 | Alcohol acetyltransferase | 6.9e−2 | 1.8 | 0.95 | 1.0 | 0.10 | 2.4 | 0.24 | 2.2 | 0.32 | 1.9 |
ANOVA probability values and ratios of mean transcript level in the rox1 strain(s) to that in the wild-type strain(s) are shown.
Global results are for data pooled across all pairwise combinations of media and genetic backgrounds examined.
For OLE1 and CYC7, the array results confirm our previous findings by Northern blotting (48), which showed little effect of a rox1 null mutation on aerobic transcript levels of these genes in either JM43 or RZ53-6 grown in SSG-TEA medium. Promoter analyses of both of these genes has revealed a mosaic of regulatory sites in addition to that for Rox1 (20, 33, 65, 72, 81, 85). Given that oleic acid, a supplement in the SSG-TEA medium, is known to repress OLE1 transcription via the fatty acid response element (20), we predicted that its derepression in the absence of Rox1 may be more pronounced under fatty acid-nonrepressing conditions. Indeed, OLE1 transcript levels were significantly (P = 0.033) higher in RZ53-6Δrox1 grown in YPGal medium but not in the JM43 genetic background; the latter observation confirms differences in OLE1 expression between these strains documented previously (48). Given the regulatory complexity of these genes (20, 48, 50, 81), the degree of derepression elicited by the absence of Rox1 apparently depends on both the growth condition and genetic background of the strains used. Under the experimental conditions used here, Rox1 appears to play a minor role in dictating steady-state levels of OLE1 and no role for dictating steady-state levels of CYC7.
Similarly, aerobic transcript levels of SUT1 were not significantly (P = 0.56) different among the rox1 and wild-type strains under the same conditions. For ATF1, the results may be considered to be marginally significant (P = 0.069, ratio = 1.8) with, as predicted, higher transcript levels under fatty acid-nonrepressing conditions (36) in the JM43 genetic background. With regard to HMG2, we have found that its expression and that of its aerobic counterpart, HMG1, change little in response to oxygen availability in JM43 (49) and have obtained Northern blot results with rox1 null strains that contradict previously published reports (77). The array results mirror these findings with no detectable differences (P = 0.43) in HMG2 transcripts among the wild-type and rox1 strains yet significantly (P = 0.025, ratio = 1.5) higher HMG1 transcript levels in the rox1 strains. Although both of these genes contain putative Rox1-binding sites in their promoters (49), neither one was significantly (P ≥ 0.41) induced under anaerobiosis in strain JM43 (see below). The factors responsible for the differences in HMG1 and HMG2 expression in JM43 compared to other yeast strains (77) are currently unknown.
Overall, the cDNA array results for these Rox1-regulated genes are consistent with previous Northern blotting results (48). They suggest that the arrays allow sufficient sensitivity to identify many of the genes that are differentially expressed with respect to this transcription factor. They also reveal significant strain and medium effects on the expression of some, which the experimental design is intended to reveal. From these results it is clear that our experimental approach should allow us to identify a substantial fraction of genes that are regulated by Rox1 but certainly not all of them.
Identification of oxygen-responsive genes.
In order to further restrict the analysis to those genes that are most likely to be directly regulated by Rox1, we examined the response of these genes to oxygen availability. Although a previous study examined the genomic response of S. cerevisiae to oxygen availability (76), differences in experimental design, microarray platforms, strains, and growth conditions preclude a direct comparison with the rox1 data we have collected. Rather, for these analyses it was essential to measure this response with replication and under the conditions used for the rox1 comparison. Moreover, given that many more genes were up-regulated in the rox1 strains than are reported to be up-regulated under anoxia (76), the number of anaerobically induced genes may be much larger than is currently appreciated and/or a large fraction of genes induced in the rox1 strains may not be directly regulated by Rox1.
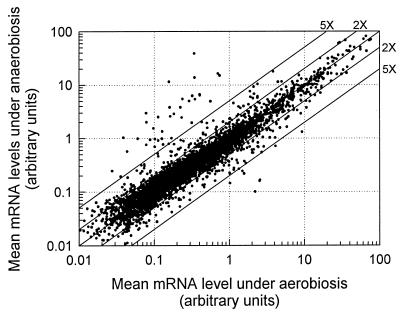
Figure 2 compares the mean hybridization intensities of the 6,150 ORFs between anaerobic and aerobic batch cultures of strain JM43 grown in SSG-TEA medium. As predicted, the majority of genes were expressed at similar levels under aerobiosis and anaerobiosis, with by far the largest differences in expression observed for a number of anaerobically induced genes. Given the large differences in transcript abundance for a number of these genes between aerobiosis and anaerobiosis, the correlation coefficient (0.90) is predictably less than that observed for the rox1-versus-wild-type comparison (Fig. 1). Statistical analyses revealed that nearly one-sixth of the genome was differentially expressed with respect to oxygen availability, with 719 genes being significantly (P < 0.05) down-regulated under anaerobiosis and 346 genes being significantly up-regulated. Although there is considerable overlap in terms of the oxygen-responsive genes we have identified and those identified by ter Linde et al. (76), the number of differentially expressed genes we identified is much larger. Moreover, there are substantive differences in some of the classes of genes affected between these two studies, as is discussed in more detail below. A direct comparison of these two studies may not be valid, however, given the aforementioned differences in experimental design and the fact that expression ratios alone, rather than statistical analyses, were used to identify oxygen-responsive genes in the study by ter Linde et al. (76).
FIG. 2.
Mean transcript levels of 6,150 yeast ORFs from aerobic and anaerobic batch cultures. Batch cultures of strain JM43 were grown in SSG-TEA medium under aerobic or anaerobic conditions. Probe generation and hybridization were as described in the legend to Fig. 1. Each point represents the mean mRNA abundance for anaerobic cultures (n = 4) compared to aerobic cultures (n = 8). The diagonal lines indicate ratios of anaerobic to aerobic transcript levels.
Because Rox1 repression is normally relieved when oxygen concentrations fall to levels that limit heme biosynthesis, we further restricted the Rox1 data set to include only genes significantly (P < 0.05) induced under anaerobiosis in the wild-type strain. Although this restriction likely eliminates some genes that are directly controlled by this factor—for example, genes that are transiently expressed under anaerobiosis or are controlled on balance by levels of anaerobic metabolites as well as transcription—it yields a manageable data set of higher confidence for further analyses. Rox1-dependent repression also requires the recruitment of the Tup1-Ssn6 repression complex. However, a comparison of putative Rox1-regulated genes we identified with those derepressed in a tup1− strain (29) reveals surprisingly poor overlap. For example, only two genes in Table 1 were induced >2-fold in the tup1− strain and five were induced >1.5-fold (29). To include all of the Rox1-regulated genes from Table 1 and those we have identified, the expression ratio (tup1−/wild type) cutoff would be less than 1. Given the poor overlap, no further restrictions were placed on the Rox1 data set. The final Rox1 set contains a total of 106 genes that were significantly (P < 0.05) up-regulated in the rox1 strains during aerobiosis, contain putative Rox1-binding sites in their promoters, and were significantly (P < 0.05) up-regulated under anoxia in a wild-type strain. In this paper, we refer to these genes as Rox1-regulated genes. They are contained in a separate downloadable file at the web site referenced above and are discussed here in terms of the functional role they play in mediating the anoxic response.
Potentially non-Rox1-regulated genes induced in the rox1 null strains.
The number of genes significantly up-regulated in the rox1 null strains is surprisingly large compared to the number of genes induced under anoxia. Presumably, a number of these genes are up-regulated in response to the derepression of Rox1-regulated genes under aerobiosis and/or the resulting changes in metabolic signals in these strains. From a functional standpoint, the aerobic derepression of one or more Rox1-regulated genes likely induces oxidative stress. Several genes involved in mitigating oxidative stress, including CTT1 (catalase T), SOD1 (Cu/Zn superoxide dismutase) and TSA1 (thioredoxin peroxidase), are up-regulated in these strains. Given that these genes are down-regulated under anoxia, it is unlikely that Rox1 directly regulates their expression. Genes associated with other stress conditions, including HSP12, HSP26, and DDR2, were also aerobically up-regulated in the rox1 strains, as were 200 other genes that were not differentially expressed with respect to oxygen availability. Overall, these results suggest that the derepression of Rox1-regulated genes during aerobiosis, genes that are normally derepressed under anoxia, likely affects a number of other regulatory networks. They also point to the need to apply additional expression and/or sequence criteria in defining targeted genes when using single-gene-knockout models that may affect the balance of other regulatory networks.
Functional classification of oxygen-responsive and Rox1-regulated genes.
