Abstract
Acyl-homoserine lactone (acyl-HSL) quorum sensing is common to many Proteobacteria including a clinical isolate of Burkholderia cepacia. The B. cepacia isolate produces low levels of octanoyl-HSL. We have examined an environmental isolate of Burkholderia vietnamiensis. This isolate produced several acyl-HSLs. The most abundant species was decanoyl-HSL. Decanoyl-HSL in B. vietnamiensis cultures reached concentrations in excess of 20 μM. We isolated a B. vietnamiensis DNA fragment containing a gene for the synthesis of decanoyl-HSL (bviI) and an open reading frame that codes for a putative signal receptor (bviR). A B. vietnamiensis bviI mutant did not produce detectable levels of decanoyl-HSL.
Many Proteobacteria produce N-acyl homoserine lactone (acyl-HSL) quorum-sensing signals. Examples include Pseudomonas aeruginosa, which uses N-butyryl-HSL (C4-HSL) and N-3-oxodocanoyl-HSL (3OC12-HSL) as signals to control the expression of dozens of genes, many of which encode virulence factors (24, 28, 47). Ralstonia solanacearum, a plant pathogen, uses N-octanoyl-HSL (C8-HSL) in quorum sensing, Agrobacterium tumefaciens uses N-3-oxooctanoyl-HSL (3OC8-HSL) to control conjugal transfer, and Erwinia carotovora uses N-3-oxohexanoyl-HSL (3OC6-HSL) to control antibiotic production and other factors (2, 4, 9, 25, 37). Generally, acyl-HSL quorum sensing involves a member of the LuxI family of signal generators and a member of the LuxR family of signal receptor transcription factors (for a recent review, see reference 3).
The genus Burkholderia has been a subject of recent attention. Species classifications in this genus remain in flux. Some members of the genus have emerged as serious opportunistic pathogens. For example, some strains of Burkholderia cepacia and perhaps other Burkholderia species can colonize the lungs of people with cystic fibrosis. In part because B. cepacia is resistant to antibiotic therapy, it can be a major health problem in colonized cystic fibrosis patients (10). The fact that Burkholderia spp. have multiple chromosomes and very plastic genomes (13) has also brought a research focus to the genus. Furthermore, some members of the genus are capable of degrading environmental pollutants and have been the subject of studies aimed at developing tools for bioremediation (29, 30, 31). For example, the subject of this study, Burkholderia vietnamiensis G4, was isolated from a holding pond at an industrial waste treatment facility, and it can degrade trichloroethylene and toluene (18).
A previous report shows that a clinical isolate of B. cepacia makes low levels of C8-HSL in laboratory cultures and that quorum sensing is involved in the regulation of virulence factors including lipases, protease, and siderophores (14). Another strain of B. cepacia produces multiple acyl-HSLs, but these signals have not been identified, nor have their concentrations been determined (17). A recent study shows that B. vietnamiensis has at least two acyl-HSL generator and receptor gene pairs (16). Here we show that B. vietnamiensis G4 produces several acyl-HSLs and we define the genetic element responsible for the production of the most abundant of these acyl-HSLs. This study provides the groundwork for identification of quorum sensing-controlled genes in B. vietnamiensis and investigations of the role of quorum sensing in the degradation of environmental pollutants like trichloroethylene.
