Abstract
Tn5 is a composite bacterial transposon that encodes a protein, transposase (Tnp), required for movement of the transposon. The initial step in the transposition pathway involves specific binding of Tnp to 19-bp end recognition sequences. Tn5 contains two different specific end sequences, termed outside end (OE) and inside end (IE). In Escherichia coli, IE is methylated by Dam methylase (IEME). This methylation greatly inhibits recognition by Tnp and greatly reduces the ability of transposase to facilitate movement of IE defined transposons. Through use of a combinatorial random mutagenesis technique (DNA shuffling), we have isolated an IEME-specific hyperactive form of Tnp, Tnp sC7v.2.0, that is able to promote high levels of transposition of IEME defined transposons in vivo and in vitro while functioning at wild-type levels with OE transposons. This protein contains a critical glutamate-to-valine mutation at amino acid 58 that is responsible for this change in end specificity.
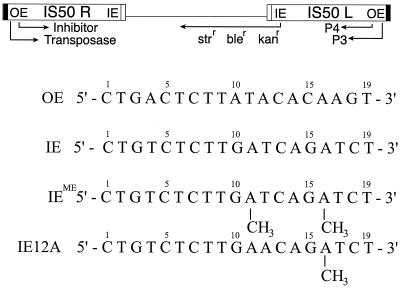
The Tn5 bacterial transposon (Fig. 1) is a conservative “cut and paste” transposon of the IS4 family (12). It encodes a 53-kDa transposase protein (Tnp) that is responsible for its movement. Tnp facilitates movement of the entire Tn5 transposon by initially binding to each of two 19-bp specific binding sequences termed outside end (OE), followed by formation of a nucleoprotein structure termed a synapse, blunt-ended cleavage of each end, association with a target DNA, and then strand transfer (11). Transposase can also promote movement of a single IS50 insertion sequence by using a combination of OE and a 19-bp inside end sequence (IE). Artificially constructed transposons defined by two IEs are also substrates for transposase. The 19-bp IE is identical to OE at 12 positions (Fig. 1). In Escherichia coli, four adenines (positions 11 and 16 of the nontransferred strand and 12 and 17 of the transferred strand) in IE are methylated by Dam methylase to yield IEME. This methylation of the DNA inhibits Tnp binding (7, 18).
FIG. 1.
Prokaryotic transposon Tn5. The transposon consists of two insertion sequences that bracket an internal sequence containing three antibiotic resistance genes. IS50R encodes the 53.3-kDa transposase protein that is responsible for its movement as well as an N-terminally truncated form, the inhibitor protein, that functions to downregulate transposition. Each insertion sequence is flanked by an outside end (OE) and an inside end (IE) that act as specific binding sites for transposase (nontransferred strand shown). The IEs contain two target sites for Dam methylation (GATC). This methylation inhibits binding of transposase. Changing position 12 of IE from thymine to adenine (IE12A) results in loss of recognition by transposase. (Nontransferred strand of each sequence shown.)
Wild-type Tn5 Tnp (Tnp WT) maintains a low level of activity in the cell, and purified protein has no activity in vitro. Random mutagenesis studies performed on the Tnp gene have resulted in the isolation of Tnp point mutants that have increased in vivo activity (4, 8, 16, 17, 20). By combining two of these point mutations, E54K and L372K, a double mutant of Tnp was created that is active in vitro (6).
In this study we used a combinatorial, random mutagenesis technique called DNA shuffling (14, 15) to create mutant forms of transposase that could function on transposons with a mutated end binding sequence called IE12A (Fig. ). Previous study has shown that Tnp WT is unable to recognize this specific end binding sequence as a substrate (21). By testing individual mutant Tnps from this library for the ability to mediate movement of transposons flanked by either two OEs or two IEs in a dam mutant background, we identified one transposase mutant, Tnp sC7, that has very high activity with IE defined transposons while retaining nearly Tnp WT levels of activity with OE transposons. We further discovered that Tnp sC7 has even higher IE-related activity when tested in a dam+ strain (IEME).
Through the use of a “minus ones/plus ones” analysis, we discovered that removal of three of the seven mutations in Tnp sC7 led to a more active form with an increased IEME:OE activity ratio. This quadruple mutant form of Tnp, Tnp sC7v2.0, contains a mutation that inhibits OE-related activity (R8C), two mutations that specifically increase IEME-related activity (E58V and E344K), and a mutation that increases transposition of transposons flanked by either IEME or OE (L372Q). In addition, Tnp sC7v2.0 has also been demonstrated to bind with increased affinity to IEME ends by gel shift assay (10). In a recent Tnp-OE DNA cocrystallographic structure analysis, it has been shown that amino acid 58 is in direct contact with DNA in the region of primary recognition.
From this structural analysis, we propose a model to explain how E58V directly enhances Tnp’s IEME binding activity. The structure also gives evidence that amino acid 372 is in a region where mutations can cause conformational alterations that change the distance between the amino and carboxy termini of Tnp and that this alteration relieves an N-terminal-C-terminal interaction that reduces the activity of Tnp WT (3).
MATERIALS AND METHODS
Media and reagents.