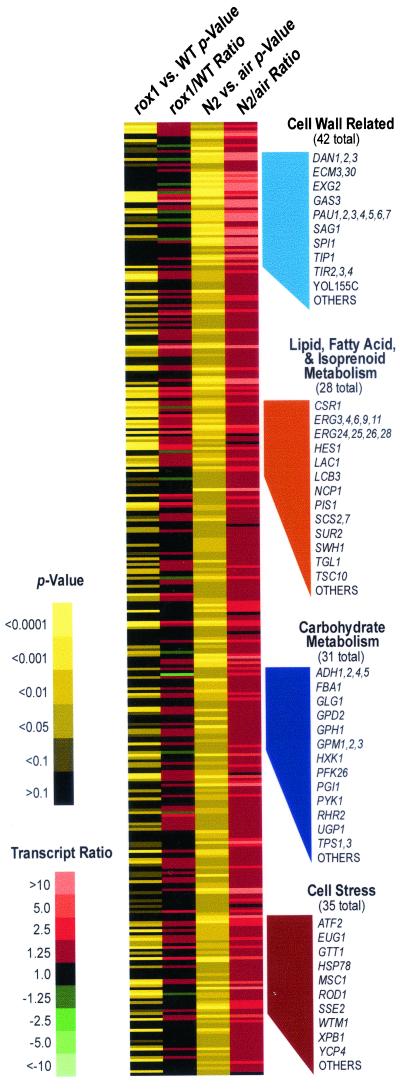
The changes in steady-state gene expression between aerobiosis and anaerobiosis are complex and result from the integration of multiple metabolic and environmental signals. Functional categorization of the anaerobically induced genes can provide insight into the genomic remodeling of gene expression that occurs and has revealed a number of interesting features. As illustrated in Fig. 3, nearly half of the anaerobically induced genes fit into four functional categories: cell wall related; lipid, fatty acid, and isoprenoid metabolism; carbohydrate metabolism; and cell stress. From Fig. 3 it is also apparent that the cell wall-related genes are up-regulated to a much larger extent than other functional groups. Figure 3 also shows that a substantial fraction (75%) of the anaerobically induced genes involved in lipid metabolism are Rox1 regulated but that Rox1 plays less of a role in regulating those involved in carbohydrate metabolism (32%), cell stress (36%), and cell wall maintenance (16%). A substantive fraction of other anaerobically induced genes have currently unknown cellular roles, and these results should therefore contribute to further functional annotation of the genome. A major goal of this study was to determine the role that Rox1 and other transcription factors play in regulating the physiological response to anoxia, so these functional categories are discussed separately below.
FIG. 3.
Functional classification of anaerobically induced genes. The 346 genes that were significantly (P < 0.05) up-regulated under anaerobiosis were grouped into major functional categories using both YPD and MIPS classifications. The ORFs are listed in alphabetical order except those appearing in specific functional groups for which gene names are provided. The ANOVA results (P values) and expression ratios (rox1/wild-type [WT] and anaerobic/aerobic [N2/air]) are shown for both comparisons and are represented by different colors, as indicated in the key. The 35 genes listed under “Cell Stress” are those left after overlap with the other functional categories had been eliminated.
Respiration, carbohydrate metabolism, and redox balance.
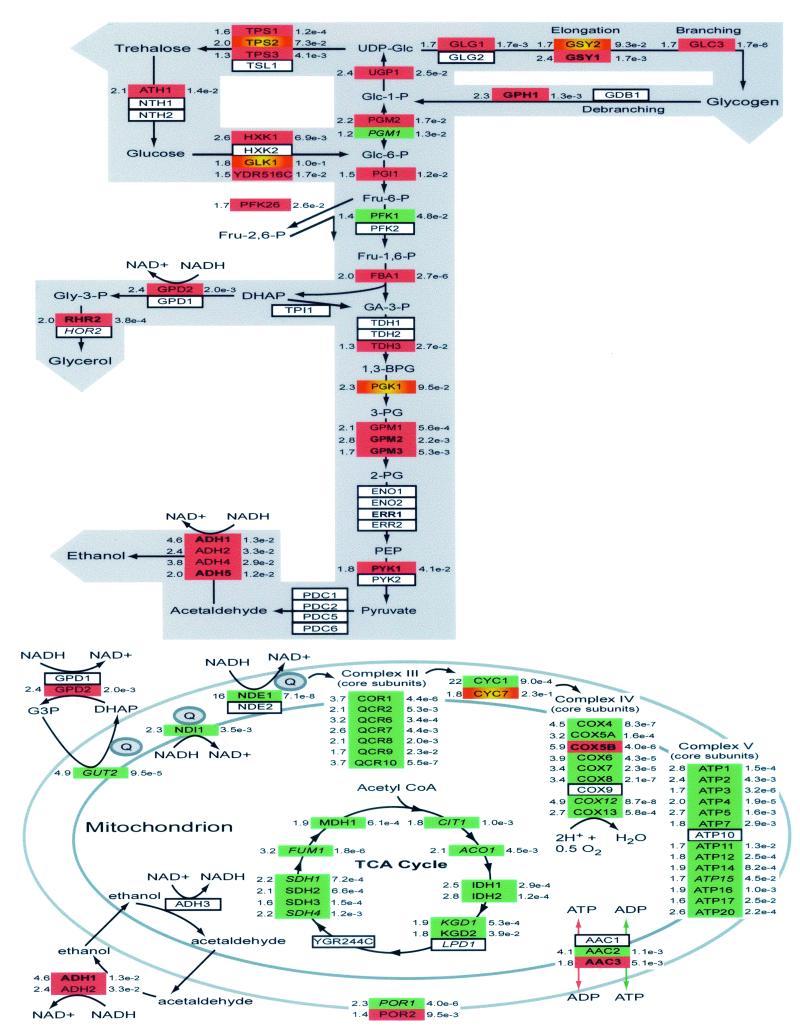
Many of the genes involved in redox balance and the dissimilatory pathways that utilize reducing equivalents were predictably altered in response to oxygen availability. Mapping the changes in mRNA levels of these genes onto metabolic pathways provides a means of assessing the molecular events that are responsible, in part, for altering metabolism (Fig. 4). As predicted, nearly all of the nuclear genes that encode respiratory complexes III, IV, and V and the tricarboxylic acid (TCA) cycle, as well as those responsible for intracellular translocation of reducing equivalents to mitochondria, are down-regulated during anoxia (Fig. 4, bottom). Several aerobic-anaerobic isoform pairs were also identified (e.g., CYC1/CYC7, COX5a/COX5b, AAC2/AAC3, and POR1/POR2). Overall, these results agree well with those obtained by DeRisi et al. (29) during a switch from fermentative to respiratory growth (diauxic transition). Moreover, given that our study represents a switch from respiro-fermentative (aerobic) to strictly fermentative (anoxic) growth on galactose, the absolute difference in transcript abundance for many of these genes is predictably less than that observed across the diauxic transition. Our results both confirm and extend those from a large number of previous studies showing that the effect of oxygen availability on respiratory and TCA cycle genes is exerted at the transcriptional level (reviewed in reference 30). However, they are in stark contrast to the results obtained by ter Linde et al. (76), who found that transcript levels of the majority of these genes showed little or no change in response to oxygen availability in glucose-limited chemostat cultures.
FIG. 4.
Schematic representation of oxygen-responsive and Rox1-regulated genes involved in reserve carbohydrate metabolism, glycolysis, redox balance, and mitochondrial function. The yeast genes encoding proteins involved in each step of the metabolic pathways are identified by name in the boxes, with bold lettering indicating significant (P < 0.05) induction in the rox1 strains and italics indicating significant repression. Red boxes indicate genes significantly up-regulated under anaerobiosis, red and yellow indicate those marginally up-regulated, green indicates those significantly up-regulated under aerobiosis, and white indicates those not affected. The magnitude of induction or repression with respect to oxygen availability is indicated to the left of each box, and the P value is shown to the right. The inferred metabolic reprogramming in the upper portion of the figure is indicated by the broad gray arrows. The direction of the arrows connecting reversible enzymatic steps indicates the inferred direction of the flow based upon expression levels with respect to oxygen availability. Abbreviations: UDP-glc, UDP glucose; Glc-1-P, glucose 1-phosphate, Glc-6-P, glucose 6-phosphate; Fru-6-P, fructose 6-phosphate; Fru-1,6-P, fructose 1,6-bisphosphate; DHAP, dihydroxyacetone phosphate; Gly-3-P, glycerol 3-phosphate; GA-3-P, glyceraldehyde 3-phosphate; 1,3-BPG, 1,3-bisphosphoglycerate; 3-PG, 3-phosphoglycerate; 2-PG, 2-phosphoglycerate; PEP, phosphoenolpyruvate; Q, ubiquinone.
In addition to the mitochondrially targeted genes shown, a number of genes associated with mitochondrial import systems and transporters were up-regulated under anoxia. Rather than playing a direct functional role during anaerobiosis, some of these may poise the organelle to respond to the reintroduction of oxygen. For example, it is possible that iso-2-cytochrome c (CYC7), whose expression is induced under anoxia but more strongly upon reoxygenation (15), plays a protective role upon reoxygenation, providing a sink for electrons from complex III in the face of partially assembled respiratory complexes and low levels of enzymes involved in mitigating oxidative stress (13). In addition, the anaerobic induction of EGD1, which is involved in targeting polypeptides from the ribosome to the mitochondrion, may allow rapid targeting of mitochondrially destined proteins upon the reintroduction of oxygen. The anaerobic induction of COT1, encoding a Zn transporter, may be associated with the anaerobic accumulation of Zn protoporphyrins noted previously (51). Finally, MDL1, a putative heme transporter, may transport heme from the mitochondrial matrix to the cytoplasm for the few b-type cytochromes that remain under anaerobiosis (51) or for the rapid mobilization of newly synthesized heme upon the reoxygenation.
Genes involved in anaerobic redox balance, reserve carbohydrate metabolism, and glycolysis were also predictably up-regulated under anoxia (Fig. 4, top). Transcript levels of the redox-regulated genes GPD2 and RHR2, which promote glycerol synthesis (and NAD+ regeneration), were anaerobically induced, whereas the isoform genes GPD1 and HOR2, which are involved in osmotic balance (6), were not. Similarly, cytoplasmic isoform genes encoding alcohol dehydrogenase were anaerobically induced whereas the mitochondrial isoform gene (ADH3) was not. A large number of genes involved in trehalose (TPS1, TPS2, and TPS3) and glycogen (GLG1, GSY1, GSY2, GLC3, and GIP2) synthesis were also induced concomitantly with some involved in their catabolism (ATH1 and GPH1, respectively), suggesting some cycling through these pathways during anaerobiosis as observed under other stress conditions (3, 63, 67). The induction of trehalose synthesis likely has a critical function, protecting membranes and integral-membrane-protein function in the face of increasing ethanol concentrations in anaerobically grown batch cultures (35).
