Abstract
The ability of neurons to alter their transcriptional programs in response to synaptic input is of fundamental importance to the neuroplastic mechanisms underlying learning and memory. Because of technical limitations of conventional gene detection methods, the current view of activity-dependent neural transcription derives from experiments in which neurons are assumed quiescent until a signaling stimulus is given. The present study was designed to move beyond this static model by examining how earlier episodes of neural activity influence transcription of the immediate–early gene Arc. Using a sensitive FISH method that detects primary transcript at genomic alleles, the proportion of hippocampal CA1 neurons that activate transcription of Arc RNA was constant at ≈40% in response to both a single novel exploration session and daily sessions repeated over 9 days. This proportion is similar to the percentage of active neurons defined electrophysiologically. However, this close correspondence was disrupted in rats exposed briefly, but repeatedly, to the same environment within a single day. Arc transcription in CA1 neurons declined dramatically after as few as four 5-min sessions, despite stable electrophysiological activity during all sessions. Additional experiments indicate that the decrement in Arc transcription occurred at the cellular, rather than synaptic level, and was not simply linked to habituation to novelty. Thus, the neural genomic response is governed by recent, but not remote, cell firing history in the behaving animal. This state-dependence of neuronal transcriptional coupling provides a mechanism of metaplasticity and may regulate capacity for synaptic modification in neural networks.
Keywords: hippocampus, immediate-early, learning, place field, memory
Studies over the past 30 years have shown that de novo transcription, in the minutes to hours after learning, is essential for long-term memory formation (reviewed in ref. 1). In recent years, immediate-early genes (IEGs) have been identified as critical components of RNA and protein synthesis-dependent synaptic plasticity and memory consolidation processes (2–5). IEG expression is induced rapidly in neurons by patterned synaptic activity that activates NMDA receptors (6). Because of technical limitations inherent in conventional steady-state gene detection methods, we currently have only a static view of IEG transcriptional induction, one in which neurons are assumed to be in a uniformly quiescent state until a stimulus is delivered. In freely behaving animals, however, individual neurons have different activity histories based on recent experience. Little is currently known about how a neuron's firing history influences subsequent rounds of synaptic activity-regulated gene transcription.
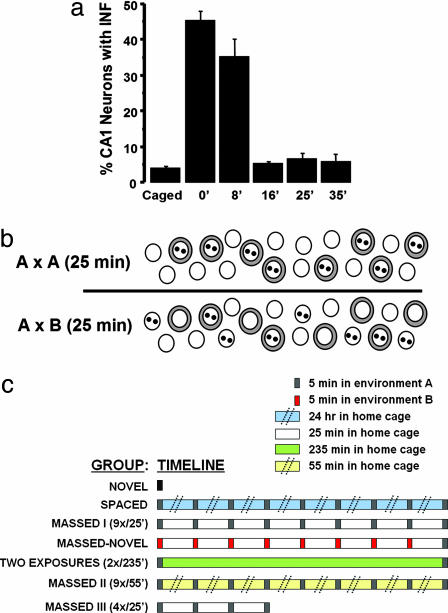
Expression of the IEG Arc (7, 8) is dynamically regulated by natural stimuli in hippocampus (9–12) and is critical for the maintenance of long-term potentiation and memory consolidation (13). Within ≈2 min of stimulation, nascent Arc RNA can be detected at the genomic sites of transcription by using high-sensitivity FISH (14, 15). These sites of Arc transcription appear as one or two intensely staining intranuclear foci (INF). The period of active transcription is brief, ending between 8 and 16 min of its initiation (ref. 16, see Fig. 1a). The processed Arc mRNA subsequently accumulates in the cytoplasm, where it can be detected ≈20-45 min after induction (14, 15). Furthermore, a second round of Arc transcription can be initiated in the same neurons when two experiences are separated by as little as 20 min (refs. 14 and 17, see Fig. 1b). These findings provide for three important advances. First, Arc INF provide a discrete marker of a temporally defined molecular event, a “snapshot” of neurons actively transcribing Arc. The rapid and transient appearance of these INF provides a specific assay for transcriptional activation in response to the behavioral epoch immediately preceding death, and is not contaminated by mRNA that accumulates in the cytoplasm from prior activity (see ref. 16). Thus, detection of Arc INF greatly increases the utility and sensitivity of in situ methods to analyze transcriptional processes per se, as compared to conventional IEG detection methodologies that measure steady-state mRNA levels. Second, detection of IEG INF provides readout of transcription in individual neurons that can be distinguished phenotypically with double or triple label FISH, making it distinct from population biochemical methods used to measure transcription such as nuclear run-off and chromatin immunoprecipitation. Finally, the subcellular localization of the Arc RNA signal can be used to detect and compare neuronal ensembles activated during two discrete behavioral experiences, and allows a unique approach to brain network mapping termed cellular compartment analysis of temporal activity by FISH (catFISH) (14).
