Abstract
The agr quorum-sensing and signal transduction system was initially described in Staphylococcus aureus, where four distinct allelic variants have been sequenced. Western blotting suggests the presence of homologous loci in many other staphylococci, and this has been confirmed for S. epidermidis and S. lugdunensis. In this study we isolated agr-like loci from a range of staphylococci by using PCR amplification from primers common to the six published agr sequences and bracketing the most variable region, associated with quorum-sensing specificity. Positive amplifications were obtained from 14 of 34 staphylococcal species or subspecies tested. Sequences of the amplicons identified 24 distinct variants which exhibited extensive sequence divergence with only 10% of the nucleotides absolutely conserved on multiple alignment. This variability involved all three open reading frames involved in quorum sensing and signal transduction. However, these variants retained several protein signatures, including the conserved cysteine residue of the autoinducing peptide, with the exception of S. intermedius of pigeon origin, which contained a serine in place of cysteine at this position. We discuss hypotheses on the mode of action and the molecular evolution of the agr locus based on comparisons between the newly determined sequences.
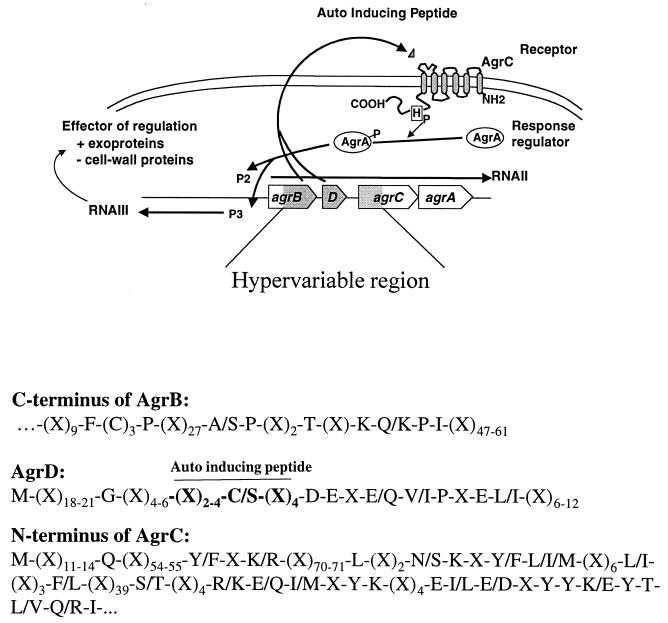
The agr locus was initially described in Staphylococcus aureus as an element controlling the production of exoproteins implicated in virulence (14, 23, 26). Its pattern of action has been shown to be complex, upregulating certain extracellular toxins and enzymes expressed postexponentially and repressing some exponential-phase surface components (17). The agr locus comprises two divergent operons expressed from promoters P2 and P3 (Fig. 1). The P2 operon includes four genes, of which two encode elements of a density-sensing cassette, agrD encodes the precursor of the autoinducing peptide (AIP), and agrB, whose product is probably involved in processing and/or secretion of the AIP (8). The two-component sensory transduction system is comprised of AgrC, the membrane sensor, and AgrA, the response regulator (11, 15, 18, 19). In brief, the AIP derived from AgrD by the action of AgrB interacts with AgrC in the membrane to activate AgrA, which upregulates transcription both from promoter P2, amplifying the response, and from P3, initiating the production of a novel effector: RNA III (6, 20). In S. aureus, delta-hemolysin is the only translation product of RNA III and is not involved in the regulatory functions of the transcript, which is therefore the primary agent for modulating the expression of other operons controlled by agr (2, 16, 20).
FIG. 1.
Functional map of the staphylococcal agr (accessory gene regulator) system and consensus alignment for AgrB, AgrD, and AgrC. The schematic map of the agr locus shows the major transcripts RNA II and RNA III (arrows), and the genes are indicated by boxes. Hypervariable regions are shaded in gray.
The agr locus in S. aureus (agrSa) has been shown to be polymorphic and can be divided into four distinct genetic groups (7, 9). Stimulation of agr expression, and hence transcription from promoters P2 and P3, is strongly dependent on the presence in the medium of the AIP derived from AgrD of the inducing strain or a member of the same group. AIPs from other groups generally inhibit agr activity, as do AIPs from other species of staphylococci (7, 9, 21). Comparison of sequences from different strains of S. aureus indicates that AgrB, AgrC, and AgrD, which are involved in the generation of the specific signal, are strikingly variable among the four groups, whereas AgrA, which codes for the response regulator, is much more conserved. Sequence variation is particularly marked in the AIP precursor, AgrD; the C-terminal two-thirds of AgrB, which may be involved in the specific cleavage and transport of the AIP; and a portion of the N-terminal half of AgrC where the extracellular sites for interaction with the AIP may be located (9). Coevolution of these sites with a specific AIP might explain the high degree of specificity for induction of the agr locus, but the mechanism for its inhibition by a broad spectrum of AIPs with little sequence similarity is not immediately obvious. Functional agr loci have also been shown to be present in S. lugdunensis (agr-1Sl) and S. epidermidis (agr-1Se). Their gene products define additional activation or inhibition groups distinct from those of S. aureus (31, 32). Sequences related to the agr regulon have also been demonstrated by hybridization in at least 14 other staphylococcal species or subspecies (3, 30), suggesting that this regulatory mechanism is widespread in the staphylococci.