Transcript levels of a large fraction of genes involved in glycolysis, as well as key allosteric modulators (e.g., PFK26), were also predictably up-regulated (Fig. 4). These results are consistent with quantitative measurements of anaerobic metabolites and stoichiometric models of flux changes during anaerobiosis (61), which show increased glycolytic flux towards ethanol and glycerol production along with an increased production and cycling of reserve carbohydrates (reviewed in reference 35). Moreover, they indicate that the redirection of metabolic flux is controlled, in part, by changes in gene expression. In terms of their regulation, Rox1 appears to play a minor yet important role, controlling some of the late steps of glycolysis (GPM2, GMP3, and PYK1 [CDC19]), ethanol formation (ADH1 and ADH5), glycogen synthesis (GSY1 and GIP2) and breakdown (GPH1), and possibly other steps (e.g., UGP1 and YDR516C). A search of the promoters of all anaerobically induced genes in this functional category reveals that over half contain one overrepresented (P < 0.05) sequence (TGTTTG), although it is currently not known what additional factors may be involved in controlling their expression.
Cell wall, lipid, and heme metabolism.
One of the most striking patterns in gene expression observed under anaerobiosis was for genes involved in remodeling the cell wall and membrane. These changes are necessitated, in part, by metabolic changes (fermentation, redox, and ethanol and glycerol production) and, in part, by lack of oxygen for the synthesis of various membrane components and enzymes. This remodeling activity is reflected as changes in the expression of genes for protein secretion, vesicle trafficking, and lipid synthesis. Moreover, the patterns observed under anaerobiosis differ substantially from those for cell wall integrity signaling (43), the unfolded-protein response (78), mitochondrial dysfunction (32), and the environmental stress response (38).
A large number of cell wall and wall-associated genes were up-regulated under anoxia and/or in the rox1 strains. As shown in Table 2, the expression of nearly the entire seripauperin (PAU) gene family was up-regulated under anoxia, with few of the genes exhibiting Rox1 regulation. All of these proteins have sites for glycosylation. In addition, members of the TIR/DAN/CWP subfamily have a C-terminal region containing a glycosyl-phosphatidylinositol (GPI) anchor site, and most of these are GPI linked into the outer layer of the cell wall (17). Surprisingly, CWP1 and CWP2, the major aerobic mannoproteins, were up-regulated in the rox1 strains, although only CWP1 was modestly up-regulated under anaerobiosis. The aerobic up-regulation of TIR2 in response to rox1 was totally strain dependent, with RZ53-6 showing no overall difference (P = 0.24) and JM43 exhibiting expressions ratios (rox1/wild type) as high as 64. Given the level of induction of the other PAU genes, it is likely that the expression of both DAN4 and TIR1 in JM43 is different from that in other yeast strains examined (1, 2, 21). Overall, these results both confirm and extend previous Northern blot studies showing that nearly all of the PAU gene family is anaerobically induced in an apparently Rox1-independent manner. As a functional group, these genes were up-regulated to an impressive extent under anaerobiosis. Changes in the expression of these genes can be expected to influence cell wall porosity (reviewed in reference 74) and may be necessitated in part by the changes in membrane fluidity that occur under anaerobiosis.
TABLE 2.
Comparison of transcript levels of the seripauperin (PAU) gene family in rox1 strains and wild-type strains grown under anaerobiosis
| ORF | Gene | Protein |
rox1 effect
|
N2 effect
|
||
|---|---|---|---|---|---|---|
| P | Ratioa | P | Ratiob | |||
| YJL223C | PAU1 | Member of the seripauperin family | 0.97 | 1.0 | 6.2e−5 | 16 |
| YIL176C | Seripauperin family member, protein identical to Pau1 | 0.28 | 0.84 | 4.9e−4 | 17 | |
| YEL049W | PAU2 | Member of the seripauperin family | 0.89 | 1.0 | 3.6e−4 | 7.8 |
| YCR104W | PAU3 | Member of the seripauperin family | 0.50 | 1.2 | 2.6e−7 | 13 |
| YLR461W | PAU4 | Member of the seripauperin family | 0.53 | 1.2 | 1.2e−4 | 6.9 |
| YFL020C | PAU5 | Member of the seripauperin family | 0.15 | 0.74 | 3.0e−5 | 24 |
| YNRO76W | PAU6 | Member of the seripauperin family | 0.61 | 0.92 | 1.9e−4 | 16 |
| YLL064C | Seripauperin family member; protein identical to Pau6 | 0.60 | 0.88 | 3.1e−4 | 23 | |
| YAR020C | PAU7 | Member of the seripauperin family | 0.25 | 1.5 | 6.7e−8 | 27 |
| YJR150C | DAN1 | Possible cell wall mannoprotein of the PAU family | 0.90 | 1.0 | 4.7e−10 | 73 |
| YLR037C | DAN2 | Member of the seripauperin family | 0.93 | 1.0 | 6.5e−4 | 16 |
| YBR301W | DAN3 | Cell wall mannoprotein of the PAU family | 5.8e−3 | 0.68 | 2.2e−4 | 19 |
| YJR151C | DAN4 | Member of the seripauperin family | 0.48 | 0.88 | 0.69 | 0.32 |
| YER011W | TIR1 | Stress-induced cell wall mannoprotein in the PAU family | 0.39 | 1.4 | 0.43 | 0.82 |
| YOR010C | TIR2 | Cold shock-induced member of the PAU family | 7.2e−16 | 30 | 1.2e−7 | 114 |
| YIL011W | TIR3 | Possible cell wall mannoprotein of the PAU family | 0.53 | 1.1 | 6.6e−10 | 38 |
| YOR009W | TIR4 | Possible cell wall mannoprotein of the PAU family | 0.11 | 1.3 | 2.3e−3 | 40 |
| YBR067C | TIP1 | Cold and heat shock-induced mannoprotein in PAU family | 1.3e−7 | 2.2 | 1.2e−3 | 2.0 |
| YAL068C | Member of the seripauperin family | 1.3e−5 | 0.67 | 1.1e−4 | 8.9 | |
| YDR542W | Member of the seripauperin family | 0.35 | 0.84 | 2.4e−4 | 16 | |
| YGL261C | Member of the seripauperin family | 0.06 | 0.80 | 2.1e−4 | 17 | |
| YGR294W | Member of the seripauperin family | 0.10 | 0.78 | 3.3e−4 | 23 | |
| YHL046C | Member of the seripauperin family | 0.41 | 0.86 | 3.1e−4 | 9.5 | |
| YIR041W | Member of the seripauperin family | 0.44 | 1.2 | 4.9e−10 | 16 | |
| YKL224C | Member of the seripauperin family | 0.07 | 0.76 | 4.6e−5 | 16 | |
| YLL025W | Member of the seripauperin family | 0.23 | 1.3 | 3.9e−5 | 11 | |
| YMR325W | Member of the seripauperin family | 0.70 | 1.2 | 3.6e−3 | 9.0 | |
| YOL161C | Member of the seripauperin family | 0.92 | 1.0 | 1.6e−4 | 17 | |
| YOR394W | Member of the seripauperin family | 0.40 | 1.3 | 2.3e−7 | 9.7 | |
| YPL282C | Member of the seripauperin family | 0.90 | 1.0 | 1.2e−4 | 8.1 | |
| YKL096W | CWP1 | Cell wall mannoprotein of the PAU family | 8.8e−5 | 1.9 | 3.0e−3 | 3.0 |
| YKL097W-A | CWP2 | Cell wall mannoprotein of the PAU family | 7.6e−3 | 1.3 | 0.02 | 0.72 |
Mean transcript level in the rox1 strains divided by that in the wild-type strains.
Mean transcript level observed under anaerobiosis divided by that under aerobiosis.
In concert with changes in the expression of structural proteins, cell wall-modifying genes were also differentially expressed with respect to oxygen availability. For example, the exoglucanase gene EXG2 and the Rox1-regulated glycosyl transferase gene GAS3 were up-regulated under anoxia, whereas the glucanase genes UTR2 and SCW10 were down-regulated. Genes for both isoforms of β-1,3-glucan synthase (FKS1 and GSC2) were down-regulated under anoxia, as was CHS2, encoding the chitin synthase responsible for normal septum synthesis (reviewed in reference 41). These changes can be expected to modify glucan linkages and may loosen the cell wall for the accommodation of additional PAU proteins under anaerobiosis. Genes involved in the processing of cell wall, plasma membrane, and secreted proteins in the endoplasmic reticulum (ER) were also differentially expressed. These include those involved in posttranslational translocation (SEC63), protein folding (SCC3) and modification (EUG1 and RCE1), and especially O- and N-linked glycosylation (KTR1, KTR4, KTR6, MNN11, OST2, PMT3, PMT5, and SWP1). Of these, SCC3, RCE1, OST2, PMT3, and PMT5 are Rox1 regulated.
Heme is a necessary cofactor for the cytochromes, catalases, and P450 enzymes of sterol synthesis, and its synthesis requires oxygen (51). Half of the heme-biosynthetic genes, including HEM4, HEM15, and the Rox1-regulated genes HEM13 and HEM15, were up-regulated under anoxia. Despite their up-regulation, heme cannot be synthesized without molecular oxygen. Rather, it is possible that under anaerobiosis, the heme liberated from the degradation of respiratory cytochromes (51) is recycled for use in the cytoplasm. It is conceivable that MDL1, a putative mitochondrial heme transporter of the ABC class (54), could be involved in such a function, as mentioned above.