Fig. 1.
Nuclear Arc FISH signal provides a temporally precise readout for recent transcriptional activity. (a) Time course of Arc transcription in CA1 neurons after a 5-min exposure to a novel environment. The time points given indicate time after removal from the environment and have been published (16). Note that the proportion of CA1 neurons with Arc INF returns to baseline levels within 16 min after removal from the environment. (b) Activation of Arc transcription in CA1 neuronal ensembles is environmental context specific. Nuclei are indicated by open circles; Arc transcription foci (INF) are indicated by two dark spots in the nuclei; cytoplasmic Arc mRNA is indicated by gray shading around nuclei. In rats exposed to the same environment twice, with each exposure separated by 25 min (A × A group; Upper), the majority of Arc positive cells contain both cytoplasmic Arc signal and INF, indicating activation of Arc transcription in the same cell population during each exposure. By contrast, in rats exposed sequentially to two different environments (A × B group; Lower), a more heterogeneous pattern of Arc staining was seen, with roughly equal proportions of cells containing only Arc cytoplasmic staining or Arc INF staining, and a smaller proportion containing both Arc cytoplasmic and INF staining. This observation indicated that the different environments activated statistically independent populations of neurons. This diagram is based on data from ref. 14. (c) Behavioral timeline for the experimental groups of experiments 1 and 2. The diagram approximates the timing of the handling of the groups on a linear scale, with the exception of the “Spaced” and “Massed-II” groups where the double hatch markings indicate longer periods between the environment exposures. [Image in a reproduced with permission from ref. 16 (Copyright 2002, Society for Neuroscience).]
Previously, the specificity of the Arc transcriptional response in CA1 neurons was investigated by using catFISH (14, 15). In rats exposed sequentially to the same environment twice, Arc mRNA transcriptional induction for each experience occurred predominantly in a single population of neurons. By contrast, in rats exposed sequentially to two different environments, Arc transcription occurred in statistically independent neural populations (see Fig. 1b). These findings are qualitatively and quantitatively comparable to results obtained from parallel cell recording experiments of rats in similar behavioral conditions (18–20), suggesting that the firing activity associated with expression of a “place field” is sufficient to induce Arc transcription in CA1 neurons.
The objective of the current study was to determine whether the coupling between neural activity and Arc transcription is static or plastic. This question was addressed by presenting rats to the same environmental context repeatedly over multiple days or repeatedly within a single day and determining the effect on Arc transcription and neural activity in CA1 neurons, using FISH and parallel cell recording methods, respectively. We find that, although the relationship between behavioral exploration and CA1 neural activity remains constant, the relationship between behavior and Arc transcription is highly plastic for repeated behavioral repetitions given within a single day. These studies indicate that the coupling between neural activity and Arc transcription is plastic and dependent on behavioral history, and identify a form of metaplasticity (21) that could impact memory consolidation processes.
Results
Similar Proportion of Arc Expressing CA1 Neurons with Repeated Daily Exposures to an Environmental Context: Activation of Arc Transcription Does Not Require Novel Experience. Past studies have shown that there is a brief period of active Arc transcription in CA1 neurons after exposure of rats to a novel context (≈10 min; ref. 16; see Fig. 1a), and that Arc catFISH can be used to map hippocampal networks activated by the same or two different environments (ref. 14, see Fig. 1b). The current studies investigated the influence of repeated neural activity on Arc transcription in CA1 neurons by exposing rats to the same environment, either over multiple days or on a single day (Fig. 1c). Each exposure lasted 5 min. The groups for experiment 1 were as follows: CAGED, rats were killed directly from their home cages; NOVEL, rats were exposed to environment A for the first time; SPACED, rats were exposed to environment A once a day for 9 consecutive days; MASSED-I, rats were exposed to environment A for nine times in a single day, with each exposure separated by 25 min; MASSED–NOVEL, rats were exposed to environment B for eight times in a single day, with each exposure separated by 25 min, and then given a final exposure to a novel environment A. The rats were killed immediately after their respective treatment, and active transcription of Arc was detected as discrete foci of in situ hybridization signal within the nuclei of CA1 neurons (14, 16).
ANOVA revealed a significant overall effect of experimental condition (F4,19 = 58.04, P < 0.0001). In agreement with past studies (14, 16), ≈40% of CA1 neurons contained Arc INF in the NOVEL group, whereas CAGED control rats showed only ≈5% of CA1 neurons with Arc INF (Fig. 2f; CAGED vs. NOVEL, P < 0.0001). Similarly, the SPACED group showed ≈40% of CA1 neurons with Arc INF (Fig. 2f; SPACED vs. NOVEL, P = 0.56; SPACED vs. CAGED, P < 0.0001).
