Abstract
The binding of Ca2+ to the 70–80 loop of protein C inhibits protein C activation by thrombin in the absence of thrombomodulin (TM), but the metal ion is required for activation in the presence of TM. Structural data suggests that the 70–80 loop is located between two antiparallel β strands comprised of residues 64–69 and 81–91 on the protease domain of protein C. To test the hypothesis that a salt-bridge/hydrogen bond interaction between Arg-67 of the former strand and Asp-82 of the latter strand modulates the unique Ca2+-binding properties of protein C, we engineered a disulfide bond between the two strands by substituting both Arg-67 and Asp-82 with Cys residues. The activation of this mutant was enhanced 40- to 50-fold independent of TM and Ca2+. Furthermore, the Arg-67 to Ala mutant of protein C was activated in the absence of TM by the Arg-35 to Glu mutant of thrombin with the same efficiency as wild-type protein C by wild-type thrombin–TM complex. These results suggest that TM functions by alleviating the Ca2+-dependent inhibitory interactions of Arg-67 of protein C and Arg-35 of thrombin.
Protein C is a multidomain vitamin K-dependent plasma serine protease zymogen that, upon activation by thrombin in complex with thrombomodulin (TM), down-regulates the coagulation cascade by inactivating factors Va and VIIIa by limited proteolysis (1–3). The activation of protein C by thrombin is a Ca2+-dependent reaction (4, 5). The metal ion is an obligatory cofactor for activation by thrombin in the presence of TM, but it is inhibitory for activation when TM is absent (4, 5). The Ca2+-binding site responsible for the paradoxical effect of the metal ion has been localized to the 70–80 loop (chymotrypsinogen numbering system; ref. 6) on the protease domain of protein C (7, 8), the same loop that also interacts with Ca2+ in other vitamin K-dependent coagulation proteases and in trypsin (9). TM or the TM fragment containing epidermal growth factor-like domains 4, 5, and 6 (TM456) improves the catalytic efficiency of thrombin toward protein C in the presence of Ca2+ by approximately three orders of magnitude by improving both the Km and kcat of the activation reaction (1). The mechanism by which TM and Ca2+ exert their cofactor function is not well understood. An attractive hypothesis is that the binding of Ca2+ to the 70–80 loop of protein C is associated with a conformational change in the zymogen (10, 11) that is optimal for interaction with thrombin in the presence of TM but inhibitory for interaction in the absence of the cofactor (1, 12). Based on this model of protein C activation, it is thought that the Ca2+-altered conformation of protein C is complementary to the TM-altered conformation of thrombin (1, 12). In support of a TM-induced conformational change in thrombin, in a recent study, we demonstrated that the activation of protein C by Arg-35 to Glu or Ala substitution mutants of thrombin is partially TM- and Ca2+-independent, suggesting that TM modulates the conformation of the 37 loop of thrombin (13). In addition to binding to exosite-1 of thrombin structural, molecular modeling and mutagenesis data have indicated that TM also has a “substrate presentation” function by binding to a basic exosite of protein C (14–16). The basic residues of the TM-interactive site on protein C are clustered on three exposed surface loops (37, 60, and 70–80) (8, 15, 16). The charge-reversal or Ala-scanning mutagenesis of basic residues of these loops have resulted in protein C mutants whose activations by thrombin have been dramatically impaired in the presence, but not in the absence, of TM (15, 17). Although these previous studies have assigned an important role for these basic residues in their interaction with the thrombin–TM complex, nevertheless, the results have not provided any insight into the mechanism by which Ca2+ stimulates protein C activation by the thrombin–TM complex and inhibits its activation by thrombin alone.
Among the basic residues of the protein C exosite that are thought to interact with TM, the contribution of Arg-67 to the zymogenic properties of protein C has not been studied. The reason may partially be due to the observation that the mutagenesis of this residue results in a mutant which, upon activation, exhibits impaired amidolytic and proteolytic activity and that the interpretation of the results of a “loss of function” mutant is not straightforward (17, 18). Examination of the three-dimensional position of Arg-67 in the x-ray crystal structure of activated Gla-domainless protein C (GD-PC) reveals that this residue is located on a β-strand (residues 64–69) that joins the 70–80 loop to an antiparallel β-structure comprised of residues 81–91 on the catalytic domain of the protein (8). The NH1 guanidyl group of Arg-67 is located within a salt-bridge/hydrogen-bonding distance (2.67 Å) from the OD2 carboxylic oxygen of Asp-82 on the second strand. To test the hypothesis that electrostatic interactions between these two residues modulate the unique Ca2+-binding properties of protein C, we have engineered a disulfide bond between Arg-67 and Asp-82 by substituting them with Cys residues. We have characterized this and two additional variants of protein C with Arg-67 and Asp-82 substitutions and propose an explanation for the Ca2+-dependent enhanced activation of protein C by the thrombin–TM complex.