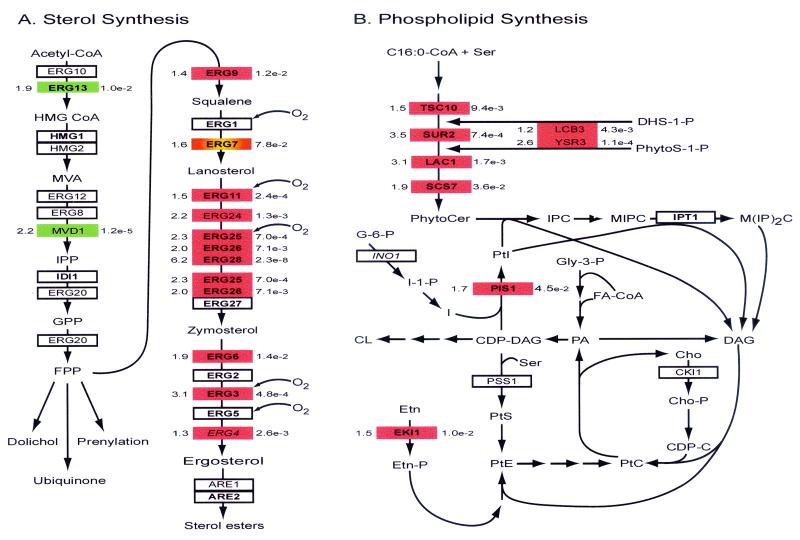
Both a sterol (at or beyond zymosterol) (Fig. 5A) and an unsaturated fatty acid are required for growth and must be exogenously supplied under anaerobiosis. The HES1 and SWH1 products (oxysterol homologs) are candidate sterol transporters (53) that were strongly induced under anaerobiosis. The genes for fatty acid import and activation (FAT1, FAA1, and FAA4) (34) were constitutively expressed, however, with respect to oxygen availability. In addition to the anaerobic up-regulation of sterol import genes, Fig. 5A shows that nearly all of the genes involved in the latter portion of sterol biosynthesis (beyond farnesylpyrophosphate [FPP]) were anaerobically induced, and many of them (ERG3, ERG6, ERG9, ERG11, ERG25, ERG26, and ERG28) are Rox1 regulated. Interestingly, a large number of other ERG genes were significantly up-regulated in the rox1 strains under aerobiosis (ERG1, ERG2, ERG5, ERG7, and ERG27) yet not under anaerobiosis.
FIG. 5.
Schematic representation of oxygen-responsive and Rox1-regulated genes involved in sterol and phospholipid biosynthesis. The yeast genes encoding proteins involved in sterol and phospholipid biosynthesis are identified by name in the boxes using the color and numbering scheme described in the legend to Fig. 4. Only key metabolic intermediates are shown. Abbreviations: HMG CoA, hydroxy-methylglutaryl coenzyme A; MVA, mevalonic acid; IPP, isopentynyl pyrophosphate; GPP, geranylpyrophosphate; FPP, farnesyl-pyrophosphate; C16:0-CoA, palmitic coenzyme A; CDP-C, CDP-choline; CDP-DAG, CDP-diacylglycerol; Cho, choline; Cho-P, cholinephosphate; CL, cardiolipin; DAG, diacylglycerol; DHS-1-P, dihydrosphingosine-1-phosphate; Etn, ethanolamine; Etn-P, ethanolaminephosphate; FA-CoA, fatty acid-CoA; G-6-P, glucose-6-phosphate; Gly-3-P, glycerol-3-phosphate; I, inositol; I-1-P, inositol-1-phosphate; IPC, inositol phosphorylceramide; MIPC, mannose-inositol-P-ceramide; M(IP)2C, phosphoinositol-mannose-inositol-P-ceramide; PA, phosphatidic acid; PtC, phosphatidylcholine; PtE, phosphatidylethanolamine; PtI, phosphatidylinositol; PtS, phosphatidylserine; PhytoCer, phytoceramide; PhytoS-1-P, phytosphingosine-1-phosphate; Ser, serine.
Genes involved in isoprenoid synthesis, including ERG10, ERG12, ERG8 and ERG20, were constitutively expressed or down-regulated under anaerobiosis in the case of ERG13 and MVD1 (Fig. 5A). Overexpression of mevalonate decarboxylase, encoded by MVD1, reduces sterol accumulation, so its down-regulation under anaerobiosis is consistent with the up-regulation of the later ERG genes observed (10). FPP is the starting point for several important synthesis pathways, including sterols, dolichol, ubiquinone, and prenylation of various membrane associated proteins. Dolichol is used as a carrier lipid or sugar donor in N glycosylation, O mannosylation, and GPI anchoring, all important for cell wall synthesis and secretion. Thus, the constitutive expression of the majority of genes involved in FPP synthesis with respect to oxygen availability is not unexpected.
Electrons for oxygen reduction by the P450 enzymes Erg5 and Erg11 and by Erg1 are usually supplied by Ncp1, whose gene is Rox1 regulated and strongly induced under anaerobiosis. Alternatively, electrons can be supplied by Cyb5, whose gene was up-regulated in the rox1 strains although not under anaerobiosis, and Cbr1, whose expression was unaltered by these treatments. This source of electrons is used by the Erg25 complex and Erg3 (52). However, flux through the latter portion of sterol synthesis is not possible without the electron acceptor oxygen (Fig. 5A). Under anaerobiosis, heme is depleted and sterol import occurs. Uptake of precursors such as zymosterol represses the expression of later ERG genes, but uptake of ergosterol does not (75). Apparently exogenous ergosterol is imported and cycles between membranes and lipid droplets but does not reach the control point in the ER, where most of the synthesis occurs (7, 75). Further inhibitory or defective intermediates may be acetylated by Atf2, whose gene was anaerobically induced, and removed by one of the ABC transporters (18). Such discrimination between imported and endogenously synthesized sterols may be retained under anaerobiosis because of the requirement of hormonal levels of specific sterols for cell cycle regulation in S. cerevisiae (24). Many sterols, such as cholesterol, are imported and used for bulk membranes, but they are not metabolized and do not regulate the expression of the later ERG genes. Overall, preference is given to endogenously synthesized ergosterol.
As shown in Fig. 5B, most of the genes encoding nonessential enzymes for sphingolipid synthesis (TSC10, SUR2, LAC1, and SCS7) are Rox1 regulated. Sphingolipid synthesis through ceramide occurs in the ER in yeast, and lipid rafts for transport of GPI proteins are evident from the ER on. The lack of early raft formation in lac1 strains accounts for the transport phenotype associated with this gene (9). In addition, the phosphatases for sphingosine-1-phosphates, encoded by LCB3 and YSR3, were up-regulated under anoxia, while their ligase (encoded by DPL1) (data not shown) was down-regulated. These sphingoid bases are thought to be involved in signaling in response to heat shock and other stresses (42) and may perform a similar function under anaerobiosis.
Much of the regulation for phospholipid synthesis is through metabolic intermediates such as phosphatidic acid (16). The lower part of Fig. 5B illustrates the metabolic regulation of phospholipid synthesis (16), showing a key regulatory point at CDP-diacylglycerol, going to phosphatidylinositol or phosphatidylserine, and feedback loops through phosphatidylcholine. Although the changes in gene expression in this pathway were minimal, they are probably important because they can be expected to alter the balance of phospholipid and sphingolipid synthesis. For example, the derepression of the Rox1-regulated PIS1 gene is consistent with the up-regulation of sphingolipid and GPI anchor synthesis observed, both of which facilitate transport of plasma membrane and cell wall proteins, independent of any signaling function. PIS1 is reported to be constitutively expressed with respect to a number of perturbations under aerobiosis (16), whereas Pss1 is regulated by inositol, protein kinase A, and transcription in order to control branching among the various phospholipids, with recycling of phosphatidylcholine providing further control (16). EKI1 regulation follows that of PIS1 in that it is also constitutively expressed as above (16) and Rox1 regulated. Similarly, the regulation of Cki1, which controls the lower synthesis pathways shown in Fig. 5B, is similar to that of Pss1. Derepression of the Rox1-regulated EKI1 gene can be expected to alter phospholipid metabolism given that alterations in any of the intermediates or enzymes affect the entire pathway (16).
Genes involved in the secretory pathway, endocytosis, trafficking to the vacuole, and vacuolar proteins were also differentially expressed with respect to oxygen availability. Multiple subunits of the vacuolar H+-ATPase (encoded by VMA4, VMA6, VMA8, and VMA10) are Rox1 regulated. In addition, some hydrolases (encoded by LAP4 and PEP4), which are also induced in the environmental stress response (38), were up-regulated under anoxia, as were other vacuolar proteins (e.g., ATH1 and the Rox1-regulated gene APE3). Anaerobic growth affects the vesicle transport system at several different stages. The small GTPases, which regulate vesicle docking and fusion, are regulated by guanine-nucleotide exchange factors (SEC12 and VPS16), GTPase-activating proteins (GYP1 and GYP6), and guanine nucleotide dissociation factors (GDI1), all of which were up-regulated under anaerobiosis (VPS16 and GYP6 are Rox1 regulated). These are involved in COPII and TRAPP trafficking from the ER to Golgi, within Golgi, and from trans-Golgi vesicles to the membrane and the vacuole. Other differentially expressed transport genes include the essential V-SNARE gene (VTI1) within the cis-Golgi; SRO77, necessary for docking and fusion at the plasma membrane; ERP5, associated with COPII; AKR2, for endocytosis; and genes for several proteins required for cytoplasm-to-vacuole transport (CVT9, CVT19, and APG12) or autophagy (APG12). Thus, anaerobiosis affects all steps in secretion and endocytosis, with the differential expression of a large number genes encoding regulatory proteins. The differential expression of these genes may very well facilitate transport of newly synthesized mannoproteins to the cell wall.