In the present study, we compare the nucleotide sequences for open reading frames (ORFs) in the agr region of a number of non-S. aureus Staphylococcus species. We confirm that recognizable agr loci are widespread in the staphylococci and are subject to considerable sequence variation. The 24 sequenced variants from 14 tested species and subspecies maintain several protein signatures that provide information concerning the mechanism by which agr diversity has arisen.
MATERIALS AND METHODS
Bacterial strains.
Bacterial strains and their origins are listed in Table 1. The agr-null strain of S. aureus RN6911 was constructed by replacement of the entire agr locus of the standard agr-positive strain RN6390 by tetM (20). Type strains were obtained from the American Type Culture Collection (ATCC), the Czechoslovak Collection of Microorganisms (CCM), the Deutsche Sammlung von Mikroorganismen (DSMZ), the Japanese Collection of Microorganisms (JCM), and the R. P. Novick Collection. Other strains were collected throughout France from human or animal infections and were referred to the Centre National de Référence des Toxémies à Staphylocoques (Lyon, France). Species were identified by colony and microscopic morphology, coagulase activity on rabbit plasma (bioMérieux, Marcy l'Étoile, France), production of clumping factor (Staphyslide; bioMérieux), and results on the ID32 Staph gallery (bioMérieux).
TABLE 1.
Bacterial strains and agr detection by PCR
| Strain | PCR agra | Nucleotide sequence denomination | GenBank accession no. | Strain | PCR agra | Nucleotide sequence denomination | GenBank accession no. | |
|---|---|---|---|---|---|---|---|---|
| S. aureus | ||||||||
| RN6911 | − | agr null | ||||||
| RN6390B | + | agr-1Sa | X52543 | |||||
| RN7690 | + | agr-1Sa | ||||||
| RN6923 | + | agr-2Sa | AF001782 | |||||
| RN9625 | + | agr-2Sa | ||||||
| RN8462 | + | agr-3Sa | AF001783 | |||||
| RN8463 | + | agr-3Sa | ||||||
| A880740 | + | agr-4Sa | AF288215 | |||||
| A890382 | + | agr-4Sa | ||||||
| S. arlettae DSM 20672 | + | agr-1ar | AF346712 | |||||
| S. auricularis | ||||||||
| ATCC 33753 | + | agr-1auri | AF346713 | |||||
| N910219 | + | agr-1auri | ||||||
| N910221 | + | agr-1auri | ||||||
| N910222 | + | agr-1auri | ||||||
| N910333 | + | agr-1auri | ||||||
| N910343 | + | agr-1auri | ||||||
| N940180 | + | agr-2auri | AF346714 | |||||
| N940327 | − | |||||||
| S. capitis subsp. capitis | ||||||||
| CCM 2734 | + | agr-1capi | AF346715 | |||||
| N900125 | + | agr-1capi | ||||||
| N930202 | + | agr-1capi | ||||||
| N930315 | + | agr-1capi | ||||||
| N940269 | + | agr-1capi | ||||||
| N950055 | + | agr-1capi | ||||||
| N910309 | + | agr-2capi | AF346716 | |||||
| N920161 | − | |||||||
| S. capitis subsp. ureolyticus ATCC 49326 | − | |||||||
| S. caprae | ||||||||
| N900362 | + | agr-1capra | AF346717 | |||||
| N9300039 | + | agr-1capra | ||||||
| N920271 | + | agr-2capra | AF346718 | |||||
| N930298 | + | agr-2capra | ||||||
| CCM3573 | − | |||||||
| N930422 | − | |||||||
| N920072 | − | |||||||
| N920021 | − | |||||||
| S. carnosus DSM 20501 | + | agr-1car | AF346719 | |||||
| S. chromogenes CCM 3387 | − | |||||||
| S. cohnii subsp. cohnii | ||||||||
| CCM 2736 | + | agr-1coco | AF346720 | |||||
| N900274 | + | agr-1coco | ||||||
| N920270 | + | agr-1coco | ||||||
| N930144 | + | agr-1coco | ||||||
| N940210 | + | agr-1coco | ||||||
| N940379 | + | agr-1coco | ||||||
| N940469 | + | agr-1coco | ||||||
| N920077 | − | |||||||
| N930306 | − | |||||||
| S. cohnii subsp. urealyticum ATCC 49330 | + | agr-1cour | AF346721 | |||||
| S. delphini DSM 20771 | − | |||||||
| S. epidermidis | ||||||||
| CCM 2124 | + | agr-1Se | Z49220 | |||||
| N910102 | + | agr-1Se | ||||||
| N910280 | + | agr-1Se | ||||||
| N910288 | + | agr-1Se | ||||||
| N910328 | + | agr-1Se | ||||||
| N910160 | + | agr-2Se | AF346724 | |||||
| N910173 | + | agr-2Se | ||||||
| N910191 | + | agr-3Se | AF346725 | |||||
| S. equorum DSM 20674 | − | |||||||
| S. felis ATCC 49168 | − | |||||||