Results
Expression. Both wild-type and the Cys-67–82 mutant of GD-PC were expressed in HEK-293 cells and purified to homogeneity as described in ref. 19. SDS/PAGE analysis under nonreducing conditions (Fig. 1 Left) indicated that both proteins expressed as two or three subforms with identical apparent molecular masses that correspond to α, β, and γ protein C that are glycosylation variants observed with this protein in refs. 10 and 20. Spectrophotometric titrations with 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) indicated that neither wild-type protein C nor Cys-67–82 mutant protein C (up to 10 μM in both native and denatured forms) increased the absorbance of DTNB at 412 nm, suggesting that both proteins lack a free sulfhydryl group (data not shown). These results suggest that a correct disulfide bond has been formed between the two engineered Cys residues in the mutant protein, although a minor band migrating at ≈110–120 kDa was also observed, suggesting that a minor fraction of the mutant has been secreted in the dimeric form. After complete activation, the concentrations of active protein C derivatives were determined by an amidolytic activity assay and active-site titrations with known concentrations of protein C inhibitor as described in ref. 21. These concentrations were within 80–100% of those expected based on zymogen concentrations, as determined from their absorbance at 280 nm. The Cys-67–82 protein C mutant protease reacted normally with protein C inhibitor and exhibited near normal amidolytic activity toward Spectrozyme PCa (normal kcat with only ≈2-fold elevated Km). The elevated Km toward the chromogenic substrate is likely due to the inability of the mutant protease to interact with Ca2+ because the metal ion is known to stimulate the amidolytic activity of activated protein C (APC) (7).
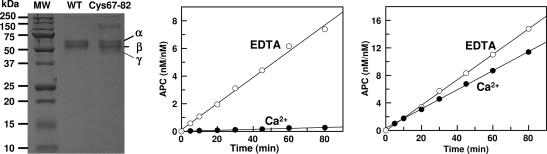
Fig. 1.
SDS/PAGE analysis of wild-type and mutant GD-PC and the time course of the initial rate of zymogen activation by thrombin in the presence of EDTA and Ca2+. (Left) One microgram of each protein was applied to 10% SDS/PAGE under nonreducing conditions. The subforms α (≈55 kDa), β (≈52 kDa), and γ (≈50 kDa) are glycosylation variants of protein C (10). (Center) The time course of activation of wild-type GD-PC (1 μM) by thrombin (10–50 nM) was monitored in TBS/EDTA (○) or TBS/Ca2+ (•) as described in Methods. The activation rates were as follows: 0.003 nM/min in Ca2+ and 0.09 nM/min in EDTA. (Right) The same as above except that the activation of Cys-67–82 protein C by thrombin (10 nM) was monitored in either EDTA (○) or Ca2+ (•). The activation rates were as follows 0.13 nM/min in Ca2+ and 0.18 nM/min in EDTA.
Effect of Ca2+ on Protein C Activation by Thrombin. It is known that Ca2+ functions as a cofactor to stimulate the activation of protein C by thrombin in the presence of TM, but the metal ion is a potent inhibitor when TM is absent (1, 5). As shown in Fig. 1 Center, the initial rate of wild-type GD-PC activation by thrombin is at least 20-fold higher in EDTA than in Ca2+. Unlike wild-type protein C, the activation of the Cys-67–82 protein C mutant by thrombin was insensitive to the presence of Ca2+ in the reaction buffer, because relative to wild-type protein C, the activation of Cys-67–82 protein C by thrombin was similar in either EDTA or Ca2+ (Fig. 1 Right). Comparisons of the activation rates suggested that, relative to wild-type protein C, the activation of Cys-67–82 protein C by thrombin was improved 40- to 50-fold in the presence of Ca2+.