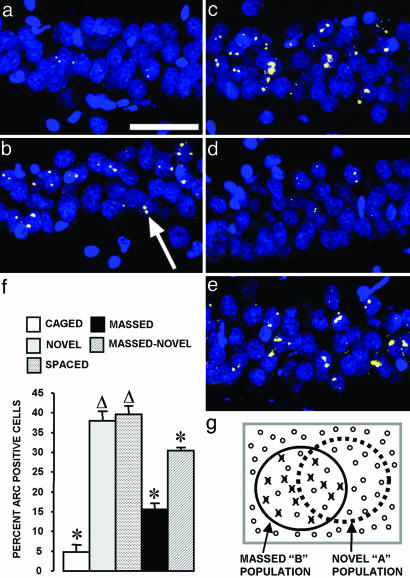
Fig. 2.
Nine exposures to an environmental context separated by 25 min, but not 24 h, decrease the proportion of Arc+ CA1 neurons. Confocal projection images from the CA1 region for each experimental condition: CAGED group (a); NOVEL group (b); SPACED group (c); MASSED-I group (d); MASSED–NOVEL group (e). The arrow in b indicates a cell containing two Arc transcription foci (yellow color) within a cell nucleus (blue color). (Scale bar, 50 μm.) (f) The percentage of Arc+ CA1 neurons (mean ± SEM) for the five behavioral groups is indicated. *, Different from all other behavioral groups (P < 0.02). Δ, different from CAGED, MASSED, and MASSED–NOVEL groups (P < 0.02). n = 4–6 rats per group. (g) Illustration explaining results from the MASSED–NOVEL group. Repeated exposure to environment B decreases the probability of Arc transcription by ≈63% in the “MASSED-B” cell population, these refractory cells are indicated with an “X” over a closed circle. The open circles indicate cells capable of Arc transcription. Based on the principle of random selection with replacement, 16% of the 40% of CA1 cells active in environment A would have also been active in environment B. At 63% failure rate, however, only 6% of the “AB overlap” population would be capable of activating Arc transcription. Therefore, the percentage of cells activating Arc transcription in the final “NOVEL-A” exposure is predicted to be 30% (24% + 6%), essentially identical to the observed value for the MASSED–NOVEL group.
Massed Exposure to a Single Environment Inhibits the Arc Transcriptional Response in a Single Population of CA1 Neurons. In contrast to the NOVEL and SPACED groups, 15% of CA1 neurons from the MASSED-I group contained Arc INF (Fig. 2f). This value was less than both the NOVEL and SPACED (P < 0.001) groups, but still greater than that of the CAGED control group (P < 0.001). In rats from the MASSED–NOVEL group, 30% of CA1 neurons were Arc INF+ (Fig. 2f). This value was different from all other groups (P < 0.02) and is quantitatively predicted if the neural population active in environment A is selected by a nonsubtractive stochastic process, as evidenced in electrophysiological studies of place cells, that includes 16% of neurons that were previously active in environment B and 24% from non-B. Because the repeated exposure reduces the percentage of Arc INF+ neurons from 40% to 15% (15 of 40 = 0.375), the contribution of Arc INF from the B representation is predicted as 16% × 0.375 = 6%. Thus, the predicted sum of Arc INF is 24% + 6% = 30%. This close correspondence to the observed value suggests that the reduction of Arc transcriptional responsiveness was limited to the cell population repeatedly activated in environment B (Fig. 2g).
As Few as Four Context Exposures, Each Separated by 25 min, Is Sufficient to Inhibit Arc Transcriptional Activation. In Experiment 2, the effect of repeated behavioral experience on Arc transcription was explored by altering two parameters of the MASSED-I exposure paradigm: the number of exposures and time interval between exposures (intersession interval). In these experiments, we also used a refined method to detect Arc INF. Whereas the FISH for experiment 1 used a full-length Arc cDNA probe, thus necessitating the use of confocal image stacks to differentiate primary transcript at INF from processed mRNA that may be present in both cytoplasm and nucleus, FISH for experiment 2 was performed by using an Arc intron enriched probe. As this probe hybridizes efficiently only to primary transcript that is present exclusively in the nucleus, and not the processed mRNA, the use of confocal stacks was not necessary and analysis could be performed with single-plane wide field images. As seen for the CAGED, NOVEL, and MASSED-I groups, this more rapid analysis produced values consistent with those from experiment 1 for the same groups, thus validating this more rapid analysis (compare Figs. 2f and 3g).
Fig. 3.
Four context exposures, separated by 25 min, are sufficient to inhibit Arc transcriptional activation. Confocal projection images of Arc intronic RNA signal (yellow color) and cell nuclei (blue color) from the CA1 region for each experimental condition: CAGED controls (a); NOVEL group (b); MASSED-I group (c); MASSED-II group (d); MASSED-III group (e); and TWO EXPOSURES group (f). (g) The number of Arc+ CA1 neurons (mean ± SEM) per field for each behavioral group. Overall ANOVA indicated a significant difference among the groups (F5,214 = 10.99, P < 0.0001). *, Different from caged, MASSED-I, MASSED-II, and MASSED-III groups (P < 0.01). (h) The comparison of average integrated fluorescent intensities per Arc+ cell did not reveal a significant difference among the various behavioral groups. n = 5 rats per group.