The bacterial strains and plasmids used in this study are described in Table 1. For acyl-HSL bioassays and for partial purification of B. vietnamiensis-produced acyl-HSLs, cultures were grown to an optical density at 600 nm of 1.6 to 2.0 (the late-logarithmic phase of growth) in Difco tryptic soy broth (pH 7.0) at 30°C with shaking (250 rpm). For 14C labeling of acyl-HSLs, cultures were grown as above in basal salts medium (pH 7.0) with 20 mM lactate as a carbon source (8). For conjugation, B. vietnamiensis was grown in Luria-Bertani broth (L broth) (6). Antibiotics were included as appropriate. For Escherichia coli, the antibiotic concentrations were as follows: ampicillin, 100 μg/ml; chloramphenicol, 35 μg/ml; gentamicin, 10 μg/ml; kanamycin, 35 μg/ml; tetracycline, 10 μg/ml. The antibiotic concentrations for B. vietnamiensis G4 were as follows: chloramphenicol, 25 to 35 μg/ml; gentamicin, 10 μg/ml; kanamycin, 35 μg/ml. Plating was carried out on media solidified with 1.5 % agar. E. coli was grown in L broth or on Luria-Bertani agar using standard procedures (6).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or characteristics | Source or reference |
|---|---|---|
| B. (cepacia) vietnamiensis | ||
| G4 | Typed as B. vietnamiensis by J. LiPuma (personal communication) | 15 |
| FMT-05 | fabF mutant derived from G4 | This study |
| IMT-61 | bviI mutant derived from G4 | This study |
| RMT-14 | bviR mutant derived from G4 | This study |
| E. coli | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 27 |
| MG4 | Δ(argF-lac)U169 zah-735::Tn 10 recA56 srl::Tn 10 | 26 |
| S17-1 (λpir) | recA thi pro hsdR | 32 |
| VJS533 | recΔA56 ara Δ(lac-proAB)X111 rpsL (φ80 lacZΔM15) | 35 |
| XL-1 Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac[F" proAB lacIqZΔM15 Tn 10 (Tetr)] | Stratagene |
| R. solanacearum | ||
| AW1-AI8 | sol18::SP | 2 |
| Plasmids | ||
| p395B | sol reporter; aidA::lacZ, Tcr | 2 |
| pBBR1MCS-2 | Mobilizable broad-host-range cloning vector, Kmr | 11 |
| pBBR1MCS-5 | Mobilizable broad-host-range cloning vector, Gmr | 11 |
| pBCL5-2 | 8.5-kb HindIII fragment containing the bviI, bviR, and fabF-like gene from B. vietnamiensis G4 in pUC19 | This study |
| pBCSFM | A 952-bp Rsr II fragment of the fabF-like gene from pBCL-2 was replaced with the pBBR1MCS-5 Gmr marker; the resulting DNA was digested with PstI, treated with the Klenow fragment, and cloned into the EcoRV site in pSUP102 | This study |
| pBCSIM | bviI, fabF-like gene PstI fragment from pBCL5-2 treated with Klenow fragment and blunt-end cloned into EcoRV-digested pSUP102; a 570-bp BsiW bviI fragment replaced with a Gmr fragment PCR amplified from pBBR1MCS-5 | This study |
| pBCSRM | 4.4-kb bviR bviI EcoRV fragment from pBCL5-2 cloned into the EcoRV site of pSUP102, 1-kb Gmr marker from pBBR1MCS-5 cloned into unique SfiI site | This study |
| pECP61.5 | rhl reporter; rhlR rhlA::lacZ Apr | 24 |
| pHV200I− | lux reporter; luxR luxI"CDABE Apr | 22 |
| pHV300I− | lux reporter; luxR luxI"CDABE Cmr | 7 |
| pKDT17 | las reporter; lasB::lacZ plac-lasR Apr | 21 |
| pSUP102 | E. coli-specific mobilizable vector Cmr Tcr | 32 |
To detect acyl-HSLs and to determine their relative abundance, we used a radiotracer procedure as described elsewhere (33). Unless otherwise specified, B. vietnamiensis was grown in 5 ml of basal salts plus lactate medium. When the culture reached late logarithmic phase, 5.0 μCi of l-[1-14C]methionine was added. After a 10-min incubation, the culture was extracted twice with equal volumes of acidified ethyl acetate and the extract was evaporated to dryness. The radiolabeled acyl-HSLs were dissolved in 20% methanol and fractionated by C18 reverse-phase high-performance liquid chromatography (HPLC). The amount of radiolabel in each fraction was determined by standard scintillation counting procedures. The retention times of the radioactive peaks corresponded to the retention times of synthetic acyl-HSL standards (Quorum Sciences Inc, Coralville, Iowa).