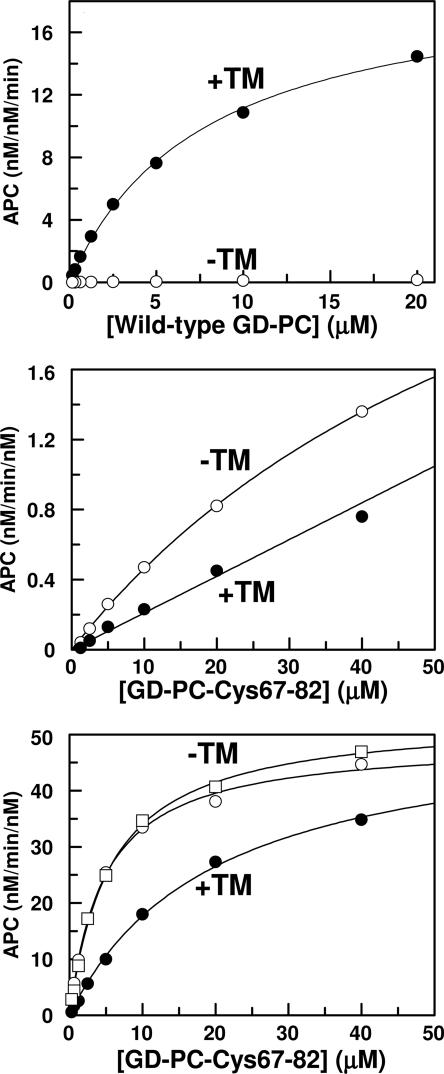
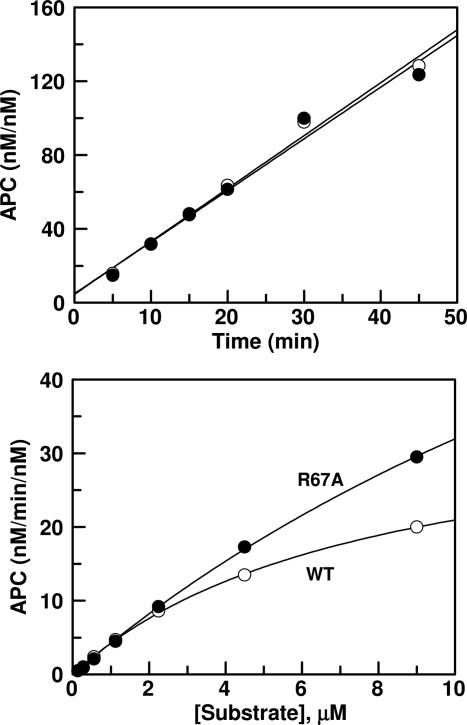
TM is known to enhance the activation of wild-type GD-PC by thrombin by approximately three orders of magnitude in the presence of Ca2+ (1). Consistent with the literature, the substrate concentration dependence of the activations indicated that TM456 dramatically improves the rate of GD-PC activation by thrombin in Ca2+ (Fig. 2 Top). However, the Cys-67–82 protein C mutant was activated by thrombin in the presence of Ca2+, but the rate was not enhanced, rather inhibited, ≈2- to 3-fold in the presence of TM (Fig. 2 Middle). Comparing Fig. 2 Top and Bottom, it can be seen that activation of the mutant by the thrombin–TM456 complex remained ≈20-fold slower than that of wild-type GD-PC. We previously demonstrated that the substitution of Arg-35 of thrombin with Glu (R35E) diminishes the requirement for Ca2+ in protein C activation in the presence of TM and improves the rate of activation ≈25-fold independent of TM (13). To determine whether the 20-fold lower activation rate of the Cys-67–82 protein C mutant is due to the inhibitory effect of Arg-35 of thrombin, the activation of the protein C mutant was also studied by R35E and R35A thrombins in both the absence and presence of TM456. The concentration dependence of the activation studies indicated that the activation of Cys-67–82 protein C by either R35E or R35A thrombins (Fig. 2 Bottom shown for R35E thrombin only) was entirely independent of both TM and Ca2+. The activation of the Cys-67–82 protein C mutant by R35E thrombin in the absence of TM exceeded that of wild-type protein C in the presence of TM (compare Fig. 2 Top and Bottom). TM slightly, ≈2- to 3-fold, impaired the activation rate with R35E thrombin by elevating Km(app) of Cys-67–82 protein C (Table 1). TM impairment of activation of Cys-67–82 protein C with R35A thrombin was even more dramatic because no saturation of the reaction was observed for up to 40 μM Cys-67–82 protein C, the highest concentration of the mutant substrate used in the experiments (Table 1). As described below, Cys-67–82 protein C is not capable of interaction with TM in the activation complex, thus the inhibitory effect of TM456 must be due to the occupancy of exosite-1 of thrombin by the cofactor.
