Abstract
The Rcs two-component pathway is involved in the regulation of capsule production in Escherichia coli. RcsC is predicted to be the sensor component of this two-component pathway, and in this study we present the first genetic data that support the role of RcsC as a hybrid sensor kinase.
Bacteria respond to changes in the environment largely through the activity of specialized signaling systems called two-component pathways. These pathways are characterized by the presence of conserved communication domains that are involved in signaling via the intermolecular transfer of phosphoryl groups. These domains, called the transmitter domain (H1) and the receiver domain (D1), contain well-conserved amino acid residues, histidine and aspartate, respectively, that are directly implicated in the phosphotransfer. This phosphorelay between transmitter and receiver domains (His→Asp) is the basis of all two-component pathways (7, 12).
In Escherichia coli K-12 the Rcs two-component pathway controls the expression of the cps operon, encoding the proteins required for the production and secretion of acidic extracellular polysaccharide colanic acid (9). Genetic studies have indicated that this pathway is composed of the RcsC sensor protein, the YojN HPt-containing protein, and the RcsB response regulator (see Fig. 2) (2, 14, 15). Analysis of the amino acid sequence of the RcsC sensor protein suggests that it is a hybrid kinase, as the protein contains well-conserved H1 and D1 domains (see Fig. 2 and 3). Only two other prokaryotic sensors with similar H1-D1 arrangements have been characterized, i.e., the VirA sensor protein of Agrobacterium tumefaciens and the LuxN sensor from Vibrio harveyi (1, 3, 8). In VirA the D1 domain is not required for signaling and this domain has a regulatory role whereby it appears to inhibit the kinase activity of VirA (4). On the other hand, the D1 domain of LuxN has been shown to play a direct role in the transfer of the phosphoryl group from LuxN to the response regulator LuxO via the HPt-containing protein LuxU (8). Interestingly, the HPt-containing protein YojN has recently been implicated in the Rcs phosphorelay, suggesting that the D1 domain of RcsC is directly involved in the Rcs phosphorelay (15). In this study we present the first genetic data to directly implicate specific amino acid residues, H463 and D859, in the activity of RcsC. In silico analysis predicts that these residues are involved in the Rcs phosphorelay, and we describe the role of RcsCH463 and RcsCD859 in the regulation of cpsB-lacZ expression. Our data support the hypothesis that RcsC has both kinase and phosphatase activities, and we discuss the implications of our data for the current model describing the complex Rcs phosphorelay, as proposed by Takeda and colleagues (15).
FIG. 2.
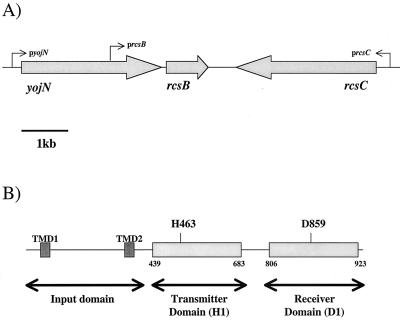
yojN rcsCB genetic locus. (A) Arrangement of genes in the yojN rcsCB locus in E. coli K-12. Arrows, approximate positions of promoters implicated in the expression of each gene. (B) The modular architecture of RcsC. The input domain spans the cytoplasmic membrane and contains two transmembrane domains (TMD). The numbers flanking the transmitter domain and receiver domain represent the predicted amino acid sequence limits of each signaling domain based on in silico homology searches.
FIG. 3.