To confirm the results of the radiotracer analysis and to gain quantitative data about each of the acyl-HSLs produced by B. vietnamiensis, we measured the levels of acyl-HSLs in HPLC-fractionated culture fluid extracts by using bioassays. Four different bioassays were employed, each being selective for different acyl-HSLs. The E. coli(pHV200I−) assay shows greatest sensitivity to 3OC6-HSL (22). The E. coli MG4(pKDT17) assay shows greatest sensitivity to 3OC12-HSL (21, 22). The E. coli(pECP61.5) assay shows greatest sensitivity to C4-HSL (20). The Ralstonia solanacearum AW1-AI8(p395B) assay shows greatest sensitivity to C8-HSL (2). The material analyzed by the bioassays was obtained as follows: cells were removed from the fluid of a 3-liter culture by centrifugation, and the culture fluid was extracted twice with equal volumes of acidified ethyl acetate. The extract was concentrated by rotary evaporation at 40 to 45°C and fractionated by C18 HPLC. Samples from each fraction eluted in HPLC were analyzed by the bioassays. Standard curves with synthetic acyl-HSLs were generated, and the amounts of acyl-HSLs in HPLC fractions were determined by comparison to the standard curves.
To confirm assignments based on HPLC retention times, fractions constituting peaks were pooled, concentrated by rotary evaporation, and subjected to further separation by HPLC in water containing methanol at a percentage of 15% lower than that in which they were eluted in the gradient. Active fractions were concentrated and analyzed by gas chromatography-mass spectrometry as described previously (23).
DNA manipulations were done by standard procedures (1, 27). For cloning the B. vietnamiensis quorum-sensing genes, an unsized library of HindIII chromosomal DNA fragments was constructed in pUC19 (38). The library was used to transform E. coli DH5α(pHV300I−). Ampicillin-resistant, luminescent colonies were selected for further study. DNA sequencing was performed at the University of Iowa DNA Facility.
For insertional mutagenesis, genes were cloned in pSUP102 and a gentamicin cassette from pBBR1MCS-5 was inserted in the cloned gene (Table 1). The pSUP102 derivatives were transferred from E. coli S17-1 (λpir) into B. vietnamiensis by conjugation using the following procedure: donors (mid-log phase) and recipients (stationary phase) were grown in L broth with appropriate antibiotics. Cells were mixed at a donor-to-recipient ratio of 4:1, washed in phosphate-buffered saline (pH 7.4), and suspended in a small volume of phosphate-buffered saline. Mating mixtures were spotted on Luria-Bertani (LB) agar and grown at 30°C overnight. Transconjugants were obtained by selection on Simmons citrate agar (SCA) (6) containing gentamicin. Screening transconjugants for chloramphenicol sensitivity allowed us to obtain insertion mutants. The constructs were confirmed by PCR and Southern blotting.
Chrome azurol S assays were used to measure siderophore activity (28). Egg yolk agar assays were used to estimate bacterial production of lecithinase (6). Difco brain heart infusion agar assays were used to measure extracellular protease activity (34).
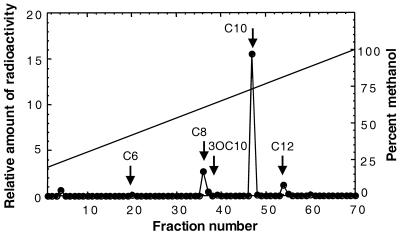
To gain information on whether B. vietnamiensis produced acyl-HSLs and, if so, in what sort of relative abundance, we incubated cultures with carboxy-labeled [14C]methionine. The cell-free culture fluid was extracted with ethyl acetate and fractionated by HPLC. As described previously, the amount of radiolabel in fractions that comigrate with known acyl-HSLs corresponds to the relative abundance of that acyl-HSL (33). There were three main peaks of radioactivity (Fig. 1). The largest peak corresponded to C10-HSL, the next largest peak corresponded to C8-HSL, and the third peak comigrated with C12-HSL.