| S. gallinarum CCM 3572 | + | agr-1gal | AF346722 | |||||
| S. haemolyticus CCM 2737 | − | |||||||
| S. hominis DSM 20328 | − | |||||||
| S. hyicus CCM 2368 | − | |||||||
| S. intermedius | ||||||||
| CCM 5739 | + | agr-1int | AF346723 | |||||
| HT20010088 | + | agr-1int | ||||||
| HT20010089 | + | agr-1int | ||||||
| HT20010090 | + | agr-1int | ||||||
| N900260 | − | |||||||
| N900269 | − | |||||||
| N900402 | − | |||||||
| N910201 | − | |||||||
| N930300 | − | |||||||
| N940276 | − | |||||||
| N940453 | − | |||||||
| N950082 | − | |||||||
| N950346 | − | |||||||
| S. kloosii DSM 20676 | − | |||||||
| S. lentus ATCC 29070 | − | |||||||
| S. lugdunensis | ||||||||
| RN8152 | + | agr-1Sl | AF173933 | |||||
| RN8153 | + | agr-1Sl | ||||||
| N940084 | + | agr-1Sl | ||||||
| N950245 | + | agr-1Sl | ||||||
| N960021 | + | agr-1Sl | ||||||
| N920217 | + | agr-2Sl | AF346728 | |||||
| N940118 | + | agr-2Sl | ||||||
| N940256 | + | agr-2Sl | ||||||
| N950199 | + | agr-2Sl | ||||||
| N960242 | + | agr-2Sl | ||||||
| S. muscae CCM 4175 | − | |||||||
| S. pasteuri ATCC 51129 | − | |||||||
| S. piscifermentans JCM 6057 | − | |||||||
| S. saprophyticus subsp. bovis CCM 4410 | − | |||||||
| S. saprophyticus subsp. saprophyticus CCM 883 | − | |||||||
| S. schleiferi subsp. coagulans JCM 7470 | − | |||||||
| S. schleiferi subsp. schleiferi ATCC 43808 | − | |||||||
| S. sciuri subsp. sciuri ATCC 29062 | − | |||||||
| S. simulans | ||||||||
| ATCC 27848 | + | agr-1sim | AF346726 | |||||
| N930284 | + | agr-1sim | ||||||
| N930309 | + | agr-1sim | ||||||
| N940200 | + | agr-1sim | ||||||
| N940232 | + | agr-1sim | ||||||
| N940279 | + | agr-1sim | ||||||
| N940377 | + | agr-1sim | ||||||
| N930143 | + | agr-2sim | AF346727 | |||||
| S. vitulinus ATCC 5145 | − | |||||||
| S. warneri CCM2 730 | − | |||||||
| S. xylosus | ||||||||
| ATCC 29971 | + | agr-1xyl | AF346729 | |||||
| N930189 | + | agr-1xyl | ||||||
| N930239 | + | agr-1xyl | ||||||
| N930408 | + | agr-1xyl | ||||||
| N930173 | − | |||||||
| N930249 | − | |||||||
| N930409 | − | |||||||
| N930409 | − |
Genomic DNA extracts were used as templates for PCR amplification with primers chosen to encompass a fragment of 1.2 kb of agr common to agr-1Sa to agr-3Sa, agr-1Se, and agr-1Sl as described in Materials and Methods. All strains tested were positive for PCR amplifying either gyrA or 16S-23S intergenic region.
DNA preparation, amplification, and sequencing.
Genomic DNA was extracted from agar plate cultures as previously described (12) and used as a template for PCR amplification with the primers AGR-1 (5"-ATGCACATGGTG CACATGCA-3") and AGR-2 (5"-CATAATCATGACGGAACTTGCTGCGCA-3") (Eurogentec) chosen to encompass a ca. 1,234-bp fragment of agr common to agr-1Sa, agr-2Sa, and agr-3Sa, agr-1Se, and agr-1Sl (GenBank accession numbers M21854, AF001782, and AF001783, AF288215, and AF173933, respectively). Amplification mixtures were denatured at 95°C for 5 min and then subjected to five low-stringency cycles (denaturation at 94°C for 1 min, annealing at 45°C for 1 min, and extension at 72°C for 1 min), followed by 30 high-stringency PCR cycles (denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1 min). Positive controls included amplification of gyrA or 16S-23S intergenic rRNA region (12). PCR products were analyzed by electrophoresis through 0.8% agarose (Sigma), purified by using the High-Pure PCR product purification kit (Boehringer Mannheim), and sequenced with the above PCR primers (Genome Express).
Sequence analysis.
Sequences were aligned by using the CLUSTAL algorithm with additional minor modifications by visual inspection and analyzed by using BLAST, GeneJockey, and CLUSTAL software (1, 35). Consensus membrane-spanning regions were determined from the aligned predicted amino acid sequences by using TMAP (http://www.mbb.ki.se/tmap/index.html) (24), and PROSITE sites and signatures were sought in AgrC by using PROSCAN (http://pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_proscan.html) (5).
Phylogenic analysis.