To determine whether the decrease of Arc transcriptional responsiveness seen in the MASSED-I group was due to the repeated exposures to environment A, or merely due to the first exposure 4 h before the final (ninth) exposure, one group of rats was exposed to environment A twice, with an intersession interval (ISI) of 4 h (Fig. 3; TWO EXPOSURES-2×/240′). The number of Arc+ CA1 neurons per field was similar to the NOVEL group (P = 0.37) and different from that of the MASSED-I group (P < 0.01), showing that the reduction of Arc transcription in the MASSED-I group cannot be attributed simply to the interval between the first and last experience. We also addressed whether an increased ISI might allow “reset” of the Arc transcriptional response by exposing one group to environment A nine times, but with an ISI of 55 min (MASSED-II group), instead of 25 min as for the MASSED-I group. The number of Arc+ cells per field was similar to that of the MASSED-I group, and different from both the NOVEL and TWO EXPOSURES groups (P < 0.01), indicating that separating the exposures by an hour was not sufficient to reset transcriptional competence in CA1 cells.
We next examined the importance of the number of behavioral repetitions on the reduction of Arc transcription. Two exposures to the same environment, separated by ≈25 min, results in a robust activation of Arc transcription during both exposures (14, 17), whereas nine repetitions, at intervals of either 25 or 55 min, leads to a dramatic loss of responsiveness in a single population of CA1 neurons. Accordingly, we tested whether four 5-min exposures to environment A, separated by 25 min (MASSED-III group), would modify Arc transcriptional competence of CA1 neurons. The MASSED-III group exhibited a similar number of Arc+ cells as the MASSED-I and MASSED-II groups (P ≥ 0.29), which was different from that of both the NOVEL and TWO EXPOSURES groups (P < 0.0005). Thus, four exposures produced the same degree of reduction of Arc transcription as nine exposures.
Because the previous analyses were threshold based (cells were either positive or negative, as determined subjectively for Fig. 2f, or by a chimeric subjective/objective approach for Fig. 3g), we asked whether any of the behavioral conditions from experiment 2 led to a change in the amount of primary transcript detected per positive cell. Analysis of integrated fluorescent intensity per Arc+ cell failed to show a significant effect of behavioral condition (Fig. 3h; F5,24 = 1.78, P = 0.15). These findings suggest that the loss of Arc transcriptional responses occurs with as few as four repetitions in an all-or-none fashion, and is not the result of a graded decrement in amount of primary transcript.
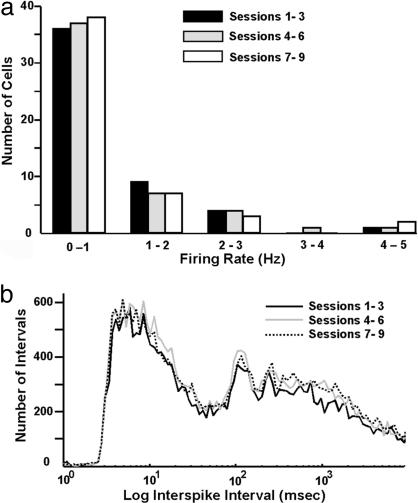
Massed Exposure to a Single Environment Does Not Alter Firing Properties of CA1 Neurons Across Repetitions. To assess whether the marked reduction of Arc transcription detected in the MASSED groups might be the result of altered neuronal firing (i.e., habituation), rats implanted with “hyperdrives” were exposed to the MASSED-I exposure protocol. Electrical activity recorded from CA1 during the exposures was grouped into blocks of three sessions (sessions 1–3, 4–6, and 7–9) and binned according to firing rate (Fig. 4a). A square-root transformation of the data were performed to satisfy the assumptions of normality and constant variance. A two-way ANOVA of these transformed data revealed no significant effect of session on firing rate (F2,149 = 0.66, P > 0.05). Log-scaled, interspike interval distributions for the session blocks 1–3, 4–6, and 7–9 (Fig. 4b) also did not reveal a significant effect of massed trials on interspike intervals, indicating that there was no change in the cells' bursting characteristics. Thus, neuronal firing properties did not change over the course of nine exposures to environment A in a single day.
Fig. 4.
Firing properties of CA1 neurons do not change with repeated exposures to an environmental context within a single day. The response properties of multiple single CA1 neurons from rats repeatedly exposed to the same environment within a single day were determined by using parallel extracellular recording methods. (a) Cell firing data from 50 cells (from three rats) were grouped into three blocks (sessions 1–3, 4–6, and 7–9) and binned according to firing rate. (b) Log scaled, interspike interval distribution for the three session blocks. Note that neither the mean firing rate (a) nor the interspike interval (b) changed over the course of the repeated exposures. The number of cells recorded from each of the three rats was 9, 14, and 27 pyramidal cells, and these electrophysiological data were pooled for subsequent analyses.