Papillation assays were performed using glucose minimal Miller medium (9) supplemented with Casamino Acids, ampicillin, chloramphenicol, 5-bromo-4-chloro-3-indolyl-β-d-galactoside, and phenyl-β-d-galactoside (Trp−-XG-PG plates) as described previously (20). All other bacterial growth was performed in Luria broth (13). When necessary, antibiotics (Sigma) were added at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 20 μg/ml; nalidixic acid, 20 μg/ml; gentamicin, 5 μg/ml; and tetracycline, 15 μg/ml. Taq DNA polymerase, T4 polymerase, T4 ligase, deoxynucleoside triphosphates (dNTPs), and all components of the Altered Sites mutagenesis kit were purchased from Promega. Restriction enzymes were purchased from either Promega or New England Biolabs. Oligonucleotides used in site-directed mutagenesis, DNA shuffling, and sequencing were purchased from Research Genetics. Radionucleotides used in sequencing were from Amersham.
Construction of plasmids.
The construction of plasmid pRZ9905, which was used in this directed-evolution study, was described earlier (10). The papillation vector, pRZ9904 (IE12A/IE12A), was constructed in the following manner. pRZ1495 (8) was digested to completion with KpnI and partially digested with EcoRI. The full-length EcoRI-KpnI transposon fragment was ligated into pUC19 to create pRZ1495/pUC. pRZ1495/pUC was digested with NotI and AflII, filled in with dNTPs and T4 DNA polymerase (Promega), and religated to form pRZΔ1495/pUC. The EcoRI/KpnI transposon-containing fragment from pRZΔ1495/pUC was then cloned into the multicloning site of pAltEX2 (Promega) to form pRZ9904 (OE/OE). The two OE binding sites of the transposon were then mutated into the sequence IE12A following the manufacturer’s protocol for the Altered Sites system (Promega). The plasmid contains a nonfunctional lacZ gene that is flanked by inverted IE12A sequences.
Plasmids pKJ1 and pKJ2 were constructed as high-copy-number plasmids consisting of pUC vector DNA flanked by either two OE (pKJ1) or two IEME (pKJ2) transposon ends separated by a kanamycin resistance gene (donor backbone).
The 14 pRZ9905 (sC7) derivatives and pRZ9905 (sC7v.2.0) were constructed by swapping restriction fragments between pRZ9905 and pRZ9905 (sC7).
Bacterial strains.
Cloning of plasmids and the directed evolution process were performed using JM109 {endA1 recA1 gyrA96 thi hsdR17 (rk− mk+) relA1 supE44 Δ(lac-proAB) [F′ traD36 proAB lacIqZΔM15]} (Promega). The site-directed mutagenesis protocol used strain ES1301 [lacZ53 mutS201::Tn5 thyA36 rha-5 metB1 deoC1 IN(rrnD-rrnE)] (Promega) and JM109. In the mating-out assay, transposase-encoding and transposon-containing plasmids were transformed into strain RZ212 [Δ(lac-proA,B) ara str recA56 srl thi/pOX38-Gen] (dam+) or JCM101 (ΔlacZ174 purF tonA2 tsx-1 thi-1 rpsL dam-3/pOX38-Gen) (dam), followed by conjugation into 14R525 (F− Nalr).
Directed-evolution process.
Directed evolution of Tn5 transposase was carried out by DNA shuffling, basically as described previously by Stemmer (14). Approximately 4 μg of pRZ9905 plasmid, prepared using a miniprep (Wizard SV preps; Promega), was partially digested in a 50-μl volume containing 100 mM Tris-HCl (pH 7.0), 5 nM MgCl, and 90 ng of DNase I. After a 7-min incubation at room temperature, the reactions were stopped by adding EDTA to 10 mM. Following the addition of loading dye, the digested DNA was electrophoresed in a 2% NuSieve gel (FMC BioProducts) next to pGEM DNA markers (Promega). A gel slice containing DNA fragments of 200 to 600 bp in size as well as a second slice containing DNA fragments from 600 to 1,000 bp in size were excised from the gel.
The DNA from each slice was purified by phenol chloroform extraction. After ethanol precipitation, the DNA pellets were dried and resuspended directly into 50 μl of an assembly reaction mix. The assembly mix contained 0.2 mM dNTPs, 2.0 mM MgCl, 50 mM KCl, 10 mM Tris-HCl (pH 9.0 at 25°C), and 0.1% Triton X-100. After addition of 0.5 U of Taq DNA polymerase, the DNA was reassembled by the following thermal cycling program: 94°C for 30 s, then 50 cycles of 94°C for 20 s, 65°C for 1 min, and 72°C for 2 min, and cooling to 4°C. A standard PCR amplification reaction using 5 μl of the assembly reaction product as a DNA template was performed for each sample (200 to 600 and 600 to 1,000 bp) to amplify the transposase gene. This transposase-encoding fragment was digested with AflII and BglII and ligated into purified AflII/BglII-digested vector DNA from pRZ9905.
Ligation products were transformed into electrocompetent JM109 cells that contained plasmid pRZ9904 (IE12A/IE12A). After outgrowth, the cells were plated on Trp−-XG-PG plates with chloramphenicol and ampicillin selection. The plates were incubated at 32°C for 14 days. At this time, pRZ9905 plasmid DNA from all colonies that exhibited at least one papilla were isolated, retransformed into JM109/pRZ9904 (IE12A/IE12A), and plated as above in order to confirm their papillation-positive phenotype. A total of five pRZ9905 derivatives (out of 20,000 original colonies screened) were confirmed to be papillation positive. Equal amounts of all five plasmid DNAs were mixed and then used as the substrate for a second round of mutagenesis and screening. This process was repeated for a total of four rounds of screening (≈20,000 colonies/round). Candidate plasmids were named according to the formula sX#, where X indicates the round of screening at which that candidate was isolated (e.g., sA indicates first round of screening) and # indicates the order in which the candidate first exhibited papillae in the particular screen (e.g., sB8 was the eighth candidate to papillate in the second round of screening).