FIG. 1.
HPLC analysis of acyl-HSLs produced by B. vietnamiensis G4. Acyl-HSLs were labeled with 14C, extracted from culture fluid, and fractionated by HPLC. Radioactivity was measured to identify peaks containing 14C-labeled acyl-HSLs. As indicated by the arrows, synthetic acyl-HSL standards were eluted as follows: C6-HSL, fraction 20; C8-HSL, fraction 36; C10-HSL, fraction 47: 3OC10-HSL, fraction 39; C12-HSL, fraction 54. The methanol concentration of the gradient is shown by the solid line.
To confirm that the radioactive peaks in the experiments described above represented acyl-HSLs and to gain information about the concentrations of acyl-HSLs in culture fluid, we performed quantitative bioassays on HPLC fractions of an ethyl acetate extract of 3 liters of fluid from an early-stationary-phase culture. We identified C6-, C8-, C10-, and C12-HSLs, and we also identified 3OC10-HSL. The concentrations of these molecules in the culture fluid were 0.12 μM C6-HSL, 2.2 μM C8-HSL, 22.4 μM C10-HSL, 0.003 μM 3OC10-HSL, and 0.65 μM C12-HSL. As expected, gas chromatography-mass spectrometry of the three most abundant peaks showed spectra identical to the spectra of synthetic C10-, C8-, and C12-HSLs (data not shown).
We screened a library of B. vietnamiensis genes in E. coli containing a reporter plasmid, pHV300I−. This plasmid contains the acyl-HSL-responsive Vibrio fischeri lux gene cluster and does not contain a functional acyl-HSL synthase gene. We obtained a luminous strain of E. coli that contained a plasmid (pBCL5-2) with an 8.5-kb insert of B. vietnamiensis DNA. In E. coli, pBCL5-2 directed the synthesis of C6-, C8-, C10-, and C12-HSL. As in B. vietnamiensis, C10-HSL was the most abundant of the acyl-HSLs. The B. vietnamiensis DNA in pBCL5-2 was sequenced, and an analysis revealed a gene that coded for a LuxI homolog and a divergently transcribed gene coding for a LuxR homolog with a rather large 1.2-kb intergenic region.
The gene coding for the LuxI homolog was 660 bp, and it was identical to the recently described bviI (16). The gene coding for the LuxR homolog was 714 bp and was identical to bviR (16). There was a gene coding for a putative transport protein downstream of bviR and a gene showing greatest similarity to fabF-like genes from P. aeruginosa and A. tumefacians (BLAST scores of 10−145) downstream of bviI. The intergenic region contained an assortment of small open reading frames, the longest of which was 384 bp. None of these open reading frames showed significant similarity to known gene products. It is possible that the intergenic region codes for a small regulatory RNA or protein.
We analyzed acyl-HSL production in the B. vietnamiensis bviR, bviI, and fabF-like gene mutants. As monitored by the radiotracer assay, the bviI mutant did not make detectable levels of any acyl-HSL except C8-HSL, which was present in trace amounts [<10 nM, compared to 2,200 nM C8-HSL in the parent as measured by the E. coli(pHV200I−) bioassay]. This, together with the analysis of acyl-HSL production by bviI-containing E. coli (see above), indicates that bviI codes for an acyl-HSL synthase that is responsible for the production of all of the abundant acyl-HSLs. The presence of traces of octanoyl-HSL in cultures of the bviI mutant suggests that there is another poorly expressed or poorly active acyl-HSL synthase, one that directs the synthesis of C8-HSL, the same signal as that produced by CepI in B. cepacia. This is consistent with a recent report by Lutter et al. (16). The parent B. vietnamiensis produces much more C8-HSL than the mutant. One explanation is that BviI is responsible for the majority of the C8-HSL produced in the parent and the hypothetical second acyl-HSL synthase contributes very little to the total C8-HSL level.