Differential expression of homologous gene pairs and other isoform genes.
S. cerevisiae is one of few yeast species that can grow in the complete absence of oxygen. This ability is thought to coincide with the gross gene duplication that occurred ∼100 million years ago in its lineage (59, 64). Thus, we might predict that a large fraction of homologous gene pairs are differentially expressed with respect to oxygen and encode proteins essential for anaerobic growth. A number of these gene pairs have been shown previously to be differentially expressed in response to a variety of other environmental conditions (19, 67).
Using the updated yeast genome duplication map of Seoighe and Wolfe (70), we examined the 455 gene pairs contained on the arrays for their response to oxygen availability and rox1. Remarkably, 117 gene pairs had at least one gene that was differentially expressed with respect to oxygen. Moreover, over one-fifth (73 genes) of the genes that were significantly up-regulated under anoxia are found in this group of 910 genes, including one-fourth of the Rox1-regulated genes. In addition, over one-eighth of the aerobically induced genes are members of this group. A number of the anaerobically induced genes from this group are shown in Fig. 3. Several are involved in reserve carbohydrate metabolism (PGM2, TPS3, and GLG1 [and GIP2; data not shown]), anaerobic redox balance (GPD2, ADH1, ADH2, and ADH5), glycolysis (TDH3, GPM2, and GPM3), and mitochondrial function (COX5b, AAC3, POR2, and CYC7). Many of these are regulated by Rox1 and perform seemingly critical functions under anaerobiosis. Although further genomic comparisons are required, these data lend themselves to the testing of the aforementioned hypothesis.
Although in the majority of cases only one of the genes in the pair was differentially expressed with respect to oxygen availability, a number of aerobic-hypoxic gene pairs were also revealed. In addition to the previously identified pairs HYP2/ANB1, COX5a/COX5b, AAC2/AAC3, and CYC1/CYC7, these included POR1/POR2, NRG1/NRG2, RSC30/RSC3, PSP1/YLR177W, PGM1/PGM2, and OYE3/OYE2. All of the previously identified hypoxic isoforms are Rox1 regulated, as are NRG2 and OYE2. Although functional analyses for many of these pairs are lacking, their differential expression in response to oxygen availability provides additional clues regarding the different functional roles they may play.
Insight into potentially branched and/or overlapping regulatory networks that Rox1 may affect.
Given that there is no evidence that Rox1 activates the transcription of any genes, the response of the 286 genes with significantly (P < 0.05) lower transcript levels in the rox1 null strains is likely the result of different metabolic and/or transcriptional signals indirectly associated with the chronic absence of this repressor. Nonetheless, an examination of this group of genes may provide insight into branched or overlapping regulatory networks that Rox1 may affect.
A large fraction (more than one-third) of the genes down-regulated in the rox1 null strains are also down-regulated under anaerobiosis in wild-type strains, which perhaps is not surprising given that the loss of this repressor and derepression of the Rox1-regulated genes is normally associated with anaerobic conditions (28). By far the largest functional group was genes associated with mitochondrial function, with the conspicuous absence, however, of the majority of nucleus-encoded respiratory subunits themselves. Table 3 is a compilation of some of these genes, specifically those that are either directly or indirectly involved in respiratory function. These include two of the major proteins (Por1 and Gut2) for import of cytoplasmic reducing equivalents, a large fraction of the TCA cycle complexes, and a number of assembly facilitators or components of respiratory complexes III, IV, and V. Although few of these, or other nucleus-encoded mitochondrial genes, were down-regulated to a large extent compared to that observed under anaerobiosis, given such a targeted set of genes it would seem that their down-regulation is an orchestrated response and one likely to have functional consequences. Indeed, as shown in Table 4, cyanide-sensitive respiration rates were significantly (P < 0.01) lower in the rox1 null strains than in their isogenic parents. This decrease in respiratory capacity is reflected as lower growth rates for the rox1 strains on YPGly (glycerol) and YPGal media (data not shown).
TABLE 3.
Selected mitochondrially targeted genes down-regulated in rox1 null strains under aerobiosis
| Functional group | ORF | Gene | Protein or function | Rox1 effect
|
Air/N2 effect
|
||
|---|---|---|---|---|---|---|---|
| P | Ratioa | P | Ratiob | ||||
| Cytoplasmic NADH import | YNL055C | POR1 | Major outer mitochondrial membrane voltage-dependent anion channel protein | 6.9e−3 | 1.5 | 4.0e−6 | 2.3 |
| YIL155C | GUT2 | FAD-dependent glycerol-3-phosphate dehydrogenase | 3.4e−2 | 1.2 | 9.5e−5 | 4.9 | |
| TCA complexes | YFL018C | LPD1 | Dihydrolipoamide dehydrogenase, (E3) component of PDH | 4.8e−2 | 1.3 | 2.4e−1 | 1.4 |
| YNR001C | CIT1 | Citrate synthetase | 3.3e−4 | 1.6 | 1.1e−3 | 1.8 | |
| YLR304C | ACO1 | Aconitase | 1.8e−2 | 1.4 | 4.5e−3 | 2.1 | |
| YDL066W | IDP1 | Isocitrate dehydrogenase (NADP+) | 4.1e−3 | 1.3 | 2.4e−2 | 1.4 | |
| YIL125W | KGD1 | α-Ketoglutarate dehydrogenase | 2.2e−3 | 1.6 | 5.3e−4 | 1.9 | |
| YKL148C | SDH1 | Succinate dehydrogenase flavoprotein subunit | 5.4e−3 | 1.5 | 7.2e−4 | 2.2 | |
| YDR178W | SDH4 | Succinate dehydrogenase membrane anchor subunit | 1.5e−2 | 1.4 | 1.2e−3 | 2.2 | |
| YPL262W | FUM1 | Fumarate hydratase | 1.4e−3 | 1.5 | 1.8e−6 | 3.2 | |
| Assembly facilitators and components of respiratory complexes III, IV, and V | YGR174C | CBP4 | Ubiquinol-cytochrome c reductase assembly factor | 4.3e−4 | 1.5 | 8.4e−3 | 1.7 |
| YML129C | COX14 | Cytochrome c oxidase assembly protein | 6.8e−3 | 1.4 | 3.0e−2 | 1.6 | |
| YLR038C | COX12 | Cytochrome c oxidase subunit VIb | 4.0e−2 | 1.3 | 8.7e−8 | 4.9 | |
| YKL087C | CYT2 | Holocytochrome c1 synthase | 7.5e−3 | 1.5 | 1.6e−3 | 2.1 | |
| YER154W | OXA1 | Assembly factor for F1F0-ATP synthase and cytochrome c oxidase | 2.2e−2 | 1.2 | 2.5e−3 | 1.5 | |
| YPL271W | ATP15 | Epsilon subunit of F1-ATP synthase | 2.4e−2 | 1.3 | 4.5e−2 | 1.7 | |
Mean transcript level in wild-type strains divided by that in rox1 strains pooled across genetic backgrounds and media.
Mean transcript level in wild-type strains grown under aerobiosis compared to that grown under anaerobiosis.
TABLE 4.
Rates of cyanide-sensitive and cyanide-insensitive respiration among wild-type and rox1 null strains
| Yeast strain | Respiration rate (nmol · min−1 · mg [wet weight]−1)a
|
|
|---|---|---|
| Cyanide sensitive | Cyanide insensitive | |
| JM43 | 8.9 ± 1.1 | 0.3 ± 0.03 |
| KKY6 (JM43Δrox1) | 5.8 ± 0.6∗ | 0.4 ± 0.07 |
| RZ53-6 | 3.8 ± 0.5 | 0.3 ± 0.03 |
| RZ53-6Δrox1 | 3.0 ± 0.5∗ | 0.3 ± 0.03 |
Values are means ± standard deviations (n ≥ 4). ∗, significantly (P < 0.01) lower than the value for the respective isogenic parent (ANOVA).
Although no single regulatory network is known to control the expression of these mitochondrially targeted genes, the heme-dependent activators Hap1 and the Hap2/3/4/5 complex control the expression of a number of them, and it is certainly plausible that a form of cross talk may exist between the heme-dependent aerobic and hypoxic regulons. In support of this hypothesis, both HAP1 and HAP4 were significantly (P < 0.005) down-regulated in the rox1 null strains; under anaerobiosis, HAP4 alone was down-regulated. Moreover, four of the TCA cycle genes listed in Table 4 (LPD1, CIT1, KGD1, and SDH1) are Hap2/3/4/5 regulated (12, 68). Although it is unlikely that lower HAP1 and HAP4 levels can solely account for the down-regulation of these genes given that other known target genes were not affected, it is clear that the derepression of Rox1-regulated genes under oxygen-replete conditions provides a signal of, or directly results in, a diminished capacity for mitochondrial function such as that normally associated with loss of this repressor under oxygen-limiting conditions.