Evolutionary distances were determined by the method of Kimura, and these values were used to construct dendrograms by the neighbor-joining method with the PHYLIP package (European Bioinformatics Institute) (27). The stability of the suggested relationships was tested by the construction of at least 1,000 bootstrap trees for each data set by using Seqboot (Phylip).
RESULTS AND DISCUSSION
Distribution of agr sequences among staphylococci.
The agr quorum-sensing and signal transduction system was first described in S. aureus, where four distinct allelic loci have been identified and sequenced. Sequences have also been determined for homologous loci in S. epidermidis and S. lugdunensis (7, 9, 31, 32). The presence of similar loci in 12 other staphylococcal species or subspecies was suggested by Southern blot hybridization with probes derived from agrSa, which produced strong positive signals for nine species and weak signals for a further three (S. hominis subsp. hominis and S. schleiferi subsp. coagulans or subsp. schleiferi) (3). We developed PCR primers common to the six determined sequences that bracketed a ca. 1.2-kbp region, including much of the agr locus. Table 1 lists the 71 strains from 14 species or subspecies of staphylococci from which a product of the expected size could be obtained with these primers, including all eight agr-positive strains of S. aureus. The agr-deleted RN6911 strain of S. aureus and 20 other staphylococcal species or subspecies did not yield a product. All of the 71 agr amplicons were sequenced and yielded 24 distinct variants. Each amplicon included the three genes expected, corresponding to agrB, agrC, and agrD. The first and last genes were truncated at each end owing to the positions of the primers. Although the agr locus has been considered to be dedicated to control of the production of virulence factors, these findings confirm that agr is in a wide range of staphylococci, including nonpathogenic species such as S. carnosus. Recent gene expression analysis by microarray (P. Dunman and S. Projan, unpublished data) has indicated that the locus is also important in the control of catabolic pathways, nutrient uptake, and energy metabolism. The lack of amplifications from 20 staphylococcal species or subspecies might indicate absence of the locus, but in at least some cases the primer sites may have diverged too far for successful amplification, particularly since three species which hybridized weakly on Southern blotting (see above) did not produce credible amplicons.
Analysis of agr sequence variability.
Of the species yielding credible agr amplicons, S. aureus yielded the previously identified four variants, S. epidermidis had three, and S. auricularis, S. capitis subsp. capitis, S. caprae, S. lugdunensis, and S. simulans had two each. Unique sequences were obtained from S. arlettae, S. carnosus, S. cohnii subsp. cohnii, S. cohnii subsp. urealyticum, S. gallinarum, S. intermedius, and S. xylosus (Table 1). Despite the conservation of the three ORFs, all 24 sequences showed considerable sequence divergence, with only 10% of the nucleotides being absolutely conserved. The agrB sequence is truncated at the N-terminal end owing to the position of the 5" primer, so the Shine-Dalgarno (SD) box and initiator codon context cannot be analyzed. The deduced amino acid sequences of the C-terminal portions aligned indicate much divergence (Fig. 1), with 8% identities and 23% conservative substitutions (the mean of the base identities in the sequence pairs was 0.44 ± 0.1). One exception to the rule of extreme divergence was the identity of AgrB sequences from the two different strains of S. simulans, although their AgrD and AgrC sequences were quite different.
The agrD sequences all possessed satisfactory SD boxes [AGGA(G/T)G-N7-10-ATG] but showed much sequence divergence throughout their lengths, including the region containing the AIP sequence. At the amino acid level, we observed only 10% identities and 26% conservative substitutions (the mean of the base identities in the sequence pairs = 0.44 ± 0.1). There is absolute conservation of two amino acid residues immediately following the AIP, and strong conservation over the next seven residues. In the upstream region, there is an absolutely conserved glycine and partial conservation at many other positions. The cysteine residue utilized in thiolactone cyclization of the AIP is absolutely conserved, with one exception: a serine residue in the AgrD from four strains of S. intermedius, all isolated from pigeons. These strains have not been shown to produce an active AIP.
AgrC sequences were also preceded by satisfactory SD boxes [AGTG(T/A)G-N3-7-ATG]. S. auricularis and S. intermedius appeared to use TTG as an initiator codon, as is commonly seen with staphylococcal proteins. Again, the alignment indicates a wide divergence of the predicted amino acid sequences for AgrC (10% identities and 22% conservative substitutions; mean of the base identities in the paired sequences = 0.35 ± 0.1), particularly in the upstream regions, with a greater degree of conservation toward the N-terminal transmembrane region, and a high degree of conservation in the C-terminal intracellular moiety, of which only the first 25 residues are present, owing to the location of the PCR primer utilized.
It might be expected that divergence of any one of these Agr peptides would require compensatory changes in the other components, because the AIP is derived from AgrD, is processed and secreted by the activity of AgrB, and then acts as a specific ligand to the extracellular portion of AgrC to induce signaling activity (9, 11). In general we observed distinct and clearly divergent sequences for all three agr ORFs for each divergent allele. The covariation of AgrB, AgrD, and AgrC was quantified by using a linear regresion method for comparing the identity for each ORF between alleles. This showed a strong correlation between the variations of the different ORFs (r = 1; analysis of variance, P < 0.001).