Fig. 2.
Concentration-dependence of GD-PC activation by thrombin. (Top) Concentration dependence of wild-type GD-PC activation by thrombin (1 nM) was monitored in the absence (○) or presence (•) of TM456 in TBS/Ca2+ as described in Methods.(Middle) The same as above except that the activation of Cys-67–82 protein C by thrombin was measured in the absence (○) or presence (•) of TM456. (Bottom) The same as above except that the activation of Cys-67–82 protein C by R35E thrombin was measured in the presence of TM456 in TBS/Ca2+ (•) or without TM456 in TBS/Ca2+ (○) and TBS/EDTA (□). Solid lines are best fit of kinetic data to the Michaelis-Menten equation. The apparent Km and kcat values are presented in Table 1.
Table 1. Kinetic parameters for activation of wild-type and mutant GD-PC by wild-type and Arg-35 mutants of thrombin in the absence and presence of TM456 and Ca2+.
| kcat, nM·min-1 nM-1 | Km, μM | |
|---|---|---|
| Thrombin | ||
| GD-PC, EDTA* | ≈5 ± 1 | ≈70 ± 5 |
| GD-PC, Ca2+† | ND | >70 |
| GD-PC, Ca2+, TM456 | 19.8 ± 0.5 | 7.7 ± 0.4 |
| R35E thrombin | ||
| Cys67-82, EDTA | 59.0 ± 1.1 | 5.6 ± 0.3 |
| Cys67-82, Ca2+ | 54.2 ± 1.0 | 4.7 ± 0.3 |
| Cys67-82, Ca2+, TM456 | 58.3 ± 2.0 | 19.6 ± 1.4 |
| R35A thrombin | ||
| Cys67-82, EDTA | 46.6 ± 3.9 | 22.1 ± 3.7 |
| Cys67-82, Ca2+ | 40.9 ± 1.0 | 16.6 ± 0.9 |
| Cys67-82, Ca2+, TM456‡ | ND | >40 |
The kinetic constants in EDTA are estimated values because 70 μM was the highest concentration of GD-PC used in the experiments.
The kinetic constants in the presence of Ca2+ could not be determined (ND) because the activation rate remained linear for up to 70 μM GD-PC, the highest concentration of substrate used in the experiments.
The kinetic constants could not be determined because the activation rate remained linear for up to 40 μM Cys67-82, the highest concentration of substrate used in the experiments.
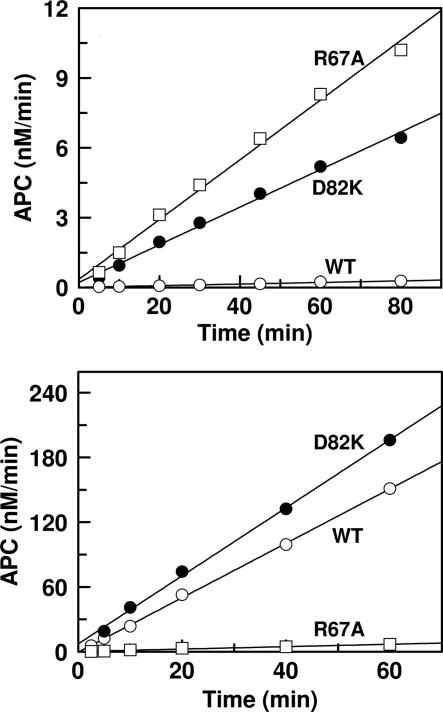
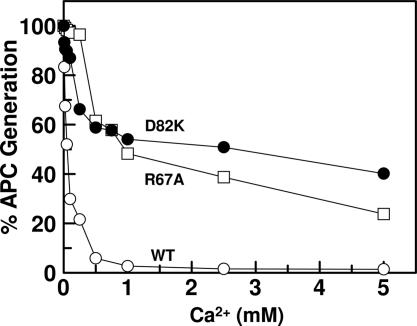
It has been demonstrated that the conformation/environment of the activation peptide of protein C is altered upon Ca2+ binding to the 70–80 loop (7, 10, 11) and that only a Ca2+-stabilized conformer of protein C can dock into the active-site groove of thrombin in the presence of TM (1). Moreover, structural and mutagenesis data have indicated that the interaction of the basic exosite of protein C with the TM4 fragment of the cofactor is required for the zymogen activation by the thrombin–TM complex (14–16). To determine whether the inability of Cys-67–82 protein C to interact with TM and/or Ca2+ is responsible for the lack of a stimulatory effect for TM in the mutant protein C activation by thrombin, two additional mutants of protein C were prepared. In the first one, Arg-67 of the zymogen was replaced with an Ala (R67A protein C) and, in the other, Asp-82 was replaced with a Lys (D82K protein C). Initial rate studies in the absence of TM indicated that activation of both R67A and D82K protein C mutants by thrombin was dramatically enhanced in the presence of Ca2+ (Fig. 3 Upper). On the other hand, although the activation of D82K protein C by thrombin was stimulated in the presence of TM, the activation of R67A protein C by thrombin was not influenced by the cofactor (Fig. 3 Lower). Thus, similar to Cys-67–82 protein C, the activation of R67A protein C by the thrombin–TM456 complex remained ≈20-fold slower than that of wild-type protein C. The thrombin–TM456 complex activated the D82K protein C mutant with apparent Km and kcat values of 3.7 ± 0.7 μM and 10.9 ± 1.0 nM·min–1·nM–1, respectively. These values are similar to the corresponding values for wild-type protein C presented in Table 1. The initial