Discussion
The present findings provide critical insights into the relationship between hippocampal neural activity and Arc transcriptional regulation. Specifically, we demonstrate that the coupling between cell firing and Arc transcription is plastic, not static, because it is influenced strongly by recent behavioral history. With quiescent rats, as in the NOVEL and SPACED groups and in earlier studies (14, 17, 22), the correspondence between Arc transcription and single unit neural activity is high. For example, the percentage of Arc+ CA1 neurons in the NOVEL and SPACED groups was 38.6 ± 1.6% (n = 10 rats; Fig. 2f). This value is indistinguishable from the percentage of complex spiking CA1 neurons that have a place field in an environment of similar size and complexity (35.6 ± 4.8%; n = 3 rats; ref. 20). The notion that Arc transcription is driven by the neural activity associated with the expression of a place field is supported further by the finding that Arc gene expression qualitatively mirrors that of place cells when rats are introduced to the same environment twice or two distinct environments (refs. 14 and 17, see Fig. 1). However, in rats exposed repeatedly to an environment within a single day (MASSED groups), the proportion of Arc+ CA1 neurons decreased dramatically as compared to the NOVEL or SPACED groups. Electrophysiological recordings demonstrated that the firing properties of CA1 neurons did not change across repeated sessions within a single day. In combination, these data indicate that the association between neural activity and Arc transcription (which we term “electrotranscriptional coupling” or ETC) is highly dynamic and linked to behavioral history.
The finding that the proportions of Arc+ CA1 neurons are similar in the NOVEL and SPACED groups (Fig. 2) demonstrates that novelty is not required for Arc transcriptional activation. Therefore, signaling pathways regulating Arc transcription in CA1 neurons do not appear to discriminate between cellular activity generated by exposure to new information as compared to that of familiar information. In close correspondence, activity patterns recorded from hippocampal neurons are not overtly different in familiar and novel environments (23, 24). Moreover, “place fields” of hippocampal neurons appear rapidly in response to an initial exposure to an environment and persist on subsequent visits (19, 20, 25). However, it is possible that Arc protein expression could be differentially regulated at a post-transcriptional level under novel or familiar conditions (26). For example, the local translation of dendritically targeted Arc mRNA (27) might be more efficient under conditions of novelty or arousal, through engagement of neuromodulatory systems (28, 29). In this view, induction of Arc transcription would be “permissive,” deferring control of functional Arc protein expression to defined synapses within the cell. Recent data indicating that Arc mRNA is subject to both positive (30) and negative translational control (31) is consistent with this notion.
The findings from the MASSED–NOVEL group are particularly instructive in understanding the mechanism of Arc ETC plasticity. As described in Results and Fig. 2g, the data from this group demonstrate that the altered association between cell firing and Arc transcription with repeated exposures was cell and experience specific, and not a generalized inhibition of Arc transcription in all CA1 neurons. Additionally, the sparsity of coding within the hippocampal network predicts that the “overlap” in CA1 neurons engaged by both environments A and B would have distinct synaptic inputs driving activity for each environment. If the failure to induce Arc transcription were manifest at the synaptic level, the NOVEL, SPACED, and MASSED–NOVEL groups would be predicted to all exhibit similar levels of Arc+ neurons, but this was not seen (Fig. 2f). Thus, the current findings indicate that the decreased ETC of the MASSED groups is a cell specific and cell-wide phenomenon, and not synapse specific. This serves to restrict possible molecular mechanisms that could underlie this form of plasticity. Two possible candidates include activation/stabilization of nuclear phosphatases and induction of transcriptional repressors, such as ICER (32); either event would be predicted to block IEG transcription.
What is the relevance of ETC plasticity to learning and memory? Interestingly, retention of spatial and contextual memories in rats is facilitated by training over multiple days as compared to administering the same amount of training within a single day, at intervals similar to those used here (33–35). The present findings suggest a possible molecular basis for this difference: massed training degrades the fidelity of coupling between neural activity and IEG expression, which is required for memory consolidation (13). Alternatively, modification of ETC by repeated activation of a neuron may increase capacity for, and decrease interference between, the encoding of multiple distinct events. In this view, ETC plasticity would aid in the separation of neural representations of different experiences by suppressing synaptic plasticity of connections onto neurons that were recently involved in encoding a given experience. Although these two possibilities might appear to be at odds, they reflect learning in relatively artificial (multiple trials of the same task within a day in the lab) versus more naturalistic (processing multiple distinct events throughout the day) conditions.