These results support the notion of a concerted coevolution of the different elements of the agr locus. Several species of staphylococci, such as S. aureus and S. epidermidis, have developed very divergent alternative alleles of the agr locus (Fig. 2) without apparent major divergence in the rest of their genomes as assessed by DNA-DNA hybridization. The agr variants are usually unable to cross-stimulate, and their bearers must communicate as “foreigners” rather than as members of the same community groups (7, 9, 21). This might represent adaptation to different microenvironments at specialized infection sites, for instance, and may indicate an incipient speciation event. Alternatively, other aspects of adaptation in the classical species might suffice to maintain these variants in the same genetic species despite an interruption in communication.
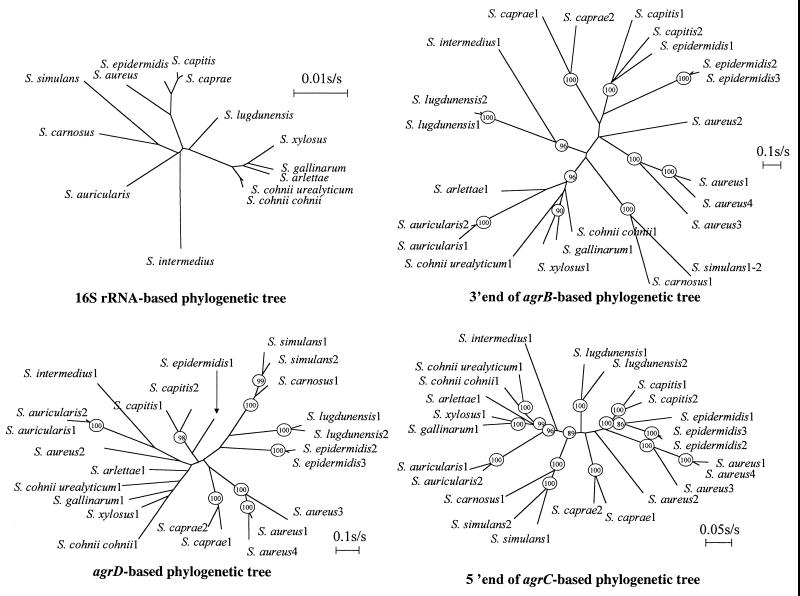
FIG. 2.
Reconstruction of staphylococcal phylogenic tree based on agrB, agrD, agrC, and 16S rRNA genes. The nucleotide sequences of 16S rRNA from staphylococci were retrieved from GenBank (S. arlettae, AB009933; S. xylosus, D83374; S. simulans, D83373; S. lugdunensis, AB009941; S. intermedius, D83369; S. gallinarum, D83366; S. epidermidis, D83362; S. cohnii urealyticum, AB009936; S. cohnii cohnii, D83361; S. carnosus, AB009934; S. caprae, AB009935; S. capitis capitis, L37599; S. auricularis, D83358; and S. aureus, X68417). They were aligned by using the multisequence alignment program CLUSTAL X. Phylogenic relationships of agr sequences and 16S rRNA genes were inferred by using the PHYLIP software package. The evolutionary distances were determined by the method of Kimura, and these values were used to construct a dendrogram by use of the neighbor-joining method. The numbers at the nodes are the proportion of 1,000 bootstrap resamplings that support the topology shown. Only bootstrap values of >80% are indicated.
Identification of conserved motifs.
The Agr proteins are highly variable in sequence, but some general motifs may be discerned. Scattered residues throughout the AgrB peptide are conserved or maintain physicochemical similarity in all variants examined (Fig. 1), and interestingly, a conserved T/K-R-V/K motif is identified by PROSITE as a potential kinase C phosphorylation site. It should also be noted that AgrB of S. cohnii subsp. cohnii contains a perfect four-element leucine zipper motif, and other sequences maintain amino acids with large hydrophobic side chains (including one to three leucines) in the corresponding positions that could correspond to a tramsmembrane hydrophobic face. This might also suggest homo- or heterophilic association involving AgrB in the bacterial membrane.
As mentioned above, AgrD has a highly conserved motif, including the absolutely conserved Glu-Asp pair just downstream of the AIP, and it is assumed that this is essential for processing the C-terminal end of the AIP. In many cases the AIP of one species or variant of Staphylococcus acts as an agr inhibitor for other staphylococci (9, 21). For S. aureus, agr inhibition increases the expression of surface proteins such as adhesins. Maintenance of this blanket inhibitory activity would be reasonable among bacteria coexisting in mixed infections or during colonization if it were to contribute to adhesiveness through interspecies communication (33). Whereas specific self-activation, at least in S. aureus and S. epidermidis, requires a thiolactone ring structure in the peptide, the more general cross-inhibition can be mediated by AIP derivatives with lactone or even lactam rings (13, 22). Interestingly, we found a single exception to the rule of a conserved cysteine five residues before the C terminus of the AIP. Four different isolates of S. intermedius from pigeons all code for serine at this position. A culture supernatant of the strain listed in Fig. 1 did not contain any peptide that inhibits agr expression in group I S. aureus nor did it contain detectable delta-hemolysin (G. Lyon and R. P. Novick, data not shown), although preliminary Northern blot hybridization tests suggest that it does produce RNA III (P. Dufour and Y. Benito, unpublished data). Moreover, a mutant of S. aureus group I agrD coding for serine at this position did not generate any detectable agr-inhibiting activity (G. Ji and R. P. Novick, data not shown). It thus appears that group I AgrB cannot process the serine-containing agrD propeptide; it is not currently known whether the S. intermedius AgrB can process such a propeptide. Two of three other strains of S. intermedius (kindly provided by W. Kloos) had agr-inhibitory activity against S. aureus (not shown); determination of the agrD sequences for these strains is currently in progress. It will be interesting to know whether a thiolactone or lactam analogue is activating or inhibitory in this species.