The acyl-HSL production pattern of the bviR mutant was essentially identical to that of the bviI mutant. We interpret this to indicate that, like many other acyl-HSL synthesis and response systems, bviI is positively autoregulated by the acyl-HSL signal it produces and the cognate R protein (5). Because of its proximity to bviR-bviI, we wondered if the fabF-like gene was involved in generation of acyl-ACPs for bviI specifically. To investigate this, we generated a null mutation in the fabF-like gene. The strain with the mutation in the fabF-like gene grew somewhat slower than the parent. That it could grow at al indicates that there are other fatty acid synthesis genes in B. vietnamiensis that can fulfill the role played by this gene. Although the absolute levels were somewhat reduced (two- to threefold lower), the mutant produced acyl-HSLs in the same relative abundance as the parent did. Therefore, we conclude that this fabF-like gene is not involved in the specificity of quorum-sensing signal generation.
BviR and BviI did not appear to regulate factors controlled by quorum sensing in B. cepacia. Previous reports showed that quorum sensing in B. cepacia controlled siderophore, protease, and lipase production (14, 17). Therefore, we examined the influence of the BviR-BviI system on these factors in B. vietnamiensis. Neither the parent, B. vietnamiensis G4, nor the bviR or bviI mutants produced detectable levels of protease or lipase. All three strains produced siderophores, as measured by the chrome azurol S assay, and the levels of the siderophores produced were indistingushable. Presumably, the genes regulated by quorum sensing in B. vietnamiensis are different from those regulated by quorum sensing in B. cepacia. In fact, B. vietnamiensis produces an antibiotic, which appears to be under quorum-sensing control (K. Lee, personal communication).
In summary we have shown that B. vietnamiensis produces detectable levels of several acyl-HSLs, the most abundant of which is C10-HSL. This is present in 10-fold excess over the second most abundant acyl-HSL, C8-HSL. Other bacteria make acyl-HSLs with acyl side chains ranging from 4 to 14 carbons. To our knowledge, this is the first report of a bacterium that makes predominantly C10-HSL. Other investigators have used gas-chromatography-mass spectrometry and have obtained similar results with respect to acyl-HSL production by B. vietnamiensis G4 (Lee, personal communication). The B. cepacia acyl-HSL is C8-HSL, and cultures of the one studied strain of B. cepacia make<0.01% of the total acyl-HSLs made by B. vietnamiensis G4 (14). Most acyl-HSL-producing bacteria make micromolar quantities of acyl-HSLs, as does B. vietnamiensis G4. The bviI gene directs E. coli to produce B. vietnamiensis acyl-HSLs, and a B. vietnamiensis bviI mutation results in the loss of C10-HSL production. The bviI mutant produced trace amounts of C8-HSLs. This is consistent with the finding of a bviI homolog in B. vietnamiensis (16).
Work by other investigators suggests that extracellular lipase, protease, and siderophore production may be controlled by quorum sensing in some B. cepacia clinical isolates (14, 17). We found that B. vietnamiensis G4 did not produce extracellular proteases or lipases that were comparable to the B. cepacia enzymes and that siderophore production did not seem to be controlled by quorum sensing. Our results are consistent with those obtained by K. Lee and colleagues (personal communication). Further studies are required to elucidate the targets of quorum-sensing control in B. vietnamiensis G4. There is a suggestion that antibiotic synthesis in B. vietnamiensis G4 may be controlled by quorum sensing (Lee, personal communication). The availability of a bviI mutant strain and the knowledge that C10-HSL is the main acyl-HSL should allow the development of screens of quorum-sensing-controlled genes in B. vietnamiensis G4.