The retrograde pathway (62) controls the expression of some of these genes (e.g., CIT1 and ACO1) and is known to activate their expression in response to diminished Hap2/3/4/5 activation or mitochondrial dysfunction (55). However, it apparently does not rescue the decrease in TCA spinning capacity despite a moderate (1.3-fold) yet significant (P < 0.005) increase in transcript levels of RTG3 alone (RTG1 and RTG2 transcripts were not affected). Moreover, considering that there is little overlap between the genes affected (up- or down-regulated) by rox1 and those regulated by the RTG pathway (32, 55), it seems unlikely that these pathways are part of a branched regulatory network that controls the expression of this subset of genes. Thus, the aerobic derepression of Rox1-regulated genes most likely affects an as-yet-unidentified regulatory network involved in controlling a number of nucleus-encoded mitochondrial genes.
Additional regulators of anaerobically induced genes.
In addition to Rox1, a number of other regulators have recently been shown to control the expression of anaerobically expressed genes in S. cerevisiae. Although searching for consensus binding sequences in the promoters of these genes can be fraught with problems associated with false positives, it may nonetheless yield insight into additional regulatory pathways involved in controlling their expression. Similarly, comparing the overlap in genes affected by oxygen availability or Rox1 with those from other genomic studies can provide insight into branched and overlapping regulatory networks. These approaches are discussed with particular reference to the PAU gene family below.
Regulation of cell wall genes.
As shown in Table 2, nearly the entire PAU gene family was significantly up-regulated under anaerobiosis. Recent reports (1, 2, 21) have implicated several factors, including the activator Upc2 (Mox4) and repressors Mox1, Mox2, Mot3, and Tup1/Ssn6, in controlling the expression of a large fraction of this gene family. From our analyses, part of the heme dependency noted previously (2, 21) for this gene family may be due to Rox1-dependent repression of UPC2; UPC2 was significantly (P < 0.01) up-regulated under aerobiosis in the rox1 null strains, and its promoter contains two consensus Rox1-binding sites (−305 TCTATTGTctTC −294 and −602 aCTATTGTgTtT −591). However, our results also suggest that the Rox1-dependent derepression of UPC2 under aerobiosis is insufficient for activation of this gene family, likely due to the repressive effects of other factors, such as Mox1 and/or Mox2, that are thought to interact with Upc2 (2, 21). Overall, these results suggest that Rox1 plays a minor role in the aerobic repression of this gene family through UPC2 but that additional regulators are primarily responsible for its induction under anaerobiosis.
Using the entire set of anaerobically induced genes, we searched for allowable permutations of the AR1 site (TCGTWHAG and TCGTTHRAG) (21) in order to gain insight into the network of genes that Upc2 may regulate. A surprisingly large number of anaerobically induced genes (106, the same number of identified Rox1 genes) contain AR1 sites, including all but one member of the PAU gene family (TIP1) (Table 2). Moreover, after Rox1-regulated genes had been eliminated from the set of anaerobically induced genes, a random search of the promoters identified several overrepresented (P < 0.01) sequences, the majority consisting of permutations of the AR1 site (e.g., TCGTTTA in 45 of 240 genes, P = 1.0e−4; ATCGTTA in 26 of 240, P = 4.9e−8; CGTTTAAG in 24 of 240, P = 7.9e−11). Remarkably, 31 of 34 cell wall-related genes that were induced under anaerobiosis have AR1 sites; Rox1 regulates only a few genes in this functional category (e.g., TIR2, PMT3, CWP1, and YMR215W). Although additional studies will be required for verification, these results suggest that nearly all of the cell wall genes that are anaerobically induced are regulated by Upc2. Based upon the occurrence of AR1 sites in other anaerobically induced genes, there is apparently substantive overlap in Rox1 and Upc2 regulation of genes involved in lipid and heme metabolism; CSR1, ERG3, ERG4, ERG9, ERG11, ERG24, ERG25, HEM14, HES1, SCC3, SCS2, MDL1, NCP1, YDR213W, and YLR380W are likely regulated by both. A large fraction (∼40%) of other AR1-containing genes encode proteins of unknown function, and these results should therefore provide additional information for genomic annotations.
Additional promoter searches of the anaerobically induced genes revealed AR2 sites (MWAAAWTGWTGA) in a few PAU genes (YHL046C, YGR294W, and PBL2) other than those identified previously (DAN1, DAN2, and DAN3) (21). We also searched for other cis regulatory sequences (e.g., STRE, LORE, and that for Mot3), but they were not significantly (P > 0.05) overrepresented in the list of anaerobically induced genes. Although Msn2/Msn4 and Mot3 likely regulate a number of these genes, the probability of finding the STRE consensus sequence (CCCCT) or that for Mot3 (HAGGYA) at the frequency observed was 100%. Ongoing studies in our laboratory are using a similar genomic approach to determine the extent to which Msn2/Msn4 and other transcription factors are involved in controlling the expression of anaerobically induced genes. Finally, a comparison of both the Rox1-regulated and anaerobically induced genes with those identified in other environmental stress conditions (19, 38) reveals surprisingly poor overlap, suggesting that the regulatory networks involved in the adaptive response to anaerobiosis do not substantially overlap with those controlling the response to other environmental challenges.
Conclusions.
In this study we focused on the physiological response of S. cerevisiae to anaerobiosis and the functional roles that Rox1 and other trans-acting factors play in mediating the response. One of the most striking patterns in expression was for genes involved in the remodeling of the cell wall and plasma membrane. This remodeling activity was reflected as changes in the expression of genes for cell wall structural proteins, modifying enzymes, protein secretion, vesicle trafficking, and phospholipid, sphingolipid, and sterol metabolism. These changes are likely necessitated by anaerobically induced changes in metabolism as well as lack of oxygen for the synthesis of various membrane components and enzymes. They can be predicted to modify cell wall porosity and combat a compromised ability to regulate membrane fluidity under anaerobiosis. Predictable changes in dissimilatory metabolism, mitochondrial function, anaerobic redox balance, and reserve carbohydrate metabolism were also observed. It is apparent that Rox1 plays an important role in regulating key steps in these processes and a predominate role in the anaerobic remodeling of sterol and lipid metabolism. Moreover, Rox1 controls the expression of a large fraction of the oxygen-requiring enzymes that are anaerobically induced. A comparison of genes affected in the rox1 null strains to those induced under anaerobiosis reveals striking differences in all functional groups, in particular for those involved in cell wall and secretion. These results argue for the involvement of multiple regulators in instigating the anaerobic program of gene expression. Moreover, a comparison of the expression ratios for Rox1-regulated genes in wild-type strains under anaerobiosis with those in the rox1 strains under aerobiosis suggests that additional regulators are involved in dictating transcript levels of Rox1-regulated genes. Although additional work clearly will be required for disentangling these regulatory networks, results from this study suggest that Rox1, in concert with Upc2, plays a dominate role in controlling the genetic program underlying the physiological response of S. cerevisiae to oxygen deprivation.
Acknowledgments
We thank R. Zitomer and R. Poyton for providing yeast strains and plasmids. We are indebted to K. Locklar and C. Russell of ResGen for their generous donation of a number of GeneFilters and useful discussions on numerical analysis. We also thank J. Hickok and K. Schutterle for their contributions to the functional analyses.
This work was supported by National Institutes of Health grant RO1-GM59826 and American Heart Association grant 9930310Z to K.E.K.