The N-terminal regions of AgrC variants shown in Fig. 1 show no convincing conserved sequence motifs; they do, however, have very similar hydropathy profiles, as previously proposed for the S. aureus AgrC variants (not shown), presumably reflecting conserved topology. The region of AgrC sequenced corresponds to a domain with transmembrane elements and the presumptive site for interaction with the AIP. TMAP membrane protein topology prediction for the 24 alleles suggests an 8- to 10-amino-acid intracellular N terminus, followed by six transmembrane segments of 21, 21, 21, 24, 29, and 29 residues, respectively, with three extracellular loops of 11 to 12, 16, and 5 residues and two intracellular loops of 12 and 7 amino acids. This prediction agrees with PhoA fusion analysis of S. aureus AgrC, except that the N terminus was found to be extracellular (11). The exact topology of the first intramembrane helix will require further investigation. The fifth transmembrane helix of AgrC from S. xylosus, S. arlettae, and S. epidermidis 1 includes a perfect four-element leucine zipper motif, and other strains conserve amino acids with large hydrophobic side chains in the same positions, suggesting either homodimerization, as seen in many histidine kinase receptors, or heterodimerization with an unidentified partner (4).
Mechanism of agr evolution.
Ji et al. (9) have suggested that the divergent agr loci have evolved by accumulation of random mutations through an unknown mechanism for the generation of hypervariability at this locus, followed by rigorous selection for functional combinations between the different elements. A limited matrix analysis between variant alleles such as agrSa-1 and agr-2Sa showed so little similarity that horizontal transfer from separate donors following divergent evolution in separate populations or species appeared to be a likely contributive mechanism. To investigate this question, we constructed phylogenetic trees from the DNA sequences of our different agr alleles and compared them with trees established from 16S rRNA loci from the same species and subspecies (Fig. 2). The branch lengths in the tree of agr sequences are unreliable because of the very high level of substitution, suggesting that there may also have been back mutations that would preclude a confident assessment of synonymous-to-nonsynonymous substitution rates. Nevertheless, the overall topology of trees established by using 16S rRNA, agrB, or agrC is remarkably similar. Less similarity was observed with agrD, but bootstrap analysis revealed that most of the interspecies relations based on agrD sequences had low confidence values (bootstrap values ranging from 12 to 65%). Variant agrB and agrC sequences from a single staphylococcal species generally occupy adjacent branches of the tree, except for agr-1Se, which reliably groups with sequences from the closely related S. capitis, rather than with the other two S. epidermidis alleles. In addition, agr-2Sa cannot be reliably grouped with other agrSa alleles by using agrB and agrD. In general, the relationships between different species and subspecies, as illustrated by 16S rRNA homologies (28), are maintained in the agr trees. S. xylosus, S. gallinarum, S. arlettae, and the two subspecies of S. cohnii group together in 16S rRNA and agrB and agrC trees, as do S. aureus, S. epidermidis, and S. capitis, although S. caprae, which was determined to be closely associated with S. capitis by using 16S rRNA, forms a separate branch by agrD and agrC sequences. In trees constructed with 16S rRNA, agrB, or agrC sequences, S. carnosus and S. simulans diverge from a single branch. This distribution provides no evidence for significant transfer of agr genes between populations of staphylococci and suggests that the wide differences observed are the result of a high rate of accumulation of mutations in this locus. However, Hayakawa et al. (29) have described two isolates of S. aureus from milk from cows with mastitis wherein the ORFs agrB and agrD were closely similar to those from agr-2Sa for one strain and from agr-3Sa for the other one, whereas the agrC ORF from the two strains was clearly an agr-1Sa type. These variants would be expected to show possible agr inhibition but not activation, and their pathogenic potential is surprising because virulence is usually associated with a functional agr system (10, 25, 34).
It is difficult to envision the precise mechanisms by which a new agr variant, in a situation where the AIP, its processing agent, or its specific receptor are rendered incompatible, can overcome the disadvantage of interrupted communication long enough to accumulate compensatory mutations in the other elements. The loss of phylogenic relationship between agrD and 16S rRNA suggests that agrD is the locus of the agr ORFs least linked to the genetic background. The driving force in agr evolution could be the aquisition of a mutation in the sequence coding for the AIP. This variant AIP would presumably act as a general inhibitor on other staphylococci, including the parent species. This activity may provide sufficient protection for the development of a suitable AgrC sequence to recover positive activation by the new self AIP, but it would still be necessary to coevolve a satisfactory AgrB to process and secrete the AIP. Suitable conditions would probably imply a rich nutrient supply to support rapid replication so as to compensate for the loss of nonadapted variants and probably a competition sufficient to engender pressure for reacquisition of a positive stimulatory communication between members of the newly formed agr group. It is not known, of course, in which particular milieu this evolution may have taken place. Given that many of the staphylococci are evidently not pathogenic for higher eukaryotes, the agr divergence would presumably represent environmental adaptations, of which pathogenesis is simply a subset.