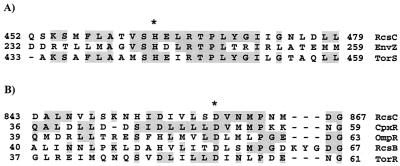
Identification of the amino acids predicted to be involved in the RcsC phosphorelay. (A) Alignment of the histidine-containing region (i.e., the H box) of the transmitter domain (H1) from E. coli sensor proteins EnvZ, TorS, and RcsC. Asterisk, position of the His residue known to be phosphorylated in EnvZ and TorS, which aligns with H463 in RcsC. (B) Alignment of the receiver domain (D1) of RcsC with the receiver domains of CpxR, OmpR, RcsB, and TorR. Asterisk, Asp residue that has been shown, either genetically or biochemically, to be phosphorylated in CpxR, OmpR, RcsB, and TorR. This residue clearly aligns with D859 in RcsC. Both alignments were carried out using the ClustalW algorithm at the European Bioinformatics Institute (http://www.ebi.ac.uk).
Induction of cpsB-lacZ expression by DjlA overproduction requires the RcsC protein
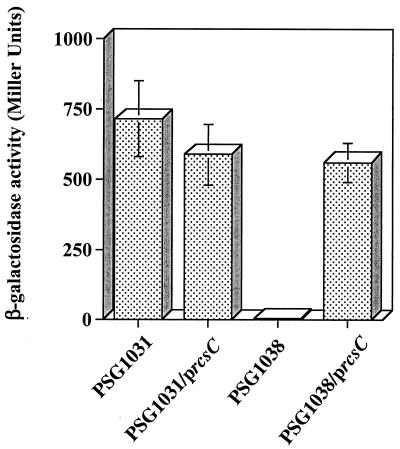
It has been previously shown that the moderate overproduction of DjlA, an inner membrane DnaJ-like protein (6), induces the expression of the cps operon in an rcsC-dependent manner (10, 16). Therefore, DjlA overproduction can be used as a signal to facilitate a genetic analysis of the activity of the RcsC protein. However, we first needed to show that a plasmid-borne copy of rcsC would respond to DjlA overproduction in the expected way. Therefore, we amplified the rcsC gene using Pfu DNA polymerase and the oligonucleotide pair DC121 and DC132. The amplified PCR product was digested with SacI and XbaI and ligated into the appropriately digested pTRC99a vector (Amersham Pharmacia Biotech). Plasmid DNA was isolated from a single transformant, and the insert was completely sequenced to confirm the presence of the rcsC gene. This plasmid, pPSG980, was then transformed into E. coli K-12 strains PSG1031 (rcsC+ cpsB-lacZ) and PSG1038 (rcsC52::Tn10 cpsB-lacZ) carrying DjlA-overproducing plasmid pPSG961-31. In pPSG961-31 DjlA production is under the control of the araBAD promoter (5). As expected, in strain PSG1031, DjlA overproduction resulted in a substantial increase in cpsB-lacZ expression (Fig. 1). In contrast, DjlA overproduction had no effect on cpsB-lacZ expression in the strain carrying mutant rcsC, PSG1038, confirming that the RcsC sensor is required for the observed increase in cpsB-lacZ expression (Fig. 1) (10). However, when a copy of the rcsC gene is provided in trans, cpsB-lacZ expression in PSG1038 was seen to increase following DjlA overproduction. This confirms that DjlA induces cps expression through the RcsC protein and permits the use of DjlA overproduction as a tool for the analysis of RcsC.
FIG. 1.
Induction of cpsB-lacZ expression by DjlA overproduction requires an intact copy of rcsC. PSG1031 (rcsC+ cpsB-lacZ) and PSG1038 (rcsC52::Tn10 cpsB-lacZ) were transformed with pPSG961-31 (paraBAD-djlA) and, where indicated, pPSG980 (p rcsC+). Cells were cultured overnight at 30°C on LB agar plates in the presence of 0.2% (wt/vol) l-arabinose. To determine the level of cpsB-lacZ expression, cells were harvested and assayed for β-galactosidase activity. The results shown are the means of five independent measurements, and the error bars represent standard deviations.