Quantitative papillation assays.
The IE12A in vivo transposition activity of Tnp WT, Tnp sA5, Tnp sB2, Tnp sC6, and Tnp sD5 was compared by a quantitative papillation assay. Competent cells of strain JM109 harboring plasmid pRZ9904 (IE12A/IE12A) were transformed with the appropriate transposase-encoding version of pRZ9905. After outgrowth, transformed cells were plated on Trp−-XG-PG plates with chloramphenicol and ampicillin. The plates were grown at 32°C until colonies began to appear (≈18 h). Individual colonies were then picked with sterile sticks and spotted onto a fresh plate in a 4 × 4 grid pattern to evenly space all colonies. One plate of 16 colonies was spotted for each mutant. Plates were incubated at 32°C and quantified for transposition by observing the appearance of papillae at 24-h intervals. Data are expressed as the average number of papillae present per colony.
Mating-out assays.
Mating-out assays were performed as described previously (5, 18). Bacterial cells with the transposon-containing plasmids pFMA52-187 (with either two OEs or two IEs) and the F factor pOX-Gen were transformed with the appropriate transposase-encoding plasmid pRZ9905. The donor used for the library screening was strain JCM101. All other mating-out assays were performed in E. coli strain RZ212. The recipient strain used was 14R525. A total of three assays were performed for each combination of transposase and end sequence. The values reported are the averages of these three data points.
In vitro transposition assays.
Substrate plasmids pKJ1 and pKJ2 were isolated from DH5α cells using a Qiafilter plasmid megakit (Qiagen). Supercoiled monomer plasmids were isolated and purified from a 1% low-melting-temperature agarose gel by phenol-chloroform extraction followed by ethanol precipitation. Purification of Tnp sC7v2.0 and Tnp EK/LP was done as described previously (10). The transposition reactions were performed at 37°C under conditions determined by Goryshin and Reznikoff (6). The concentration of plasmid was 35.5 nM, and Tnp was added to a concentration of 280 nM.
RESULTS
Directed evolution of a mutant transposase that functions with IE12A inverted repeats.
A directed-evolution approach was used in order to restore transposase activity with the end binding sequence IE12A. The technique that was used, DNA shuffling, was first described by Stemmer in 1994, when the usefulness of the method was demonstrated by altering the substrate specificity of beta-lactamase (14, 15). The technique involves a series of in vitro DNA manipulations of a gene that result in the introduction of point mutations and allows random recombination within a population of mutant sequences. The mutated genes can then be cloned into plasmids, followed by selection for increased activity in vivo. Any clones that have desirable phenotypes are then processed through another round of mutagenesis and recombination followed by selection for a further improved phenotype. It has also been demonstrated that this method can be used in conjunction with a screen (instead of a selection) in which a modest number of colonies (≈104) are analyzed per round (1, 19).
A standard screen that has been used to isolate transposase gain-of-function mutants is the papillation assay (8). In the case of Tn5, this type of screen has only been used previously to isolate single point mutations that increase the transposition frequency of wild-type (OE defined) transposons. One limitation of the papillation screen is that the standard deviation of the papillation rate for a particular Tnp-end combination is very large, and it is difficult to locate hyperactive mutants from among the many false-positives that are statistical anomalies.
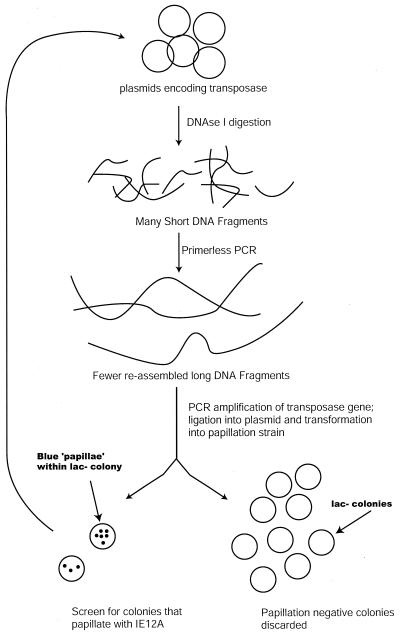
We developed a modified papillation assay for Tn5 that would allow identification of Tnp mutants that were highly increased in transposase activity compared to Tnp WT and altered in primary DNA binding. This was accomplished by constructing a transposon that contained a lacZ gene lacking transcription and translation start sites that was bracketed by defective end sequences. The end sequence chosen was IE12A (Fig. 1). This sequence is identical to the native IE sequence except that position 12 is changed from an AT to a TA. This single-base-pair change results in loss of recognition by transposase at the level of primary sequence binding (21). By using this sequence in the papillation assay, the initial background rate of transposition with Tnp WT was zero. We used this as a screen in conjunction with an sPCR mutagenesis protocol to evolve Tnp to function with this end sequence (see Materials and Methods) (Fig. 2).
FIG. 2.
Directed evolution of Tnp utilizing a screen with defective transposon end sequences. Plasmid encoding Tn5 transposase was partially digested with DNase I, and fragments were purified from an agarose gel. The fragments were then reassembled in a primerless PCR. The reassembled fragments were used as a template to amplify the Tnp gene. The PCR products were next digested with restriction endonucleases and ligated into the plasmid vector. The ligation products were transformed into E. coli cells containing the defective transposon and screened by the papillation assay. Plasmids from rare colonies that were able to move the defective transposon were then mixed and used as the substrate for the next round of mutagenesis-recombination.