Acknowledgments
We thank Lynn Teesch, University of Iowa Mass Spectrometry Facility, for performing the CI-MS analysis; Kyoung Lee for providing data in advance of publication; and David P. Speert, Department of Pediatrics, University of British Columbia, for providing materials.
This work was supported by a grant from the Cystic Fibrosis Foundation.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1989. Current protocols in molecular biology, vol. 1. John Wiley & Sons, Inc., New York, N.Y.
- 2.Flavier, A. B., L. M. Ganova-Raeva, M. A. Schell, and T. P. Denny. 1997. Hierarchical autoinduction in Ralstonia solanacearum: control of acyl-homoserine lactone production by a novel autoregulatory system responsive to 3-hydropalmitic acid methyl ester. J. Bacteriol. 179:7089-7097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuqua, C., and E. P. Greenberg. 1998. Self-perception in bacteria: quorum sensing with acylated homoserine lactones. Curr. Opin. Microbiol. 1:183-189. [DOI] [PubMed] [Google Scholar]
- 4.Fuqua, W. C., and S. C. Winans. 1994. A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J. Bacteriol. 176:2796-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuqua, W. C., S. C. Winans, and E. P. Greenberg. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerhardt, P., R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.). 1994. Methods for general and molecular bacteriology. American Society for Microbiology, Washington, D.C.
- 7.Gray, K. M., and E. P. Greenberg. 1992. Physical and functional maps of the luminescence gene cluster in an autoinducer-deficient Vibrio fischeri strain isolated from a squid light organ. J. Bacteriol. 174:4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hareland, W., R. L. Crawford, P. J. Chapman, and S. Dagley. 1975. Metabolic function and properties of 4-hydroxyphenylacetic acid 1-hydroxylase from Pseudomonas acidovorans. J. Bacteriol. 121:272-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hwang, I. Y., P. L. Li, L. H. Zhang, K. R. Piper, D. M. Cook, M. E. Tate, and S. K. Farrand. 1994. TraI, a LuxI homologue, is responsible for production of conjugation factor, the Ti plasmid N-acylhomoserine lactone autoinducer. Proc. Natl. Acad. Sci. USA 91:4639-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isles, A., I. Maclusky, M. Corey, R. Gold, C. Prober, P. Fleming, and H. Levison. 1984. Pseudomonas cepacia infection in cystic fibrosis; an emerging problem. J. Pediatr. 104:206-210. [DOI] [PubMed] [Google Scholar]
- 11.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, Jr., and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 12.Latifi, A., M. K. Winson, M. Foglino, B. W. Bycroft, G. S. Stewart, A. Lazdunski, and P. Williams. 1995. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol. Microbiol. 17:333-343. [DOI] [PubMed] [Google Scholar]
- 13.Lessie, T. G., W. Hendrickson, B. D. Manning, and R. Devereux. 1996. Genomic complexity and plasticity of Burkholderia cepacia. FEMS Microbiol. Lett. 144:117-128. [DOI] [PubMed] [Google Scholar]
- 14.Lewenza, S., B. Conway, E. P. Greenberg, and P. A. Sokol. 1999. Quorum sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J. Bacteriol. 181:748-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lipuma, J. J., S. E. Dasen, D. W. Nielson, S. E. Stern, and T. L. Stull. 1990. Person-to-person transmission of Pseudomonas cepacia between patients with cystic fibrosis. Lancet 336:527-532. [DOI] [PubMed] [Google Scholar]
- 16.Lutter, E., S. Lewenza, J. J. Dennis, M. B. Visser, and P. A. Sokol. 2001. Distribution of quorum-sensing genes in the Burkholderia cepacia complex. Infect. Immun. 69:4661-4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKenney, D., K. E. Brown, and D. G. Allison. 1995. Influence of Pseudomonas aeruginosa exoproducts on virulence factor production in Burkholderia cepacia: evidence of interspecies communication. J. Bacteriol. 177:6989-6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nelson, M. J. K., S. O. Montgomery, E. J. O'Neill, and P. H. Pritchard. 1986. Aerobic metabolism of trichloroethylene by a bacterial isolate. Appl. Environ. Microbiol. 52:383-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ochsner, U. A., A. K. Koch, A. Fiechter, and J. Reiser. 1994. Isolation and characterization of a regulatory gene affecting rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. J. Bacteriol. 176:2044-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parsek, M. R., A. L. Schaefer, and E. P. Greenberg. 1997. Analysis of random and site-directed mutations in rhlI, a Pseudomonas aeruginosa gene encoding an acylhomoserine lactone synthase. Mol. Microbiol. 26:301-310. [DOI] [PubMed] [Google Scholar]