The conserved signaling residues of RcsC, H463 and D859, are essential for induction of cpsB-lacZ expression
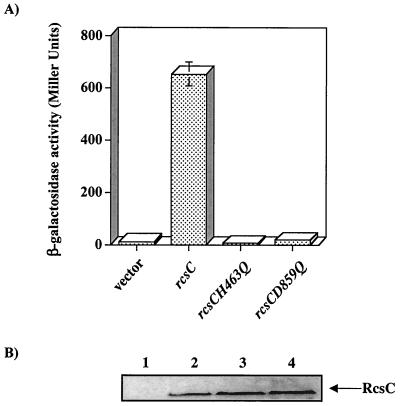
In silico examination of the RcsC amino acid sequence predicts that this protein is a hybrid sensor kinase as it contains both transmitter (H1) and receiver (D1) domains (Fig. 2B). These domains are characterized by the presence of conserved amino acids, histidine and aspartate, respectively, often associated with the transfer of phosphoryl groups during signaling. To identify these conserved residues in RcsC, we aligned the amino acid sequence of this protein with the amino acid sequences of well-characterized transmitter and receiver domains from other proteins (Fig. 3). This alignment clearly predicts that H463 and D859 are the conserved residues in the H1 and D1 domains of RcsC, respectively. Therefore, we predict that these residues are directly involved in the Rcs phosphorelay. To test this, we individually mutated H463 and D859 to the nonphosphorylatable amino acid glutamine (Q), resulting in RcsCH463Q and RcsCD859Q (encoded by plasmids pPSG980H463Q and pPSG980D859Q, respectively). Mutagenesis, using the Stratagene QuikChange kit, was carried out according to the manufacturer's instructions. To introduce point mutations into rcsC, we used pPSG980 as a template and oligonucleotide pairs DC190 and DC191 (rcsCH463Q) and DC188 and DC189 (rcsCD859Q) (Table 1). Plasmid DNA was recovered from each mutant, and the rcsC allele was recloned into pTRC99a, completely sequenced to verify the presence of the desired mutation, and then transformed into strain PSG1038 (rcsC52::Tn10 cpsB-lacZ) containing pPSG961-31. A single transformant was restreaked onto Luria-Bertani (LB) agar plates, with or without added l-arabinose, and incubated at 30°C overnight. Cells were then harvested and washed in 1× phosphate-buffered saline, and the level of cpsB-lacZ expression was determined by β-galactosidase activity assays. The results show that RcsCH463Q and RcsCD859Q could not induce cpsB-lacZ expression in response to DjlA overproduction (Fig. 4A). Therefore H463 and D859 are required for the DjlA-mediated activation of RcsC. Indeed, the inactivity of the RcsCH463Q protein is the first genetic evidence that supports the role of RcsC as a kinase. Moreover, the data obtained with the RcsCD859D protein is in agreement with recent results showing that the HPt-containing protein YojN is involved in the Rcs phosphorelay (15).
TABLE 1.
Oligonucleotides used during this study
| Oligonucleotide | Sequence |
|---|---|
| DC121 | ATAGCGAGCTCGTACAACCCTGAAAGCCTCG |
| DC132 | ATAGCTCTAGACTACGAATCCCGCGATTTCCTGACC |
| DC188 | GATATCGTGCTTAGCAACGTCAACATGCC |
| DC189 | GGCATGTTGACGTTGCTAAGCACGATATC |
| DC190 | CCACCGTCAGTCAAGAGCTGCGAACGC |
| DC191 | GCGTTCGCAGCTCTTGACTGACGGTGG |
| DC210 | CTAGAACAAAAACTCATCTCAGAAGAGGATCTGTGA |
| DC211 | AGCTTCACAGATCCTCTTCTGAGATGAGTTTTTGTT |
| DC212 | TCGAGCCATGGTCAGAGCATTGGCGTTAGTGCa |
| DC213 | ATAGCTCTAGAGAATCCCGCGATTTCCTGACCb |
The NcoI site is underlined.
The XbaI site is underlined.
FIG. 4.