Despite the fact that Tnp WT was unable to promote transposition of IE12A-flanked transposons in the papillation screen, we were able to isolate five random mutants (out of 20,000 candidates) that suppressed the end sequence mutation and yielded papillae after an initial cycle of mutagenesis-recombination (designated sA#). A pool containing equal amounts of plasmid encoding each of these mutant Tnps was then used as the initial substrate for a second round of mutagenesis-recombination. Following this second round, the mutated transposase genes were cloned into vector DNA, and 20,000 candidates were screened for transposition activity with IE12A-defined transposons via the papillation screen. From the second round, a total of six active mutants were isolated.
A third round of mutagenesis-recombination was then performed, followed by screening for activity with the mutant end sequence. This time hundreds of colonies were positive for papillation. Of these, seven were clearly more active than the others and were isolated to serve as a template for a fourth round. In the fourth round of screening, there were hundreds of colonies in which transposition was occurring. None of these, however, were as active as the most active mutant from the third round of mutagenesis-recombination (Tnp sC6).
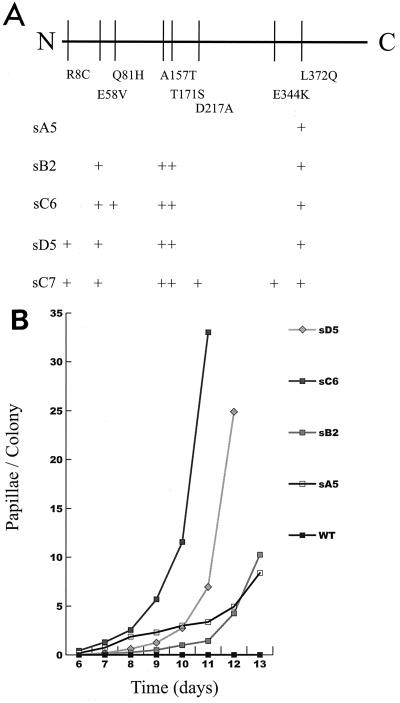
The most active isolate from each round was sequenced and tested for transposition activity with IE12A-defined transposons in a quantitative papillation assay (Fig. 3). The most active first-round isolate (sA5) contained a leucine-to-glutamine point mutation at amino acid 372 (L372Q). This mutation was also found in isolates from subsequent rounds. Transposase sB2 contains four mutations, E58V, A157T, T171S, and L372Q. Both Tnp sC6 (third-round isolate) and Tnp sD5 (fourth-round isolate) contain this combination plus one added mutation each. The mutation Y81H is the only mutation that separates the most active mutant (Tnp sC6) from the noticeably less active sB2. A second isolate from round three, Tnp sC7, is similar to sD5 except that it has two additional mutations, D217A and E344K. Tnp sC7’s activity with IE12A defined transposons is similar to that of sD5 (data not shown).
FIG. 3.
Point mutations in mutated Tnps as indicated by DNA sequencing for Tnp sA5 (best first-round papillator), sB2 (best second-round papillator), sC6 (best third-round papillator), sD5 (best fourth-round papillator), and sC7 (A). The transposition activity of four mutant Tnps was quantified by papillation assay (B). Tnp WT was also tested but failed to promote a single detectable transposition event.
Screening of library against OE and IE.
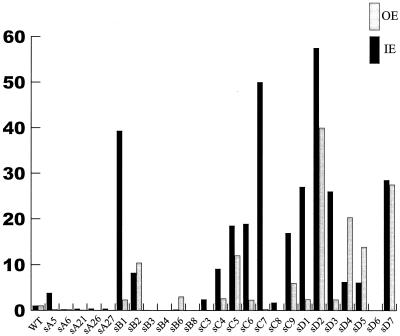
We further analyzed the mutant transposase proteins for their activity with OE defined transposons and IE defined transposons via the mating-out assay in a dam strain (5). Tnp WT activity with IE in this assay is equivalent to its activity with OE. Many of the mutants that we isolated, however, exhibited higher activity with IE than with OE (Fig. 4). One mutant in particular, Tnp sC7, is specifically hyperactive with IE transposons while exhibiting little change in the frequency of OE transposon movement. Additionally, this protein has an increased activity with inside ends when tested in a dam+ strain.
FIG. 4.
Normalized in vivo transposition activity of mutant Tnps with IE- and OE-defined transposon. The ability of each mutant Tnp to promote transposition of transposons with IE and OE sequences was quantified by the mating-out assay. While Tnp WT results in equal levels of transposition for each transposon, some mutants preferentially function with one end. Tnp sC7 is very hyperactive with IE-flanked transposons while functioning at a “normal” level when tested with OE-defined transposons. Transposition rates shown were obtained by dividing the transposition frequency for the mutant Tnp by the rate of Tnp WT activity (6.5 × 10−5).
Mating-out results, shown in Table 1, indicated that transposition frequency with OE is reduced in the dam+ strain RZ212 for both Tnp sC7 and Tnp WT. Since binding of Tnp to OE is not affected by Dam methylation, this difference merely reflects the difference in transposition activity between strains RZ212 and JCM101. Despite this reduction, the rate of IE defined transposition mediated by Tnp sC7 is even higher in RZ212 than in JCM101, presumably due to the presence of methylation in the ends. This is the opposite effect that occurs in Tnp WT, where methylation of the end sequence inhibits recognition by transposase.