REFERENCES
- 1.Abramova, N., O. Sertil, S. Mehta, and C. V. Lowry. 2001. Reciprocal regulation of anaerobic and aerobic cell wall mannoprotein gene expression in Saccharomyces cerevisiae. J. Bacteriol. 183:2881–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abramova, N. E., B. D. Cohen, O. Sertil, R. Kapoor, K. J. Davies, and C. V. Lowry. 2001. Regulatory mechanisms controlling expression of the DAN/TIR mannoprotein genes during anaerobic remodeling of the cell wall in Saccharomyces cerevisiae. Genetics 157:1169–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexandre, H., V. Ansanay-Galeote, S. Dequin, and B. Blondin. 2001. Global gene expression during short-term ethanol stress in Saccharomyces cerevisiae. FEBS Lett. 498:98–103. [DOI] [PubMed] [Google Scholar]
- 4.Andreasen, A., and T. Stier. 1953. Anaerobic nutrition of Saccharomyces cerevisiae. I. Ergosterol requirement for growth in a defined medium. J. Cell. Comp. Physiol. 41:23–36. [DOI] [PubMed] [Google Scholar]
- 5.Andreasen, A., and T. Stier. 1954. Anaerobic nutrition of Saccharomyces cerevisiae. II. Unsaturated fatty acid requirement for growth in defined medium. J. Cell. Comp. Physiol. 43:271–281. [DOI] [PubMed] [Google Scholar]
- 6.Ansell, R., K. Granath, S. Hohmann, J. M. Thevelein, and L. Adler. 1997. The two isoenzymes for yeast NAD+-dependent glycerol 3-phosphate dehydrogenase encoded by GPD1 and GPD2 have distinct roles in osmoadaptation and redox regulation. EMBO J. 16:2179–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Athenstaedt, K., D. Zweytick, A. Jandrositz, S. D. Kohlwein, and G. Daum. 1999. Identification and characterization of major lipid particle proteins of the yeast Saccharomyces cerevisiae. J. Bacteriol. 181:6441–6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balasubramanian, B., C. V. Lowry, and R. S. Zitomer. 1993. The Rox1 repressor of the Saccharomyces cerevisiae hypoxic genes is a specific DNA-binding protein with a high-mobility-group motif. Mol. Cell. Biol. 13:6071–6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barz, W. P., and P. Walter. 1999. Two endoplasmic reticulum (ER) membrane proteins that facilitate ER-to-Golgi transport of glycosylphosphatidylinositol-anchored proteins. Mol. Biol. Cell 10:1043–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berges, T., D. Guyonnet, and F. Karst. 1997. The Saccharomyces cerevisiae mevalonate diphosphate decarboxylase is essential for viability, and a single Leu-to-Pro mutation in a conserved sequence leads to thermosensitivity. J. Bacteriol. 179:4664–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bourdineaud, J. P., G. De Sampaio, and G. J. Lauquin. 2000. A Rox1-independent hypoxic pathway in yeast. Antagonistic action of the repressor Ord1 and activator Yap1 for hypoxic expression of the SRP1/TIR1 gene. Mol. Microbiol. 38:879–890. [DOI] [PubMed] [Google Scholar]
- 12.Bowman, S. B., Z. Zaman, L. P. Collinson, A. J. Brown, and I. W. Dawes. 1992. Positive regulation of the LPD1 gene of Saccharomyces cerevisiae by the HAP2/HAP3/HAP4 activation system. Mol. Gen. Genet. 231:296–303. [DOI] [PubMed] [Google Scholar]
- 13.Burke, P. V., and K. E. Kwast. 2000. Oxygen dependence of expression of cytochrome c and cytochrome c oxidase genes in S. cerevisiae. Adv. Exp. Med. Biol. 475:197–208. [DOI] [PubMed] [Google Scholar]
- 14.Burke, P. V., K. E. Kwast, F. Everts, and R. O. Poyton. 1998. A fermentor system for regulating oxygen at low concentrations in cultures of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 64:1040–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burke, P. V., D. C. Raitt, L. A. Allen, E. A. Kellogg, and R. O. Poyton. 1997. Effects of oxygen concentration on the expression of cytochrome c and cytochrome c oxidase genes in yeast. J. Biol. Chem. 272:14705–14712. [DOI] [PubMed] [Google Scholar]
- 16.Carman, G. M., and S. A. Henry. 1999. Phospholipid biosynthesis in the yeast Saccharomyces cerevisiae and interrelationship with other metabolic processes. Prog. Lipid Res. 38:361–399. [DOI] [PubMed] [Google Scholar]
- 17.Caro, L. H., H. Tettelin, J. H. Vossen, A. F. Ram, E. H. van Den, and F. M. Klis. 1997. In silico identification of glycosyl-phosphatidylinositol-anchored plasma-membrane and cell wall proteins of Saccharomyces cerevisiae. Yeast 13:1477–1489. [DOI] [PubMed] [Google Scholar]
- 18.Cauet, G., E. Degryse, C. Ledoux, R. Spagnoli, and T. Achstetter. 1999. Pregnenolone esterification in Saccharomyces cerevisiae. A potential detoxification mechanism. Eur. J. Biochem. 261:317–324. [DOI] [PubMed] [Google Scholar]
- 19.Causton, H. C., B. Ren, S. S. Koh, C. T. Harbison, E. Kanin, E. G. Jennings, T. I. Lee, H. L. True, E. S. Lander, and R. A. Young. 2001. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12:323–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi, J. Y., J. Stukey, S. Y. Hwang, and C. E. Martin. 1996. Regulatory elements that control transcription activation and unsaturated fatty acid-mediated repression of the Saccharomyces cerevisiae OLE1 gene. J. Biol. Chem. 271:3581–3589. [DOI] [PubMed] [Google Scholar]
- 21.Cohen, B. D., O. Sertil, N. E. Abramova, K. J. Davies, and C. V. Lowry. 2001. Induction and repression of DAN1 and the family of anaerobic mannoprotein genes in Saccharomyces cerevisiae occurs through a complex array of regulatory sites. Nucleic Acids Res. 29:799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crowley, J. H., F. W. Leak, Jr., K. V. Shianna, S. Tove, and L. W. Parks. 1998. A mutation in a purported regulatory gene affects control of sterol uptake in Saccharomyces cerevisiae. J. Bacteriol. 180:4177–4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cumsky, M. G., C. Ko, C. E. Trueblood, and R. O. Poyton. 1985. Two nonidentical forms of subunit V are functional in yeast cytochrome c oxidase. Proc. Natl. Acad. Sci. USA 82:2235–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dahl, C., H. P. Biemann, and J. Dahl. 1987. A protein kinase antigenically related to pp60v-src possibly involved in yeast cell cycle control: positive in vivo regulation by sterol. Proc. Natl. Acad. Sci. USA 84:4012–4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deckert, J., R. A. Khalaf, S. M. Hwang, and R. S. Zitomer. 1999. Characterization of the DNA binding and bending HMG domain of the yeast hypoxic repressor Rox1. Nucleic Acids Res. 27:3518–3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deckert, J., R. Perini, B. Balasubramanian, and R. S. Zitomer. 1995. Multiple elements and auto-repression regulate Rox1, a repressor of hypoxic genes in Saccharomyces cerevisiae. Genetics 139:1149–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deckert, J., A. M. Rodriguez Torres, J. T. Simon, and R. S. Zitomer. 1995. Mutational analysis of Rox1, a DNA-bending repressor of hypoxic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:6109–6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deckert, J., A. M. Torres, S. M. Hwang, A. J. Kastaniotis, and R. S. Zitomer. 1998. The anatomy of a hypoxic operator in Saccharomyces cerevisiae. Genetics 150:1429–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680–686. [DOI] [PubMed] [Google Scholar]
- 30.de Winde, J. H., and L. A. Grivell. 1993. Global regulation of mitochondrial biogenesis in Saccharomyces cerevisiae. Prog. Nucleic Acid Res. Mol. Biol. 46:51–91. [DOI] [PubMed] [Google Scholar]
- 31.Donzeau, M., J. P. Bourdineaud, and G. J. Lauquin. 1996. Regulation by low temperatures and anaerobiosis of a yeast gene specifying a putative GPI-anchored plasma membrane protein. Mol. Microbiol. 20:449–459. (Erratum, 21:431.) [DOI] [PubMed] [Google Scholar]
- 32.Epstein, C. B., J. A. Waddle, W. Hale, V. Dave, J. Thornton, T. L. Macatee, H. R. Garner, and R. A. Butow. 2001. Genome-wide responses to mitochondrial dysfunction. Mol. Biol. Cell 12:297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Errede, B., T. S. Cardillo, M. A. Teague, and F. Sherman. 1984. Identification of regulatory regions within the Ty1 transposable element that regulate iso-2-cytochrome c production in the CYC7- H2 yeast mutant. Mol. Cell. Biol. 4:1393–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fargeman, N. J., P. N. Black, X. D. Zhao, J. Knudsen, and C. C. DiRusso. 2001. The Acyl-CoA synthetases encoded within FAA1 and FAA4 in Saccharomyces cerevisiae function as components of the fatty acid transport system linking import, activation, and intracellular utilization. J. Biol. Chem. 276:37051–37059. [DOI] [PubMed] [Google Scholar]
- 35.Francois, J., and J. L. Parrou. 2001. Reserve carbohydrates metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25:125–145. [DOI] [PubMed] [Google Scholar]
- 36.Fujiwara, D., O. Kobayashi, H. Yoshimoto, S. Harashima, and Y. Tamai. 1999. Molecular mechanism of the multiple regulation of the Saccharomyces cerevisiae ATF1 gene encoding alcohol acetyltransferase. Yeast 15:1183–1197. [DOI] [PubMed] [Google Scholar]
- 37.Fujiwara, D. U., H. Yoshimoto, H. Sone, S. Harashima, and Y. Tamai. 1998. Transcriptional co-regulation of Saccharomyces cerevisiae alcohol acetyltransferase gene, ATF1 and delta-9 fatty acid desaturase gene, OLE1 by unsaturated fatty acids. Yeast 14:711–721. [DOI] [PubMed] [Google Scholar]
- 38.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gnaiger, E., R. Steinlechner-Maran, G. Mendez, T. Eberl, and R. Margreiter. 1995. Control of mitochondrial and cellular respiration by oxygen. J. Bioenerg. Biomembr. 27:583–596. [DOI] [PubMed] [Google Scholar]
- 40.Hardwick, J. S., F. G. Kuruvilla, J. K. Tong, A. F. Shamji, and S. L. Schreiber. 1999. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl. Acad. Sci. USA 96:14866–14870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henar, V. M., A. Duran, and C. Roncero. 1999. Chitin synthases in yeast and fungi. EXS 87:55–69. [DOI] [PubMed] [Google Scholar]
- 42.Jenkins, G. M., and Y. A. Hannun. 2001. Role for de novo sphingoid base biosynthesis in the heat-induced transient cell cycle arrest of Saccharomyces cerevisiae. J. Biol. Chem. 276:8574–8581. [DOI] [PubMed] [Google Scholar]
- 43.Jung, U. S., and D. E. Levin. 1999. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signaling pathway. Mol. Microbiol. 34:1049–1057. [DOI] [PubMed] [Google Scholar]
- 44.Kastaniotis, A. J., T. A. Mennella, C. Konrad, A. M. Torres, and R. S. Zitomer. 2000. Roles of transcription factor Mot3 and chromatin in repression of the hypoxic gene ANB1 in yeast. Mol. Cell. Biol. 20:7088–7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kastaniotis, A. J., and R. S. Zitomer. 2000. Rox1 mediated repression. Oxygen dependent repression in yeast. Adv. Exp. Med. Biol. 475:185–195. [PubMed] [Google Scholar]
- 46.Keng, T. 1992. HAP1 and ROX1 form a regulatory pathway in the repression of HEM13 transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:2616–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuhn, K. M., J. L. DeRisi, P. O. Brown, and P. Sarnow. 2001. Global and specific translational regulation in the genomic response of Saccharomyces cerevisiae to a rapid transfer from a fermentable to a nonfermentable carbon source. Mol. Cell. Biol. 21:916–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kwast, K. E., P. V. Burke, K. Brown, and R. O. Poyton. 1997. REO1 and ROX1 are alleles of the same gene which encodes a transcriptional repressor of hypoxic genes in Saccharomyces cerevisiae. Curr. Genet. 32:377–383. [DOI] [PubMed] [Google Scholar]
- 49.Kwast, K. E., P. V. Burke, and R. O. Poyton. 1998. Oxygen sensing and the transcriptional regulation of oxygen-responsive genes in yeast. J. Exp. Biol. 201:1177–1195. [DOI] [PubMed] [Google Scholar]
- 50.Kwast, K. E., P. V. Burke, B. T. Staahl, and R. O. Poyton. 1999. Oxygen sensing in yeast: evidence for the involvement of the respiratory chain in regulating the transcription of a subset of hypoxic genes. Proc. Natl. Acad. Sci. USA 96:5446–5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Labbe-Bois, R., and P. Labbe. 1990. Tetrapyrrole and heme biosynthesis in the yeast Saccharomyces cerevisiae, p.235–285. In H. A. Dailey (ed.), Biosynthesis of heme and chlorophylls. McGraw-Hill, New York, N.Y.