RcsCH463 and RcsCD859 are required for the induction of cpsB-lacZ expression associated with DjlA overproduction. (A) PSG1038 (rcsC52::Tn10 cpsB-lacZ)/pPSG961-31 (paraBAD-djlA+) was transformed with either pTRC99a (vector) or derivatives carrying different mutant alleles of rcsC,rcsC+ (pPSG980), rcsCH463Q (pPSG980H463Q), or rcsCD859Q (pPSG980D859Q). Cells were cultured for 20 h at 30°C on LB agar supplemented with 0.2% l-arabinose and assayed for β-galactosidase activity. The results shown are the means of four independent measurements, and the error bars represent standard deviations. In some cases the error bars are too small to be shown on the graph. (B) PSG1038 (rcsC52::Tn10 cpsB-lacZ) cells carrying plasmids expressing different c-myc-tagged alleles of rcsC were grown as outlined in the text and collected by centrifugation, and 0.2 A600 equivalents were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting with a monoclonal antibody that recognizes the c-myc epitope. Lane 1, pBMM101 (vector); lane 2, pBMM102 (rcsC-myc); Lane 3, pBMM103 (rcsCH463Q-myc); Lane 4, pBMM105 (rcsCD859Q-myc). The arrow indicates the RcsC band that cross-reacts with the anti-myc monoclonal antibody.
RcsC mutants accumulate in the cell to the same level as RcsC.
To verify the negative effect of RcsC mutants, we needed to confirm that the mutant proteins accumulated in the cell to the same level as wild-type RcsC. Unfortunately, anti-RcsC antibodies are not available and several attempts to raise these antibodies in our laboratories failed. Therefore, a vector that allowed the addition of a c-myc epitope tag to the C termini of the wild-type and mutant proteins was constructed. This vector, pBMM101, was constructed from expression vector pTRC99a as follows. Two complementary oligonucleotides were synthesized (DC210 and DC211; Table 1) such that when these oligonucleotides anneal they form a small region of double-stranded DNA, with a 5" XbaI-compatible end and a 3" HindIII-compatible end, that encodes the c-myc epitope, Leu Glu Gln Lys Leu Iso Ser Glu Glu Asp Leu. The annealed oligonucleotides were then ligated with pTRC99a and electroporated into E. coli XL1-Blue. The plasmid DNA from several of the resulting transformants was sequenced, and a clone was selected and named pBMM101. Thus, proteins can be C-terminally tagged with the c-myc epitope by subcloning the genes encoding these proteins into the NcoI-XbaI sites of pBMM101. The different rcsC alleles were amplified by PCR using Pfu DNA polymerase, and the PCR products were then ligated into pBMM101, resulting in plasmids pBMM102 (rcsC-myc), pBMM103 (rcsCH463Q-myc), and pBMM104 (rcsCD859Q-myc). These plasmids were transformed into PSG1038/pPSG961-31 cells and tested for their ability to induce cpsB-lacZ expression in response to DjlA overproduction. The results were identical to those in Fig. 3A, indicating that the c-myc epitope tag had no effect on the activity of the RcsC protein (data not shown). However, under the growth conditions used for our biochemical analyses, i.e., no added IPTG, it was not possible to detect any of the RcsC proteins after immunoblotting and hybridization with anti-myc monoclonal antibodies, even though it is clear from the β-galactosidase assays that RcsC-myc is being produced (data not shown). Therefore, cells were induced with 1 mM IPTG for 120 min at 37°C and samples were taken for immunoblotting. A band at the correct predicted molecular mass (approximately 100 kDa) was observed in all samples tested, except the vector control (Fig. 4B). Moreover, the intensities of this band in all lanes were similar, confirming that the mutations have no apparent affect on the inherent stability of RcsC. Interestingly, even under these IPTG-inducing conditions the mutant rcsC alleles were unable to complement the rcsC52::Tn10 mutation (data not shown). Therefore, we conclude that the mutations must be interfering with the normal signaling activity of RcsC.