TABLE 1.
In vivo transposition rate of Tnp WT and Tnp sC7
| Tnp | JCM101 (dam)
|
RZ212 (dam+)
|
||||
|---|---|---|---|---|---|---|
| Frequency
|
IE/OE | Frequency
|
IEME/OE | |||
| IE | OE | IEME | OE | |||
| WT | 6.5 × 10−5 | 6.5 × 10−5 | 1.0 | 1.0 × 10−8 | 3.1 × 10−6 | 3.2 × 10−3 |
| sC7 | 1.8 × 10−3 | 3.7 × 10−5 | 50 | 2.6 × 10−3 | 3.3 × 10−6 | 794 |
Analysis of sC7.
We next focused on studying the contribution of each amino acid mutation in Tnp sC7 to the IEME hyperactivity of the protein. The goal of this analysis was twofold: to understand how individual mutations affect the transposition activity of the protein in order to increase our understanding of transposase structure-function relationships, and to use this information to remove as many mutations as possible without a decrease in activity and without loss of IEME-OE discrimination. Since a comprehensive trial of all possible combinations of these seven mutations was cumbersome (128 possible combinations), we focused instead on two classes of seven mutants, or 14 total combinations. The first class, the minus ones, consisted of all possible combinations of six mutations (i.e., reverting a single mutation from sC7 to WT). The second set, the plus ones, consisted of each of the seven mutations alone. These mutant proteins were all assayed for in vivo transposition activity with IEME and OE by mating-out assays (Table 2).
TABLE 2.
Results of minus ones/plus ones analysisa
| Tnp | Minus ones
|
Plus ones
|
||||||
|---|---|---|---|---|---|---|---|---|
| IEME
|
OE
|
IEME
|
OE
|
|||||
| Frequency | Normalizedb | Frequency | Normalizedc | Frequency | Normalizedd | Frequency | Normalizede | |
| WT | 1.0 × 10−8 | 1.0 | 3.1 × 10−6 | 1.0 | ||||
| sC7 | 2.6 × 10−3 | 1.0 | 3.3 × 10−6 | 1.0 | ||||
| R8C | 3.1 × 10−3 | 1.2 | 6.0 × 10−6 | 1.8 | <1.3 × 10−9 | —f | 4.0 × 10−7 | 0.1 |
| E58V | 1.8 × 10−6 | 6.9 × 10−4 | 1.1 × 10−6 | 0.3 | 4.0 × 10−4 | 4 × 104 | 1.4 × 10−6 | 0.5 |
| A157T | 3.1 × 10−3 | 1.2 | 4 × 10−6 | 1.2 | <1.3 × 10−9 | — | 6.6 × 10−7 | 0.2 |
| T171S | 2.8 × 10−3 | 1.1 | 2.6 × 10−6 | 0.8 | 5.6 × 10−9 | 0.6 | 1.8 × 10−6 | 0.6 |
| D217A | 3.7 × 10−3 | 1.3 | 4.5 × 10−6 | 1.4 | <1.3 × 10−9 | — | 3.4 × 10−6 | 1.1 |
| E344K | 5.5 × 10−4 | 0.2 | 1.7 × 10−5 | 5.2 | 4.3 × 10−8 | 4.3 | 6.8 × 10−7 | 0.2 |
| L372Q | 2.6 × 10−4 | 0.1 | 2.8 × 10−7 | 0.1 | 8.9 × 10−8 | 8.9 | 1.1 × 10−5 | 3.5 |
Minus ones contain all mutations present in Tnp sC7 except at indicated positions, e.g., R8C minus one contains the wild-type arginine at position 8. Plus ones are Tnp WT except that the indicated amino acid is mutated to the residue present in Tnp sC7, e.g., R8C plus one contains a cysteine at amino acid 8.
Frequency/2.6 × 10−3.
Frequency/3.3 × 10−6.
Frequency/1.0 × 10−8.
Frequency/3.1 × 10−6.
—, below detection levels.
The results clearly reveal that E58V has the most profound effect of all the mutations on the activity of Tnp sC7. This mutation in the wild-type background (E58V plus one) increases IEME-related transposition by 40,000-fold, while removal of the mutation from Tnp sC7 (E58V minus one) drops the transposition activity by more than 1,000-fold. The mutation has comparatively little effect on OE-related activity. The mutation E344K exhibits a weaker sequence-specific effect on activity. Its removal from Tnp sC7 (E344K minus one) decreases IEME-related activity by fivefold while stimulating OE-related activity about fivefold. This result correlates with the plus-ones data, as E344K in the wild-type background stimulates IEME-related activity fourfold and decreases OE-related activity about fivefold. The third mutation that increases Tnp sC7 activity with IEME is L372Q. Unlike the mutations E58V and E344K, this stimulation is not sequence specific. Its removal from sC7 reduced both IEME- and OE-related activity to the same degree, and its plus-one phenotype reveals activity stimulation with both substrates.
Of the four remaining mutations, R8C was the most interesting. Alone, it reduced OE-related transposition nearly 10-fold, and its removal from sC7 increased OE-related activity approximately 2-fold. Its removal had little effect on IEME-related activity, while by itself it decreased IEME-related activity below detectable levels. The remaining three (A157T, T171S, and D217A) all have little effect on either IEME or OE-related activity when removed from sC7.