The numerous sequence variants of the agr locus reported here confirm that this locus is highly variable even between subpopulations of a single species. Once fixed, however, the variants appear to be quite stable (many separate isolates in our study had identical sequences) and are a good marker for species and subspecies identification. We find no evidence for horizontal transfer, which could potentially confound species identifications, and conclude that the agr alleles have evolved contemporaneously with their parent species. The causes for agr divergence within a species and whether these events precede a more general divergence, even speciation, remain unclear. It will be useful to design fresh PCR primers to recover agr sequences from species and strains which failed to produce satisfactory amplicons, and the sequences discussed here should aid in this process. It is particularly desirable to determine the generality of the serine-for-cysteine substitution in S. intermedius and to determine the biochemical and physiological characteristics of this unusual variant.
.
Acknowledgments
We thank G. Lyon, M. Gouy, and C. Mougel for helpful discussions and Y. Benito, H. Meugnier, N. Violland, C. Courtier, and C. Gardon for technical assistance.
REFERENCES
- 1.Aiyar, A. 2000. The use of CLUSTAL W and CLUSTAL X for multiple sequence alignment. Methods Mol. Biol. 132:221-241. [DOI] [PubMed] [Google Scholar]
- 2.Benito, Y., F. A. Kolb, P. Romby, G. Lina, J. Etienne, and F. Vandenesch. 2000. Probing the structure of RNAIII, the Staphylococcus aureus agr regulatory RNA, and identification of the RNA domain involved in repression of protein A expression. RNA 6:668-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donvito, B., J. Etienne, T. Greenland, C. Mouren, V. Delorme, and F. Vandenesch. 1997. Distribution of the synergistic haemolysin genes hld and slush with respect to agr in human staphylococci. FEMS Microbiol. Lett. 151:139-144. [DOI] [PubMed] [Google Scholar]
- 4.Goudreau, P. N., and A. M. Stock. 1998. Signal transduction in bacteria: molecular mechanisms of stimulus-response coupling. Curr. Opin. Microbiol. 1:160-169. [DOI] [PubMed] [Google Scholar]
- 5.Hofmann, K., P. Bucher, L. Falquet, and A. Bairoch. 1999. The PROSITE database, its status in 1999. Nucleic Acids Res. 27:215-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janzon, L., and S. Arvidson. 1990. The role of the delta-lysin gene (hld) in the regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus. EMBO J. 9:1391-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jarraud, S., G. J. Lyon, A. M. Figueiredo, G. Lina, F. Vandenesch, J. Etienne, T. W. Muir, and R. P. Novick. 2000. Exfoliatin-producing strains define a fourth agr specificity group in Staphylococcus aureus. J. Bacteriol. 182:6517-6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ji, G., R. C. Beavis, and R. P. Novick. 1995. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. USA 92:12055-12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ji, G., R. Beavis, and R. P. Novick. 1997. Bacterial interference caused by autoinducing peptide variants. Science 276:2027-2030. [DOI] [PubMed] [Google Scholar]
- 10.Lammers, A., P. J. Nuijten, and H. E. Smith. 1999. The fibronectin binding proteins of Staphylococcus aureus are required for adhesion to and invasion of bovine mammary gland cells. FEMS Microbiol. Lett. 180:103-109. [DOI] [PubMed] [Google Scholar]
- 11.Lina, G., S. Jarraud, G. Ji, T. Greenland, A. Pedraza, J. Etienne, R. P. Novick, and F. Vandenesch. 1998. Transmembrane topology and histidine protein kinase activity of AgrC, the agr signal receptor in Staphylococcus aureus. Mol. Microbiol. 28:655-662. [DOI] [PubMed] [Google Scholar]
- 12.Lina, G., A. Quaglia, M. E. Reverdy, R. Leclercq, F. Vandenesch, and J. Etienne. 1999. Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among staphylococci. Antimicrob. Agents Chemother. 43:1062-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayville, P., G. Ji, R. Beavis, H. Yang, M. Goger, R. P. Novick, and T. W. Muir. 1999. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc. Natl. Acad. Sci. USA 96:1218-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morfeldt, E., L. Janzon, S. Arvidson, and S. Lofdahl. 1988. Cloning of a chromosomal locus (exp) which regulates the expression of several exoprotein genes in Staphylococcus aureus. Mol. Gen. Genet. 211:435-440. [DOI] [PubMed] [Google Scholar]
- 15.Morfeldt, E., I. Panova-Sapundjieva, B. Gustafsson, and S. Arvidson. 1996. Detection of the response regulator AgrA in the cytosolic fraction of Staphylococcus aureus by monoclonal antibodies. FEMS Microbiol. Lett. 143:195-201. [DOI] [PubMed] [Google Scholar]
- 16.Morfeldt, E., K. Tegmark, and S. Arvidson. 1996. Transcriptional control of the agr-dependent virulence gene regulator, RNAIII, in Staphylococcus aureus. Mol. Microbiol. 21:1227-1237. [DOI] [PubMed] [Google Scholar]
- 17.Novick, R. P. 1999. Regulation of pathogenicity in Staphylococcus aureus by a peptide-based density-sensing system, p. 129-146. In G. M. Dunny and S. C. Winans (ed.), Cell-cell signaling in bacteria. ASM Press, Washington, D.C.