RcsC phosphatase activity requires D859 but is largely independent of H463
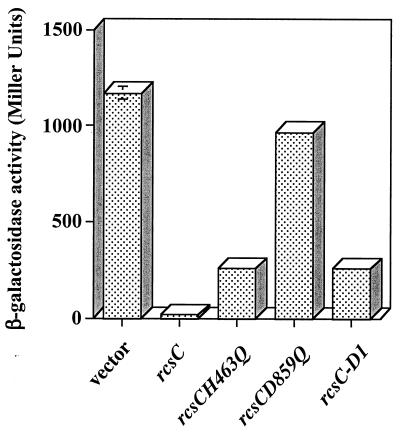
Previous studies have suggested that RcsC has both positive and negative regulatory effects on the expression of the cps operon (2, 14). This is consistent with the RcsC sensor having both kinase and phosphatase activities. Therefore, in the presence of an activating signal, RcsC kinase activity would result in an increase in the level of phospho-RcsB and an increase in cps expression. On the other hand, in the absence of an activating signal, the phosphatase activity of RcsC would result in a net dephosphorylation of phospho-RcsB, leading to low levels of cps expression. It is possible to isolate rcsC mutant strains that have high levels of cps expression in the absence of a signal, e.g., strains carrying rcsC137, and it has been suggested that these mutants have lost the phosphatase activity associated with RcsC (2). This is supported by the observation that the rcsC137 allele is a recessive mutation that can be complemented with a wild-type copy of rcsC supplied in trans. We can use this observation to determine whether the rcsC mutants constructed during this study have retained some phosphatase activity. Strain SG20907 (rcsC137 cpsB-lacZ) (a gift from Susan Gottesman, National Institutes of Health) was transformed with either pPSG980 or the plasmids encoding the different rcsC alleles, and the cells were assayed for β-galactosidase activity. As expected, in the presence of the vector alone there is a high level of cpsB-lacZ expression, i.e., 1,175 U (Fig. 5). However, when rcsC is supplied in trans from pPSG980, the level of cpsB-lacZ expression is reduced 70-fold, confirming that wild-type rcsC can complement the rcsC137 mutation. Expression of the rcsCH463Q allele also reduced the level of cpsB-lacZ expression, indicating that RcsCH463Q still retains some phosphatase activity, although the levels of phosphatase are apparently reduced compared to those for the wild-type protein. In contrast, expression of rcsCD859Q failed to complement rcsC137, suggesting that RcsCD859 does not have phosphatase activity. This indicates that D859 is important for RcsC phosphatase activity. In support of this, we cloned the coding sequence for the D1 domain of RcsC (RcsC-D1) into pTRC99a, resulting in plasmid pMMG104, and found that the RcsC-D1 domain, alone, is sufficient for complementation of the rcsC137 allele, suggesting that the phosphatase activity of RcsC is localized to the receiver domain (Fig. 5).
FIG. 5.
RcsCD859 is required for complementation of the rcsC137 allele. SG20907 (rcsC137 cpsB-lacZ) was transformed with either pTRC99a (vector) or derivatives carrying different rcsC alleles, as indicated. Cells were grown on LB agar at 30°C for 20 h and assayed for β-galactosidase activity. The results shown are the means of two independent measurements, and the error bars represent standard deviations. In some cases the error bars are too small to be shown on the graph.