We made all three combinations of reverting two of the three mutations as well as a mutant that reverts all three of these mutations back to wild type to see if these combinations would lead to an increase in overall IEME-related activity without sacrificing specificity for IEME over OE. The construct with all three mutations removed had the second highest IEME-related activity and the best IEME-OE discrimination (Table 3). The four-mutation construct, Tnp (R8C, E58V, E344K, L372Q), was named Tnp sC7v2.0.
TABLE 3.
In vivo frequency of Tnp sC7 with indicated mutations reverted to wild typea
| Tnp | IEME
|
OE
|
IEME/OE | ||
|---|---|---|---|---|---|
| Frequency | Normalizedc | Frequency | Normalizedd | ||
| sC7 | 2.6 × 10−3 | 1.0 | 3.3 × 10−6 | 1.0 | 794 |
| sC7 (157, 171) | 6.5 × 10−3 | 2.5 | 7.0 × 10−6 | 2.1 | 933 |
| sC7 (157, 217) | 8.4 × 10−3 | 3.3 | 2.25 × 10−5 | 6.8 | 373 |
| sC7 (171, 217) | 5.9 × 10−3 | 2.3 | 1.18 × 10−5 | 3.6 | 502 |
| sC7 (157, 171, 217)b | 7.4 × 10−3 | 2.8 | 7.1 × 10−6 | 2.1 | 1,042 |
Numbers in parentheses indicate positions changed to wild-type residues.
Renamed sC7v2.0.
Frequency/2.6 × 10−3.
Frequency/3.3 × 10−6.
In vitro transposition of IEME- and OE-defined transposons with Tnp sC7v2.0 and Tnp EK/LP.
Tnp sC7v2.0 promotes high levels of movement of transposons with IEME-bracketed transposons in vivo. However, its ability to move OE-defined transposons is much lower. This is the opposite phenotype that is displayed by a previously constructed hyperactive Tnp mutant, Tnp EK/LP. To determine if this discrimination also exists in a purified system, we performed transposition reactions with purified Tnp sC7v2.0 and Tnp EK/LP on transposon-containing plasmid DNAs that were isogenic except for the presence of either OE or IEME sequences (Fig. 5).
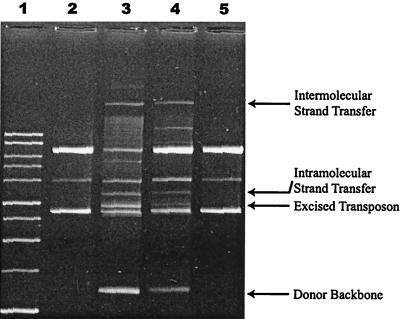
FIG. 5.
In vitro transposition of OE- and IEME-defined transposons. Tnp sC7v2.0 and Tnp EK/LP activity with plasmid DNA containing transposons with either two OE or two IEME ends. Lane 1, 1-kb DNA ladder (Promega); lane 2, Tnp sC7v2.0 with OE-defined transposons; lane 3, Tnp EK/LP with OE-defined transposon; lane 4, Tnp sC7v2.0 with IEME transposons; lane 5, Tnp EK/LP with IEME transposons.
Purified plasmid DNA containing an OE-defined transposon was incubated with either purified Tnp sC7v2.0 (lane 2) or Tnp EK/LP (lane 3). As demonstrated previously (6), purified Tnp EK/LP catalyzes cleavage and strand transfer reactions of OE-defined transposons. Tnp sC7v2.0, which does not exhibit a hyperactive phenotype with OE transposons in the cell, is nonfunctional with these transposons in vitro.
Purified plasmid DNA containing IEME-defined transposons was also incubated with either Tnp sC7v2.0 (lane 4) or Tnp EK/LP (lane 5). Despite its inability to function with OE transposons, Tnp sC7v2.0 converts plasmids with IEME transposon substrate into transposition products and intermediates. Tnp EK/LP is unable to do so despite its functionality on OE transposons. Therefore, the discrimination of each protein between IEME and OE in vivo is also observed in vitro.
DISCUSSION
Bacterial transposons such as Tn5 have evolved within the cell by maintaining a low level of mobility (11). The main roadblock to understanding how Tn5 transposes is the fact that purified Tnp WT has no detectable in vitro activity. This barrier was recently overcome for Tn5 by the development of a double mutant hyperactive form of the transposase (Tnp EK/LP) that is able to promote all steps of the transposition reaction in vitro (6). This system has defined many aspects of Tn5 transposition that either could not be addressed by or could only be alluded to by in vivo experiments. Here we report a new, hyperactive mutant form of Tnp, Tnp sC7v.2.0, that has an altered end sequence specificity. This new protein has been used in conjunction with Tnp EK/LP to address basic questions about transposition that are fundamental to our understanding of the overall process but were not easily examined (10). Furthermore, the detailed study of the protein that we report in this paper has led to a greater understanding of the structure and function of Tn5 transposase.
Screening of mutant library with alternative end substrates.
We initially used a directed-evolution approach in conjunction with a screen for restoration of transposition activity with the defective end sequence IE12A. We chose this end sequence because Tnp WT had no detectable activity with transposons defined by IE12A, making it easier to identify hyperactive Tnp mutants (colonies producing papillae) among the background (no papillae). This led to the isolation of a library of mutant Tnps that were able to function with this end substrate. The mutant Tnps were increased in their transposition activity with IE12A through each of the first three rounds. The most active mutant protein, Tnp sC6, is able to function on the mutant end sequence as well as Tnp WT with OE (data not shown).