- 52.Lamb, D. C., D. E. Kelly, N. J. Manning, M. A. Kaderbhai, and S. L. Kelly. 1999. Biodiversity of the P450 catalytic cycle: yeast cytochrome b5/NADH cytochrome b5 reductase complex efficiently drives the entire sterol 14-demethylation (CYP51) reaction. FEBS Lett. 462:283–288. [DOI] [PubMed] [Google Scholar]
- 53.Levine, T. P., and S. Munro. 2001. Dual targeting of Osh1p, a yeast homologue of oxysterol-binding protein, to both the Golgi and the nucleus-vacuole junction. Mol. Biol. Cell 12:1633–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lill, R., and G. Kispal. 2001. Mitochondrial ABC transporters. Res. Microbiol. 152:331–340. [DOI] [PubMed] [Google Scholar]
- 55.Liu, Z., and R. A. Butow. 1999. A transcriptional switch in the expression of yeast tricarboxylic acid cycle genes in response to a reduction or loss of respiratory function. Mol. Cell. Biol. 19:6720–6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lowry, C. V., M. E. Cerdan, and R. S. Zitomer. 1990. A hypoxic consensus operator and a constitutive activation region regulate the ANB1 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 10:5921–5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lowry, C. V., and R. S. Zitomer. 1984. Oxygen regulation of anaerobic and aerobic genes mediated by a common factor in yeast. Proc. Natl. Acad. Sci. USA 81:6129–6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lowry, C. V., and R. S. Zitomer. 1988. ROX1 encodes a heme-induced repression factor regulating ANB1 and CYC7 of Saccharomyces cerevisiae. Mol. Cell. Biol. 8:4651–4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moller, K., L. Olsson, and J. Piskur. 2001. Ability for anaerobic growth is not sufficient for development of the petite phenotype in Saccharomyces kluyveri. J. Bacteriol. 183:2485–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ness, F., S. Bourot, M. Regnacq, R. Spagnoli, T. Berges, and F. Karst. 2001. SUT1 is a putative Zn[II]2Cys6-transcription factor whose upregulation enhances both sterol uptake and synthesis in aerobically growing Saccharomyces cerevisiae cells. Eur. J. Biochem. 268:1585–1595. [PubMed] [Google Scholar]
- 61.Nissen, T. L., U. Schulze, J. Nielsen, and J. Villadsen. 1997. Flux distributions in anaerobic, glucose-limited continuous cultures of Saccharomyces cerevisiae. Microbiology 143:203–218. [DOI] [PubMed] [Google Scholar]
- 62.Parikh, V. S., M. M. Morgan, R. Scott, L. S. Clements, and R. A. Butow. 1987. The mitochondrial genotype can influence nuclear gene expression in yeast. Science 235:576–580. [DOI] [PubMed] [Google Scholar]
- 63.Parrou, J. L., M. A. Teste, and J. Francois. 1997. Effects of various types of stress on the metabolism of reserve carbohydrates in Saccharomyces cerevisiae: genetic evidence for a stress-induced recycling of glycogen and trehalose. Microbiology 143:1891–1900. [DOI] [PubMed] [Google Scholar]
- 64.Piskur, J. 2001. Origin of the duplicated regions in the yeast genomes. Trends Genet. 17:302–303. [DOI] [PubMed] [Google Scholar]
- 65.Prezant, T., K. Pfeifer, and L. Guarente. 1987. Organization of the regulatory region of the yeast CYC7 gene: multiple factors are involved in regulation. Mol. Cell. Biol. 7:3252–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Regnacq, M., P. Alimardani, B. E. Moudni, and T. Berges. 2001. Sut1p interaction with Cyc8p(Ssn6p) relieves hypoxic genes from Cyc8p-Tup1p repression in Saccharomyces cerevisiae. Mol. Microbiol. 40:1085–1096. [DOI] [PubMed] [Google Scholar]
- 67.Rep, M., M. Krantz, J. M. Thevelein, and S. Hohmann. 2000. The transcriptional response of Saccharomyces cerevisiae to osmotic shock. Hot1p and Msn2p/Msn4p are required for the induction of subsets of high osmolarity glycerol pathway-dependent genes. J. Biol. Chem. 275:8290–8300. [DOI] [PubMed] [Google Scholar]
- 68.Repetto, B., and A. Tzagoloff. 1989. Structure and regulation of KGD1, the structural gene for yeast alpha-ketoglutarate dehydrogenase. Mol. Cell. Biol. 9:2695–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sabova, L., I. Zeman, F. Supek, and J. Kolarov. 1993. Transcriptional control of AAC3 gene encoding mitochondrial ADP/ATP translocator in Saccharomyces cerevisiae by oxygen, heme and ROX1 factor. Eur. J. Biochem. 213:547–553. [DOI] [PubMed] [Google Scholar]
- 70.Seoighe, C., and K. H. Wolfe. 1999. Updated map of duplicated regions in the yeast genome. Gene 238:253–261. [DOI] [PubMed] [Google Scholar]
- 71.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3–21. [DOI] [PubMed] [Google Scholar]
- 72.Sherman, F., J. W. Stewart, C. Helms, and J. A. Downie. 1978. Chromosome mapping of the CYC7 gene determining yeast iso-2-cytochrome c: structural and regulatory regions. Proc. Natl. Acad. Sci. USA 75:1437–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smith, R. L., and A. D. Johnson. 2000. Turning genes off by Ssn6-Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem. Sci. 25:325–330. [DOI] [PubMed] [Google Scholar]
- 74.Smits, G. J., J. C. Kapteyn, E. H. van den, and F. M. Klis. 1999. Cell wall dynamics in yeast. Curr. Opin. Microbiol. 2:348–352. [DOI] [PubMed] [Google Scholar]
- 75.Soustre, I., P. H. Dupuy, S. Silve, F. Karst, and G. Loison. 2000. Sterol metabolism and ERG2 gene regulation in the yeast Saccharomyces cerevisiae. FEBS Lett. 470:102–106. [DOI] [PubMed] [Google Scholar]
- 76.ter Linde, J. J., H. Liang, R. W. Davis, H. Y. Steensma, J. P. van Dijken, and J. T. Pronk. 1999. Genome-wide transcriptional analysis of aerobic and anaerobic chemostat cultures of Saccharomyces cerevisiae. J. Bacteriol. 181:7409–7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thorsness, M., W. Schafer, L. D’Ari, and J. Rine. 1989. Positive and negative transcriptional control by heme of genes encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase in Saccharomyces cerevisiae. Mol. Cell. Biol. 9:5702–5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Travers, K. J., C. K. Patil, L. Wodicka, D. J. Lockhart, J. S. Weissman, and P. Walter. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101:249–258. [DOI] [PubMed] [Google Scholar]
- 79.Trueblood, C. E., R. M. Wright, and R. O. Poyton. 1988. Differential regulation of the two genes encoding Saccharomyces cerevisiae cytochrome c oxidase subunit V by heme and the HAP2 and REO1 genes. Mol. Cell. Biol. 8:4537–4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Turi, T. G., and J. C. Loper. 1992. Multiple regulatory elements control expression of the gene encoding the Saccharomyces cerevisiae cytochrome P450, lanosterol 14 alpha-demethylase (ERG11). J. Biol. Chem. 267:2046–2056. [PubMed] [Google Scholar]
- 81.Vasconcelles, M. J., Y. Jiang, K. McDaid, L. Gilooly, S. Wretzel, D. L. Porter, C. E. Martin, and M. A. Goldberg. 2001. Identification and characterization of a low oxygen response element involved in the hypoxic induction of a family of Saccharomyces cerevisiae genes. Implications for the conservation of oxygen sensing in eukaryotes. J. Biol. Chem. 276:14374–14384. [DOI] [PubMed] [Google Scholar]
- 82.Visser, W., W. A. Scheffers, W. H. Batenburg-van der Vegte, and J. P. van Dijken. 1990. Oxygen requirements of yeasts. Appl. Environ. Microbiol. 56:3785–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zitomer, R. S., P. Carrico, and J. Deckert. 1997. Regulation of hypoxic gene expression in yeast. Kidney Int. 51:507–513. [DOI] [PubMed] [Google Scholar]
- 84.Zitomer, R. S., and C. V. Lowry. 1992. Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol. Rev. 56:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zitomer, R. S., J. W. Sellers, D. W. McCarter, G. A. Hastings, P. Wick, and C. V. Lowry. 1987. Elements involved in oxygen regulation of the Saccharomyces cerevisiae CYC7 gene. Mol. Cell. Biol. 7:2212–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]