Effect of RcsCH463Q and RcsCD859Q on in vivo signaling by the wild-type RcsC protein
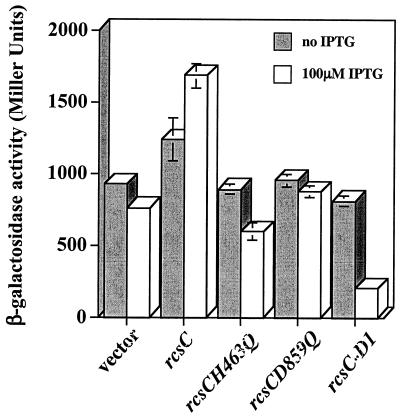
Our results indicate that RcsCH463Q and RcsC-D1 have a phosphatase activity that is unaffected by DjlA overproduction (Fig. 4A and data not shown). Therefore, these mutant RcsC proteins should have a negative effect on signaling by the wild-type RcsC protein. PSG1031 (rcsC+ cpsB-lacZ) was transformed with plasmids expressing the different alleles of rcsC, and the level of β-galactosidase activity in the cell was assayed in the presence of DjlA overproduction. The expression of the different rcsC alleles is controlled by the trc promoter, and, under noninducing conditions (i.e., in the absence of IPTG), the expression of rcsCH463Q, rcsCD859Q, and rcsC-D1 had no effect on RcsC signaling (Fig. 6). This is in contrast to the complementation results obtained using rcsC137, where the presence of either rcsCH463Q or rcsC-D1 complemented rcsC137 under noninducing conditions (Fig. 5). However, when IPTG (isopropyl-β-d-thiogalactopyranoside) was added to 100 μM, the expression of rcsCH463Q and rcsC-D1 significantly reduced the level of cpsB-lacZ expression, supporting our hypothesis that both RcsCH463Q and RcsC-D1 retain the phosphatase activity of RcsC (Fig. 6). However, the expression of rcsCD859Q failed to reduce cpsB-lacZ expression in the presence or absence of IPTG, confirming that RcsCD859Q has no phosphatase activity.
FIG. 6.
RcsCH463Q and RcsC-D1 interfere with normal signaling from the RcsC protein. PSG1031 (rcsC+ cpsB-lacZ)/pPSG961-31 (paraBAD-djlA) was transformed with either pTRC99a (vector) or derivatives carrying different mutant alleles of rcsC, as indicated. Cells were cultured for 20 h at 30°C on LB agar supplemented with 0.2% l-arabinose and assayed for β-galactosidase. The results shown are the means of four independent measurements, and the error bars represent standard deviations. In some cases the error bars are too small to be shown on the graph.
We used both rcsC137 and DjlA overproduction as tools to determine whether mutant derivatives of RcsC can interfere with normal signaling. From these results it is interesting that the level of mutant RcsC proteins required to perturb signaling is dependent on the signal used to activate the pathway. In the rcsC137 background, basal levels of expression of the rcsC mutant alleles from plasmids pPSG980H463Q and pMMG104 affected signaling (Fig. 5). On the other hand, when DjlA overproduction is used as the activating signal, higher levels of expression of rcsCH859Q and rcsC-D1 (achieved by the addition of 100 μM IPTG) are required before any effect on cpsB-lacZ expression is observed (Fig. 6). This difference in sensitivity can be simply explained by the fact that, in the latter case, DjlA (the signal) is overproduced. Therefore, assuming that DjlA interacts with RcsC, it is not unreasonable to expect that higher levels of both RcsCH463Q and RcsC-D1 will be required to interfere with the normal interaction between DjlA and RcsC. The fact that the overproduction of RcsCD859Q has no effect on cpsB-lacZ expression (Fig. 6) can be attributed to the fact that RcsCD859Q does not appear to have any phosphatase activity (Fig. 5).