- 18.Novick, R. P., S. J. Projan, J. Kornblum, H. F. Ross, B. Kreiswirth, F. Vandenesch, and S. Moghazed. 1995. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol. Gen. Genet. 248:446-458. [DOI] [PubMed] [Google Scholar]
- 19.Novick, R. P., H. F. Ross, A. M. S. Figueiredo, G. Abramochkin, and T. Muir. 2000. Activation and inhibition of the staphylococcal AGR Syst. Science 287:391a. [Google Scholar]
- 20.Novick, R. P., H. F. Ross, S. J. Projan, J. Kornblum, B. Kreiswirth, and S. Moghazeh. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulartory RNA molecule. EMBO J. 12:3967-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otto, M., H. Echner, W. Voelter, and F. Gotz. 2001. Pheromone cross-inhibition between Staphylococcus aureus and Staphylococcus epidermidis. Infect. Immun. 69:1957-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otto, M., R. Sussmuth, C. Vuong, G. Jung, and F. Gotz. 1999. Inhibition of virulence factor expression in Staphylococcus aureus by the Staphylococcus epidermidis agr pheromone and derivatives. FEBS Lett. 1999:257-262. [DOI] [PubMed] [Google Scholar]
- 23.Peng, H. L., R. P. Novick, B. Kreiswirth, J. Kornblum, and P. M. Schlievert. 1988. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 170:4365-4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Persson, B., and P. Argos. 1997. Prediction of membrane protein topology utilizing multiple sequence alignments. J. Protein Chem. 16:453-457. [DOI] [PubMed] [Google Scholar]
- 25.Projan, S. J., and R. P. Novick. 1997. The molecular basis of pathogenicity, p. 55-82. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human disease. Churchill Livingstone, New York, N.Y.
- 26.Recsei, P., B. Kreiswirth, M. O'Reilly, P. Schlievert, A. Gruss, and R. P. Novick. 1986. Regulation of exoprotein gene expression in Staphylococcus aureus by agr. Mol. Gen. Genet. 202:58-61. [DOI] [PubMed] [Google Scholar]
- 27.Retief, J. D. 2000. Phylogenetic analysis using PHYLIP. Methods Mol. Biol. 132:243-258. [DOI] [PubMed] [Google Scholar]
- 28.Takahashi, T., M. Kaneko, Y. Mori, M. Tsuji, N. Kikuchi, and T. Hiramune. 1997. Phylogenetic analyses of Staphylococcus based on the 16S rDNA sequence and assignment of clinical isolates from animals. J. Vet. Med. Sci. 59:775-783. [DOI] [PubMed] [Google Scholar]
- 29.Takeuchi, S., T. Maeda, N. Hashimoto, K. Imaizumi, T. Kaidoh, and Y. Hayakawa. 2001. Variation of the agr locus in Staphylococcus aureus isolates from cows with mastitis. Vet. Microbiol. 79:267-274. [DOI] [PubMed] [Google Scholar]
- 30.Tegmark, K., E. Morfeldt, and S. Arvidson. 1998. Regulation of agr-dependent virulence genes in Staphylococcus aureus by RNAIII from coagulase-negative staphylococci. J. Bacteriol. 180:3181-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Wamel, W. J., G. van Rossum, J. Verhoef, C. M. Vandenbroucke-Grauls, and A. C. Fluit. 1998. Cloning and characterization of an accessory gene regulator (agr)-like locus from Staphylococcus epidermidis. FEMS Microbiol. Lett. 163:1-9. [DOI] [PubMed] [Google Scholar]
- 32.Vandenesch, F., S. J. Projan, B. Kreiswirth, J. Etienne, and R. P. Novick. 1993. Agr-related sequences in Staphylococcus lugdunensis. FEMS Microbiol. Lett. 111:115-122. [DOI] [PubMed] [Google Scholar]
- 33.Vuong, C., H. L. Saenz, F. Gotz, and M. Otto. 2000. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J. Infect. Dis. 182:1688-1693. [DOI] [PubMed] [Google Scholar]
- 34.Wesson, C. A., L. E. Liou, K. M. Todd, G. A. Bohach, W. R. Trumble, and K. W. Bayles. 1998. Staphylococcus aureus Agr and Sar global regulators influence internalization and induction of apoptosis. Infect. Immun. 66:5238-5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wheeler, D. L., C. Chappey, A. E. Lash, D. D. Leipe, T. L. Madden, G. D. Schuler, T. A. Tatusova, and B. A. Rapp. 2000. Database resources of the National Center for Bio/Technology Information. Nucleic Acids Res. 28:10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]