Quantification of the transposition levels of the isolated mutants with the native end sequences IE and OE revealed that most mutants were also hyperactive with transposons defined by at least one of the native end sequences (IE or OE) and many isolates functioned preferentially with IE-flanked transposons, and in the case of Tnp sC7, the difference in activity levels was markedly high. Even more fortuitous was the discovery that Tnp sC7 not only is not inhibited for transposition activity by methylation of IE (which reduces Tnp WT levels by ≈102) but even prefers IEME transposons to those bracketed by IE.
Binding of Tnp to OE and IEME.
Hyperactive Tnp EK/LP and Tnp sC7v2.0 exhibit opposite preferences for binding of OE and IEME. Each of these proteins contains a single mutation that is primarily responsible for the difference in end sequence preference. In the case of Tnp EK/LP, the mutation E54K is responsible for binding preference. Tnp E54K was isolated as a point mutation that increases transposition of OE transposons (20). Further study showed that the mutation has the opposite effect on IE transposons (in a dam background). A more detailed study of the interactions of Tnp E54K with DNA ends found that its DNA recognition ability was dependent on the sequence at positions 10, 11, and 12.
Tnp sC7v2.0, which prefers IE- or IEME-flanked transposons to those bordered by OE, contains the mutation E58V, which is predominantly responsible for its hyperactivity. Tnp E58V (E58V plus one) exhibits a very high level of transposition activity on IEME transposons while functioning at Tnp WT levels on those bordered by OE. This Tnp mutation was isolated from a screen for activity with a transposon bordered by the defective end sequence IE12A. Additionally, Tnp mutants containing this mutation have a higher activity when the inside end sequences are modified by Dam methylase, which adds two methyl groups to the major groove of this region (10, 11, 12) of IE.
Recently, a Tnp-DNA cocrystallographic structure of a preintegration synaptic complex of Tnp EK/LP with OE has been reported (3). In this structure, K54 and E58 reside on an α-helix that contains extensive protein-DNA contacts. This helix interacts with OE DNA in the major groove. K54 makes a base-specific contact with position 10 of the OE transferred strand. The wild-type glutamate at amino acid 58 also interacts with OE DNA in this region. It makes a base-specific contact to nucleotide 10 of the nontransferred strand and also, by a water-mediated interaction, with nucleotide 11. In IEME DNA, position 11 of the nontransferred strand and position 12 of the transferred strand DNA would be methylated. Since the inhibition of Tnp activity by Dam methylase can be changed to a favorable interaction by mutation of a single amino acid, it is logical to assume that the side chain of E58 interacts unfavorably with the added methyl group(s), inhibiting binding of Tnp.
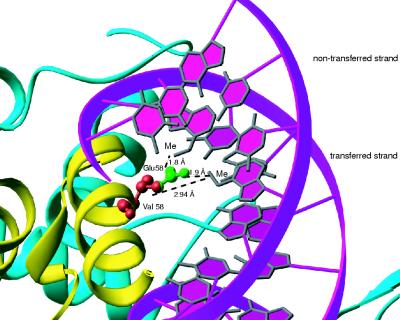
Based on the structure of precleaved OE complexed with Tnp EK/LP, we designed a model of the interaction between E58 and the two added methyl groups. We removed bp 11 and 12 from the structure and replaced them with the base pairs of the IE sequence. Next, the adenine bases were modified by the addition of methyl groups to the N6 positions (Fig. 6). The distance between amino acid residue 58 and each of the added methyl groups is unfavorable (1.8 and 1.9 Å), suggesting that binding of wild-type Tnp to IEME is not allowed due to steric hindrance. Replacement of E58 with the shorter valine side chain (red) increases these distances and suggests the existence of a favorable hydrophobic interaction between valine and the added methyl groups. Removal of the added DNA methyl groups (not shown) would lead to a hole in the structure that would be occupied by water molecules, resulting in loss of the favorable hydrophobic interaction between valine and the methylated adenines. An X-ray crystallographic structure of Tnp sC7v2.0 complexed with IEME DNA would make an interesting comparison to the existing Tnp EK/LP-OE structure and further understanding of how proteins are able to favorably interact with methylated DNA.
FIG. 6.
Molecular modeling of E58 and V58 with IEME DNA. Based on an existing crystallographic structure of Tnp EK/LP complexed with precleaved OE DNA, we modeled the possible interaction between glutamate 58 and IEME. Bp 11 and 12 of OE were removed and changed to IE sequence. Methyl groups were modeled at N6 of the nontransferred strand adenine at position 11 and the transferred strand adenine at position 12. The model suggests that Tnp is blocked from binding to IEME because of steric hindrance between E58 (red plus green) and the added methyl groups. Mutation of amino acid 58 to valine (red) increases the distance between the two added DNA methyl groups and the amino acid side chain and relieves the inhibition.
L372Q.