Intermolecular complementation of RcsC mutants in vivo
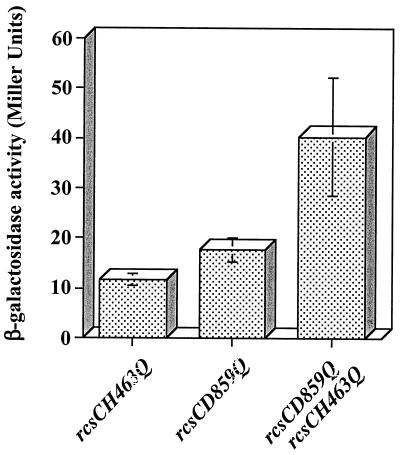
We were interested in testing whether the rcsCH463Q and rcsCD859Q alleles could complement each other in vivo. To carry out this experiment, both alleles need to be expressed at the same time in PSG1038 (rcsC52::Tn10 cpsB-lacZ). Plasmid pTRC99a, used to construct both pPSG980H463Q and pPSG980D859Q, carries the bla gene, encoding ampicillin resistance. Therefore, the rcsCD859Q allele was cloned into the pEH3 vector, which carries the cat gene and which encodes resistance to chloramphenicol, resulting in plasmid pBMM110. Both pPSG980H463Q and pBMM110 have pUC-based origins, and cells carrying both plasmids can be selected on medium containing both ampicillin and chloramphenicol. In this experiment, constitutive 5- to 10-fold overproduction of DjlA was achieved using kanamycin resistance-encoding pACYC-based plasmid pPSG957 (unpublished data). Therefore, PSG1038 carrying the pPSG957 plasmid was transformed with pPSG980H463Q, pBMM110, or both plasmids together, and the cells were tested for their ability to induce cpsB-lacZ expression in the presence of DjlA overproduction. As expected the presence of either the rcsCH463Q or the rcsCD859Q allele resulted in no increase in cpsB-lacZ expression (Fig. 7). However, when both alleles were expressed together, there was a small increase (approximately threefold) in the level of cpsB-lacZ expression, suggesting that RcsCH463Q and RcsCD859Q show some intermolecular complementation (Fig. 7). Unfortunately, due to the fact that both pPSG980H463Q and pBMM110 have the same type of origin, this system does not permit quantitative estimations of the level at which intermolecular complementation occurs. Moreover, the relatively large error bars seen in Fig. 7 might be indicative of bacterial populations that contain different relative levels of each plasmid. However, these data can be used to infer that RcsC is probably active as a multimer (most likely a dimer) and that, during signaling, phosphoryl groups can be transferred between H463 and D859 on different RcsC monomers.
FIG.7.
In vivo intermolecular complementation between rcsCH463Q and rcsCD859Q. PSG1038 (rcsC52::Tn10 cpsB-lacZ)/pPSG957 (djlA+) was transformed with either pPSG980H463Q (rcsCH463Q), pBMM110 (rcsCD859Q), or both plasmids together (rcsCH463Q rcsCD859Q). Cells were cultured for 20 h at 30°C on LB agar and assayed for β-galactosidase activity. The results shown are the means of four independent measurements, and the error bars represent standard deviations.
Conclusions
RcsC is predicted to be a hybrid sensor kinase, and computer analyses implicates RcsCH463 and RcsCD859 in signaling. In this study we show, for the first time, that RcsCH463 and RcsCD859 are required for the induction of cps expression. Therefore, as for LuxN in Vibrio harveyi, both the transmitter domain (H1) and the receiver domain (D1) of RcsC are required for signaling, and this supports the role of RcsC as a hybrid kinase. We propose that under activating conditions, e.g., DjlA overproduction, dessication, or increased osmolarity (11, 13), RcsC autophosphorylates on the conserved His residue (H463) in its transmitter domain, H1 (Fig. 5). This phosphoryl group is then transferred to the conserved Asp residue (D859) in the receiver domain, D1, of RcsC. RcsC has also been shown to repress cps expression, and we have shown that this negative activity appears to be localized to the D1 receiver domain and requires the D859 residue. We propose that this negative effect on cpsB-lacZ expression is, in fact, a phosphatase activity associated with the RcsC sensor protein, and biochemical studies to support this model are under way.
Acknowledgments
Funding for part of this work was provided by a fellowship from the Fondation pour la Recherche Medical (to D.J.C.).
We thank Susan Gottesman for the gift of SG20907. D.J.C. thanks the Department of Biology at the NUI Maynooth in Ireland, the Department of Biology and Biochemistry at the University of Bath, the C.N.R.S., and the Universite Paris-Sud for their support during parts of this work.