Molecular and cellular studies of effector IEGs such as Arc, Homer, CPG2, and Narp, which target the excitatory synapse, are providing insights into the mechanisms of synaptic modification (4, 36). Moreover, IEG transcription factors such as zif268 are now recognized to be required for late-phase synaptic plasticity (37). Thus, there is emerging support for the notion that IEGs, working in an orchestrated fashion with each other and existing synaptic proteins, are essential for protein synthesis-dependent synaptic plasticity (3). If individual IEGs exhibit distinct properties of ETC plasticity, their differential presence in neurons could create a remarkable combinatorial complexity of molecules that influence the ability of neurons to modify synaptic connections. The current findings provide insight into a form of neural metaplasticity that provides a framework for understanding mechanisms of behavioral plasticity and cognition.
Materials and Methods
Behavioral Handling Procedures: Arc FISH Experiments. For experiment 1, 24 9-month-old male F344 rats (Harlan Sprague–Dawley) were handled before the experiment to familiarize them to the experimenter and to the handling procedures in general. Environment A was a square platform surrounded by 10-cm-high walls. Environment B was a rectangular platform without walls, elevated 10 cm above a supporting table, and was located in an adjacent room. Lighting intensity and access to distal cues, unique to each room, was similar in the two environments. Each of the 3,600-cm2 environments was divided into nine 400-cm2 grids. During each 5-min exploration session, the rat was picked up, wherever it was, and released into the center of a different grid square every 15 s on a semirandom schedule. By the end of an exploration session, the rats were placed in each of the nine grid squares at least twice. This procedure ensured that the rats sampled the entire environment during each exploration session. For experiment 2, the exact same apparatus and handling procedure were used, but subjects were 30 3-month old male Sprague–Dawley rats (Harlan Sprague–Dawley). The intervals between exploration sessions and death for the different behavioral groups are described in Results and shown graphically in Fig. 1c.
All animal handling and surgical procedures were in accord with National Institutes of Health guidelines and approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Arizona and at the University of New Mexico.
FISH. The rats were killed by decapitation, and brains were processed for FISH as described in detail (14, 16, 38). Briefly, digoxigenin-labeled antisense cRNA riboprobes were hybridized on tissue sections overnight at 56°C and detected with the use of a commercial cyanine-3 (CY3) tyramide signal amplification kit (DirectFISH; PerkinElmer Life Sciences). Sections were counterstained with either YOYO-1 (experiment 1; Molecular Probes) or DAPI (experiment 2; Molecular Probes). For experiment 1, the Arc riboprobe was generated from a full-length cDNA (7); for experiment 2, FISH was performed by using an Arc intron-enriched riboprobe (see below). As reported (14), slides run with the Arc sense riboprobe failed to produce any specific signal (data not shown).
Primers spanning introns 1 and 2 (and the small exon 2) of the rat Arc gene were selected by using commercial software (vector nti). The exact sequence of the primers is as follows, with the base pair designations matching those of GenBank accession no NM_019361: forward, 5′-GCCAGTCTTGGGCAGCATAGCT-3′ (bp 1923–1944); reverse, 5′-TCAGCTCTGAGGCTGAGCTG-3′ (bp 2331–2312).
PCR was performed with these primers and Sprague–Dawley rat genomic DNA using the Expand High-Fidelity PCR System (Roche Diagnostics). The gel-purified PCR fragment was cloned by using the PCR-Script Amp Cloning kit (Stratagene) and the identity of the insert was confirmed by bidirectional sequencing of the resulting clones, followed by blast alignment to the rat genome.
Microscopy, Image Acquisition, and Image Analysis. For experiment 1, confocal image stacks from CA1 (anteroposterior, ≈-3.6 mm from bregma) were collected at a Z frequency of 1 μm with a Leica TCS-4D confocal microscope using either a ×100 oil or a ×20 objective. Laser and PMT settings were optimized for the detection of Arc INF for each slide, but kept constant for all images acquired within a slide. Manual cell counts were performed as detailed (16, 17) by an experimenter blind to the behavioral conditions. The designation Arc+ was given to nuclei that contained Arc INF. Three slides were analyzed for each rat, and the average number of cells analyzed was 163 per rat. The values are reported as the percentage of Arc+ cells per total counted cells for each rat.
For experiment 2, images of the CA1 region (anteroposterior, approximately – 3.6 mm from bregma) were acquired with a Nikon TE2000U epifluorescence microscope and ×10 objective, and digitized by using a CoolSNAP-Hq charge-coupled device camera (Roper Scientific). Wide-field images of DAPI (cell nuclei) and CY3 (Arc transcription foci) were acquired and combined by using metamorph software (Universal Imaging) for three fields per slide, and three slides per rat. A single optimized acquisition exposure time was used for all images acquired from a particular slide. In the offline analysis, a chimeric subjective/quantitative approach was used. First, nuclei containing any level of visible Arc signal were traced and information (integrated, average, maximum, and minimum intensities) of Arc signal was exported to excel. Then, cell nuclei were automatically defined as Arc INF positive or negative based on input threshold values for integrated and maximum single intensities, which were applied to all images from a given slide. The number of Arc+ nuclei was then compiled for the CA1 region of each rat.