rate studies in the presence of increasing concentrations of Ca2+ also suggested that both R67A and D82K protein C mutants could interact with Ca2+, although with ≈5- to 20-fold lower affinities. In contrast to an apparent Kd of 50 ± 5 μM for interaction of wild-type protein C with Ca2+, the corresponding value was elevated to 935 ± 135 μM for R67A protein C and 230 ± 40 μM for D82K protein C (Fig. 4). Thus, the improvements in the activation of both R67A and D82K protein C mutants by thrombin in the absence of TM were accompanied by decreased affinities of mutants for Ca2+.
Fig. 3.
Time course of protein C activation by thrombin in the absence and presence of TM. (Upper) The time course of activation of wild-type protein C (○), R67A protein C (□), and D82K protein C (•) (1 μM each) by thrombin (10–50 nM) was monitored in TBS/Ca2+, and the rate of activation was determined by an amidolytic activity assay described in Methods.(Lower) The same as above except that the activation rates by thrombin (1 nM) was monitored in the presence of TM456 (200 nM).
Fig. 4.
The inhibitory effect of Ca2+ on the activation of protein C derivatives by thrombin in the absence of TM. The activation of zymogens (1 μM each) by thrombin (10 nM) was monitored in TBS containing 1 mg/ml BSA, 0.1% PEG 8000, and indicated concentrations of Ca2+. After 30 min of incubation at room temperature and inhibition of thrombin activity by antithrombin, the rate of activation was measured by an amidolytic activity assay described in Methods. The symbols are as follows: protein C (○), R67A protein C (□), and D82K protein C (•).
These results suggest that either Arg-67 of protein C is required for the accelerating effect of TM and/or activation peptides of both Cys-67–82 and R67A (but not D82K) protein C mutants are trapped in a conformation that is not complementary to the active-site groove of the thrombin–TM456 complex. To investigate this question further, the activation of the protein C mutants by trypsin was analyzed in the presence of either 1 mM EDTA or 5 mM Ca2+. Interestingly, in contrast to ≈15-fold stimulatory effect of EDTA on the activation of protein C, the activation of all three protein C mutants (Cys-67–82, R67A, and D82K) by trypsin was insensitive to either Ca2+ or EDTA (data not shown). Because both R67A and D82K protein C mutants can interact with Ca2+, a complete lack of Ca2+ effect on their activation by trypsin strongly suggests that the energetic coupling between the activation peptide and the 70–80 loop of protein C (7, 10, 11) has been disrupted in mutant zymogens. Thus, the mutants may not be capable of responding normally to effect of Ca2+ even at a saturating concentration of the metal ion. This hypothesis may explain the lower inhibitory effect of Ca2+ and the basis for the marked improvement in activation of protein C mutants by thrombin in the absence of TM. These results also suggest that the allosteric effect of Ca2+ on the activation peptide of protein C is regulated by a salt bridge/hydrogen bond between residues Arg-67 and Asp-82 of protein C. Thus, the inability of TM to enhance the activation of R67A and Cys-67–82 protein C mutants by thrombin is most likely due to the inability of these mutants to interact with the cofactor. These results further suggest that interaction of the Ca2+-stablized conformer of the activation peptide of protein C with the active-site groove of thrombin plays a dominant role in recognition of the substrate by the protease. This point has been nicely demonstrated by two recent studies monitoring the competitive effect of p-aminobenzamidine on protein C activation by the thrombin–TM complex (22, 23).