REFERENCES
- 1.Bassler, B. L., M. Wright, R. E. Showalter, and M. R. Silverman. 1993. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9:773-786. [DOI] [PubMed] [Google Scholar]
- 2.Brill, J. A., C. Quinlan-Walshe, and S. Gottesman. 1988. Fine-structure mapping and identification of two regulators of capsule synthesis in Escherichia coli K-12. J. Bacteriol. 170:2599-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, C.-H., and S. C. Winans. 1992. Functional roles assigned to the periplasmic, linker, and receiver domains of the Agrobacterium tumefaciens VirA protein. J. Bacteriol. 174:7033-7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, C.-H., J. Zhu, and S. C. Winans. 1996. Pleiotropic phenotypes caused by genetic ablation of the receiver module of the Agrobacterium tumefaciens VirA protein. J. Bacteriol. 178:4710-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarke, D. J., I. B. Holland, and A. Jacq. 1997. Point mutations in the transmembrane domain of DjlA, a membrane-linked DnaJ-like protein, abolish its function in promoting colanic acid production via the Rcs signal transduction pathway. Mol. Microbiol. 25:933-944. [DOI] [PubMed] [Google Scholar]
- 6.Clarke, D. J., A. Jacq, and I. B. Holland. 1996. A novel DnaJ-like protein in Escherichia coli inserts into the cytoplasmic membrane with a type III topology. Mol. Microbiol. 20:1273-1286. [DOI] [PubMed] [Google Scholar]
- 7.Dutta, R., L. Qin, and M. Inouye. 1999. Histidine kinases: diversity of domain organisation. Mol. Microbiol. 34:633-640. [DOI] [PubMed] [Google Scholar]
- 8.Freeman, J. A., B. N. Lilley, and B. L. Bassler. 2000. A genetic analysis of the functions of LuxN: a two-component hybrid sensor kinase that regulates quorum sensing in Vibrio harveyi. Mol. Microbiol. 35:139-149. [DOI] [PubMed] [Google Scholar]
- 9.Gottesman, S. 1995. Regulation of capsule synthesis: modification of the two-component paradigm by an accessory unstable regulator, p. 253-262. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. ASM Press, Washington, D.C.
- 10.Kelley, W., and C. Georgopoulos. 1997. Positive control of the two-component RcsC/B signal transduction network by DjlA: a member of the DnaJ family of molecular chaperones in Escherichia coli. Mol. Microbiol. 25:913-931. [DOI] [PubMed] [Google Scholar]
- 11.Ophir, T., and D. L. Gutnick. 1994. A role for exopolysaccharides in the protection of microorganisms from dessication. Appl. Environ. Microbiol. 60:740-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parkinson, J. S., and E. C. Kofoid. 1992. Communication modules in bacterial signalling proteins. Annu. Rev. Genet. 26:71-112. [DOI] [PubMed] [Google Scholar]
- 13.Sledjeski, D. D., and S. Gottesman. 1996. Osmotic shock induction of capsule synthesis in Escherichia coli K-12. J. Bacteriol. 178:1204-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stout, V., and S. Gottesman. 1990. RcsC and RcsB: a two-component regulator of capsule synthesis in Escherichia coli. J. Bacteriol. 172:659-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takeda, S.-I., Y. Fujisawa, M. Matsubara, H. Aiba, and T. Mizuno. 2001. A novel feature of the multistep phosphorelay in Escherichia coli: a revised model of the RcsC→YojN→RcsB signalling pathway implicated in capsular synthesis and swarming behaviour. Mol. Microbiol. 40:440-450. [DOI] [PubMed] [Google Scholar]
- 16.Zuber, M., T. A. Hoover, and D. L. Court. 1995. Analysis of a Coxiella burnetti gene product that activates capsule synthesis in Escherichia coli: requirement for the heat shock chaperone DnaK and the two-component regulator RcsC. J. Bacteriol. 177:4238-4244. [DOI] [PMC free article] [PubMed] [Google Scholar]