Hyperdrive Implantation Surgeries and Electrophysiological Recording. Three 12-month-old male F344 retired breeder rats (Harlan Sprague–Dawley) were used for single unit recording. The rats were housed in Plexiglas guinea pig tubs, placed on a reverse 12-h/12-h light/dark cycle (lights off at 10 a.m.), and given access to food and water ad libitum. The rats underwent surgical implantation of an adjustable, 48-channel, 12-microelectrode array (Hyperdrive) as detailed in previous studies (20). After recovery from surgery, the electrodes of the Hyperdrive were gradually lowered to the CA1 region of the hippocampus. Twelve of the tetrode probes were used for recording unit activity. One additional probe was placed in the corpus callosum as a reference for differential recordings, and another was used to acquire electroencephalogram (EEG) activity near the hippocampal fissure (optimized to record the ≈7-Hz theta rhythm). Neuronal output was passed through a unit gain field effect transistor and then to a computer-interfaced Cheetah amplifier system (Neuralynx, Tucson, AZ). Single unit responses were amplified between 500 and 5,000 times and bandpass filtered between 600 Hz and 6 KHz. EEG activity was acquired at a gain of 2,000, with bandpass filters set between 5 and 500 Hz. Proximity to the CA1 region was verified by the presence of sharp wave-ripples/complexes within the EEG during slow-wave sleep for each probe. Once in the layer, electrodes were optimized for maximum cell counts. Recording sessions were conducted as the rats were given the MASSED-I behavioral protocol as described for the Arc studies.
After the recording sessions were completed, cells were separated by using a multidimensional “cluster-cutting” technique (20, 39). This method uses the different parameter characteristics of the individual spikes on each of the four separate channels of the tetrode. Units were classified as pyramidal cells and included in the study if they had a spike width of at least 300 μs, a mean firing rate <5 Hz during behavior, and was recorded on the same tetrode as other complex spiking cells.
Statistical Analyses. The main effect of behavioral treatment was determined by one-way ANOVA. When the main effect was significant at the α < 0.05 level, further comparisons between groups were conducted with Fisher's probably least-squares difference post hoc tests (statview software).
Acknowledgments
We thank J. Bohanick, K. Bohne, K. Marjon, M. Papapavlou, and K. Olson for technical assistance. We also thank Dr. Gail Lewandowski for the generation of software to assist in the data reduction and analysis for experiment 2. This work was supported by National Institutes of Health Grants MH060123 (to J.F.G.), AG023309 (to C.A.B., B.L.M., P.F.W., and J.F.G.), AG09219 (to C.A.B. and P.F.W.), and MH46823 (to B.L.M.).
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Davis, H. & Squire, L. R. (1984) Psychol. Bull. 96, 518–559. [PubMed] [Google Scholar]
- 2.Clayton, D. F. (2000) Neurobiol. Learn. Mem. 74, 185–216. [DOI] [PubMed] [Google Scholar]
- 3.Guzowski, J. F. (2002) Hippocampus 12, 86–104. [DOI] [PubMed] [Google Scholar]
- 4.Lanahan, A. & Worley, P. (1998) Neurobiol. Learn. Mem. 70, 37–43. [DOI] [PubMed] [Google Scholar]
- 5.Tischmeyer, W. & Grimm, R. (1999) Cell. Mol. Life Sci. 55, 564–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole, A. J., Saffen, D. W., Baraban, J. M. & Worley, P. F. (1989) Nature 340, 474–476. [DOI] [PubMed] [Google Scholar]
- 7.Lyford, G. L., Yamagata, K., Kaufmann, W. E., Barnes, C. A., Sanders, L. K., Copeland, N. G., Gilbert, D. J., Jenkins, N. A., Lanahan, A. A. & Worley, P. F. (1995) Neuron 14, 433–445. [DOI] [PubMed] [Google Scholar]
- 8.Link, W., Konietsko, U., Kauselmann, G., Krug, M., Schwanke, B., Frey, U. & Kuhl, D. (1995) Proc. Natl. Acad. Sci. USA 92, 5734–5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guzowski, J. F., Setlow, B., Wagner, E. K. & McGaugh, J. L. (2001) J. Neurosci. 21, 5089–5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelly, M. P. & Deadwyler, S. A. (2002) Neuroscience 110, 617–626. [DOI] [PubMed] [Google Scholar]
- 11.Montag-Sallaz, M., Welzl, H., Kuhl, D., Montag, D. & Schachner, M. (1999) J. Neurobiol. 38, 234–246. [PubMed] [Google Scholar]
- 12.Pinaud, R., Penner, M. R., Robertson, H. A. & Currie, R. W. (2001) Brain Res. Mol. Brain Res. 91, 50–56. [DOI] [PubMed] [Google Scholar]
- 13.Guzowski, J. F., Lyford, G. L., Stevenson, G. D., Houston, F. P., McGaugh, J. L., Worley, P. F. & Barnes, C. A. (2000) J. Neurosci. 20, 3993–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guzowski, J. F., McNaughton, B. L., Barnes, C. A. & Worley, P. F. (1999) Nat. Neurosci. 2, 1120–1124. [DOI] [PubMed] [Google Scholar]
- 15.Guzowski, J. F., McNaughton, B. L., Barnes, C. A. & Worley, P. F. (2001) Curr. Opin. Neurobiol. 11, 579–584. [DOI] [PubMed] [Google Scholar]
- 16.Vazdarjanova, A., McNaughton, B. L., Barnes, C. A., Worley, P. F. & Guzowski, J. F. (2002) J. Neurosci. 22, 10067–10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vazdarjanova, A. & Guzowski, J. F. (2004) J. Neurosci. 24, 6489–6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kubie, J. L. & Ranck, J. B. J. (1983) in Neurobiology of the Hippocampus, ed. Seifert, W. (Academic, New York), pp. 433–447.