- 21.Passador, L., J. M. Cook, M. J. Gambello, L. Rust, and B. H. Iglewski. 1993. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science 260:1127-1130. [DOI] [PubMed] [Google Scholar]
- 22.Pearson, J. P., K. M. Gray, L. Passador, K. D. Tucker, A. Eberhard, B. H. Iglewski, and E. P. Greenberg. 1994. Structure of the autoinducer required for the expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. USA 91:197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pearson, J. P., L. Passador, B. H. Iglewski, and E. P. Greenberg. 1995. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 92:1490-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pesci, E. C., J. P. Pearson, P. C. Seed, and B. H. Iglewski. 1997. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3127-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pirhonen, M., D. Flego, R. Heikinheimo, and E. T. Palva. 1993. A small diffusible signal molecule is responsible for the global control of virulence and exoenzyme production in the plant pathogen Erwinia carotovora. EMBO J. 12:2467-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ralling, G., S. Bodrug, and T. Linn. 1985. Growth rate-dependent regulation of RNA polymerase synthesis in Escherichia coli. Mol. Gen. Genet. 201:379-386. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Schwyn, B., and J. B. Neilands. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160:47-56. [DOI] [PubMed] [Google Scholar]
- 29.Shields, M. S., S. O. Montgomery, P. J. Chapman, S. M. Cuskey, and P. H. Pritchard. 1989. Novel pathway of toluene catabolism in the trichloroethylene-degrading bacterium G4. Appl. Environ. Microbiol. 55:1624-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shields, M. S., and M. J. Reagin. 1992. Selection of a Pseudomonas cepacia strain constitutive for the degradation of trichloroethylene. Appl. Environ. Microbiol. 58:3977-3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shields, M. S., M. J. Reagin, R. R. Gerger, R. Campbell, and C. Somerville. 1995. TOM, a new aromatic degradative plasmid from Burkholderia (Pseudomonas)cepacia G4. Appl. Environ. Microbiol. 61:1352-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simon, R., M. O'Connell, M. Labes, and A. Pühler. 1986. Plasmid vectors for the genetic analysis and manipulation of Rhizobia and other Gram-negative bacteria. Methods Enzymol. 118:640-659. [DOI] [PubMed] [Google Scholar]
- 33.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 34.Sokol, P. A., D. E. Ohman and B. H. Iglewski. 1979. A more sensitive plate assay for detection of protease production by Pseudomonas aeruginosa. J. Clin. Microbiol. 9:538-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stewart, V., and J. V. Parales, Jr. 1988. Identification and expression of genes narL and narX of the nar (nitrate reductase) locus in Escherichia coli K-12. J. Bacteriol. 170:1589-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whiteley, M., K. M. Lee, and E. P. Greenberg. 1999. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:13904-13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams, P., N. J. Bainton, S. Swift, S. R. Chhabra, M. K. Winson, G. S. A. B. Stewart, G. P. C. Salmond, and B. W. Bycroft. 1992. Small molecule-mediated density-dependent control of expression in prokaryotes: bioluminescence and the biosynthesis of carbapenem antibiotics. FEMS Microbiol. Lett. 100:161-168. [DOI] [PubMed] [Google Scholar]
- 38.Yannisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]