The leucine-to-glutamine mutation (L372Q) that was present in all mutants sequenced (sA5, sB2, sC6, sC7, and sD5) was both familiar and surprising. It was familiar because a hyperactive mutation at this position was isolated previously. It was surprising because the previously isolated mutation causes a leucine-to-proline (L372P) substitution at this position (16). The L372P mutation results in two consecutive proline residues in an α-helix. In the cocrystal of Tnp EK/LP with OE, the L372P mutation appears to result in a destabilization of amino acids 372 to 390. It has been proposed that the mutation changes the conformation of the catalytic domain in relation to the C-terminal dimerization domain (3). This alteration can improve activity by increasing the distance between the N and C termini. The close proximity of these termini in Tnp WT is thought to decrease the rate of transposition (11). The wild-type leucine residue, visible in an earlier partial Tnp structure (2), is buried in a hydrophobic pocket, and it is likely that substitution with a glutamine destabilizes this hydrophobic packaging. Given that two different hyperactive mutations have been isolated by PCR-based random mutagenesis techniques that only result in single-nucleotide changes in the codon and can only make five different amino acid substitutions at this residue (methionine, valine, arginine, proline, or glutamine), it is likely that a more direct mutagenesis approach such as codon randomization could yield other interesting mutations at position 372.
Acknowledgments
We thank Barb Schriver for her efforts producing the large number of agar plates used for the papillation screens. We also thank Igor Goryshin and Archna Bhasin for critical reading of the manuscript. We additionally thank Doug Davies for helpful discussions and Scott Lovell for assistance with crystallographic figures.
This work was funded by National Institutes of Health (NIH) grant GM50692 awarded to W.S.R. Partial support for T.A.N. was provided by NIH training grant GM08349.
REFERENCES
- 1.Crameri, A., E. Whitehorn, and W. P. Stemmer. 1996. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat. Biotechnol. 14:315–319. [DOI] [PubMed] [Google Scholar]
- 2.Davies, D. R., Braam, L. M., Reznikoff, W. S., and I. Rayment. 1999. The three-dimensional structureof a Tn5 transposase-related protein determined to 2.9-Å resolution. J. Biol. Chem. 274:11904–11913. [DOI] [PubMed] [Google Scholar]
- 3.Davies, D. R., I. Goryshin, W. Reznikoff, and I. Rayment. 2000. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science 289:77–85. [DOI] [PubMed] [Google Scholar]
- 4.DeLong, A., and M. Syvanen. 1991. Trans-acting transposase mutant from Tn5. Proc. Natl. Acad. Sci. USA 88:6072–6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goryshin, I. Y., Y. Kil, and W. S. Reznikoff. 1994. DNA length, and twisting constraints on IS50 transposition. Proc. Natl. Acad. Sci. USA 91:10834–10838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goryshin, I. Y., and W. S. Reznikoff. 1998. Tn5 in vitro transposition. J. Biol. Chem. 273:7367–7374. [DOI] [PubMed] [Google Scholar]
- 7.Jilk, R. A., D. York, and W. S. Reznikoff. 1996. The organization of the outside end of transposon Tn5. J. Bacteriol. 178:1671–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krebs, M. P., and W. S. Reznikoff. 1988. Use of a Tn5 derivative that creates lacZ translational fusions to obtain a transposition mutant. Gene 63:277–285. [DOI] [PubMed] [Google Scholar]
- 9.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 10.Naumann, T. A., and W. S. Reznikoff. 2000. Trans catalysis in Tn5 transposition. Proc. Natl. Acad. Sci. USA 97:8944–8949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reznikoff, W. S., A. Bhasin, D. Davies, I. Goryshin, L. Mahnke, T. Naumann, I. Rayment, M. Steiniger-White, and S. Twining. 1999. Tn5: a molecular window on transposition. Biochem. Biophys. Res. Commun. 266:729–734. [DOI] [PubMed] [Google Scholar]
- 12.Rezsohazy, R., B. Hallet, J. Delcour, and J. Mahillon. 1993. The IS4 family of insertion sequences: evidence for a conserved transposase motif. Mol. Microbiol. 9:1283–1295. [DOI] [PubMed] [Google Scholar]
- 13.Sambrook, J., E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 14.Stemmer, W. P. 1994. Rapid evolution of a protein in vitro by DNA shuffling. Nature 370:389–391. [DOI] [PubMed] [Google Scholar]
- 15.Stemmer, W. P. 1994. DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Proc. Natl. Acad. Sci. USA 91:10747–10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weinreich, M. D., A. Gasch, and W. S. Reznikoff. 1994. Evidence that the cis preference of the Tn5 transposase is caused by nonproductive multimerization. Genes Dev. 8:2363–2374. [DOI] [PubMed] [Google Scholar]
- 17.Wiegand, T. W., and W. S. Reznikoff. 1992. Characterization of two hypertransposing Tn5 mutants. J. Bacteriol. 174:1229–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin, J. C. P., M. Krebs, and W. S. Reznikoff. 1988. Effect of dam methylation on Tn5 transposition. J. Mol. Biol. 199:35–45. [DOI] [PubMed] [Google Scholar]
- 19.Zhang, J. H., G. Dawes, and W. P. Stemmer. 1997. Directed evolution of a fucosidase from a galactosidase by DNA shuffling and screening. Proc. Natl. Acad. Sci. USA 94:4504–4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou, M., and W. S. Reznikoff. 1997. Tn5 mutants that alter DNA binding specificity. J. Mol. Biol. 271:362–373. [DOI] [PubMed] [Google Scholar]
- 21.Zhou, M., A. Bhasin, and W. S. Reznikoff. 1997. Molecular genetic analysis of transposase-end DNA sequence recognition: cooperativity of three adjacent base-pairs in specific interaction with a mutant Tn5 transposase. J. Mol. Biol. 276:913–925. [DOI] [PubMed] [Google Scholar]