- 19.Thompson, L. T. & Best, P. J. (1990) Brain Res. 509, 299–308. [DOI] [PubMed] [Google Scholar]
- 20.Wilson, M. A. & McNaughton, B. L. (1993) Science 261, 1055–1058. [DOI] [PubMed] [Google Scholar]
- 21.Abraham, W. C. & Bear, M. F. (1996) Trends Neurosci. 19, 126–130. [DOI] [PubMed] [Google Scholar]
- 22.Guzowski, J. F., Knierim, J. J. & Moser, E. I. (2004) Neuron 44, 581–584. [DOI] [PubMed] [Google Scholar]
- 23.Muller, R. U. & Kubie, J. L. (1987) J. Neurosci. 7, 1951–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Keefe, J. (1976) Exp. Neurol. 51, 78–109. [DOI] [PubMed] [Google Scholar]
- 25.Hill, A. J. (1978) Exp. Neurol. 62, 282–297. [DOI] [PubMed] [Google Scholar]
- 26.Kelly, M. P. & Deadwyler, S. A. (2003) J. Neurosci. 23, 6443–6451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steward, O. & Worley, P. F. (2001) Neuron 30, 227–240. [DOI] [PubMed] [Google Scholar]
- 28.McGaugh, J. L. (2000) Science 287, 248–251. [DOI] [PubMed] [Google Scholar]
- 29.McIntyre, C. K., Miyashita, T., Setlow, B., Marjon, K. D., Steward, O., Guzowski, J. F. & McGaugh, J. L. (2005) Proc. Natl. Acad. Sci. USA 102, 10718–10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin, Y., Edelman, G. M. & Vanderklish, P. W. (2002) Proc. Natl. Acad. Sci. USA 99, 2368–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zalfa, F., Giorgi, M., Primerano, B., Moro, A., Di Penta, A., Reis, S., Oostra, B. & Bagni, C. (2003) Cell 112, 317–327. [DOI] [PubMed] [Google Scholar]
- 32.De Cesare, D. & Sassone-Corsi, P. (2000) Prog. Nucleic Acid Res. Mol. Biol. 64, 343–369. [DOI] [PubMed] [Google Scholar]
- 33.Commins, S., Cunningham, L., Harvey, D. & Walsh, D. (2003) Behav. Brain Res. 139, 215–223. [DOI] [PubMed] [Google Scholar]
- 34.Morris, R. G. M. & Doyle, J. (1985) in Electrical Activity of the Archicortex, eds. Buzsaki, G. & Vanderwolf, C. H. (Akademiai Kiado, Budapest).
- 35.Spreng, M., Rossier, J. & Schenk, F. (2002) Behav. Brain Res. 128, 103–108. [DOI] [PubMed] [Google Scholar]
- 36.Cottrell, J. R., Borok, E., Horvath, T. L. & Nedivi, E. (2004) Neuron 44, 677–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones, M. W., Errington, M. L., French, P. J., Fine, A., Bliss, T. V., Garel, S., Charnay, P., Bozon, B., Laroche, S. & Davis, S. (2001) Nat. Neurosci. 4, 289–296. [DOI] [PubMed] [Google Scholar]
- 38.Guzowski, J. F. & Worley, P. F. (2001) in Current Protocols in Neuroscience (Wiley, New York), Vol. 1, pp. 1.8.1–1.8.16. [DOI] [PubMed] [Google Scholar]
- 39.McNaughton, B. L., Barnes, C. A., Meltzer, J. & Sutherland, R. J. (1989) Exp. Brain Res. 76, 485–496. [DOI] [PubMed] [Google Scholar]