Similar to Cys-67–82 protein C, the activation of R67A protein C by the thrombin mutants was also independent of TM. Interestingly, as presented in Fig. 5, the rate of protein C activation by thrombin in the presence of TM456 in Ca2+-containing buffer (wild-type system) was comparable to the rate of R67A protein C activation by R35A thrombin in the same buffer in the absence of TM456 (mutant system). These results clearly suggest that TM functions primarily by alleviating the inhibitory effects of Arg-67 of protein C and Arg-35 of thrombin in the ternary complex in the presence of Ca2+.
Fig. 5.
Activation of wild-type protein C and R67A mutant of protein C by wild-type and R35A thrombins. (Upper) The time course of activation of 1 μM of either wild-type protein C (○) by wild-type thrombin in the presence of TM456 or the activation of the R67A protein C mutant (•) by R35A thrombin in the absence of TM456 was monitored in TBS/Ca2+. At the indicated time intervals, the activity of thrombin was inhibited by antithrombin, and the rate of APC generation was determined as described in Methods.(Lower) The same as above except that the activation reactions were performed as a function of increasing concentrations of the zymogens.
Discussion
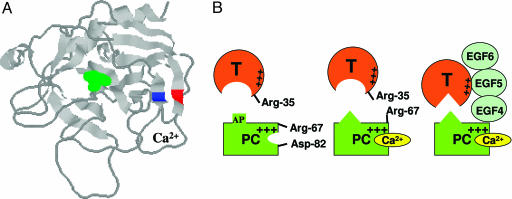
Protein C has a basic exosite that is topologically similar to the anion binding exosite-1 of thrombin (8, 24). The basic residues Arg-67 and Arg-75 and the acidic residues Glu-77 and Glu-80 are conserved on exosites of both proteins. A distinct difference between the two exosites is that instead of a Lys at position 70 of thrombin, this residue is a Glu in protein C. Because of this difference, thrombin is not capable of interaction with Ca2+, and the charged groups of four residues, Arg-67, Lys-70, Glu-77 and Glu-80, are cross-connected with one another to stabilize the 70–80 loop and exosite-1 of thrombin (24). On the other hand, the 70–80 loop of protein C is stabilized by Ca2+, presumably ligated by six oxygen atoms (24), three of which are contributed by the carboxylic group of the acidic residues Glu-70, Glu-77, and Glu-80, similar to that in trypsin (9). Because previous mutagenesis of Arg-67 of protein C has resulted in a mutant, which upon activation exhibited impaired amidolytic and proteolytic activities (17, 18), we speculated that Arg-67 may also be critical for the stabilization of the structure of the 70–80 loop of protein C/activated protein C. The examination of the three-dimensional position of Arg-67 in the x-ray crystal structure of activated GD-PC (8) suggested that the 70–80 loop is located between two antiparallel β strands comprised of residues 64–69 and 81–91 (Fig. 6A). The NH1 guanidyl group of Arg-67 of the former strand is located 2.67 Å from the OD2 carboxylic oxygen of Asp-82 on the latter strand (8). Assuming that this structural feature is also true for the zymogen GD-PC, we thought that the substitution of both Arg-67 and Asp-82 of the substrate with Cys residues may lead to formation of a disulfide bond between the two residues, thereby stabilizing the 70–80 loop and possibly providing some information about the role of Arg-67 in the zymogenic and Ca2+-binding properties of the protein. It appears that this goal has been achieved because the mutant was expressed primarily as a monomeric protein with improved zymogenic properties and no reactivity with DTNB. Moreover, upon activation, the mutant protease exhibited near normal amidolytic activity and reactivity with protein C inhibitor. This mutant, together with the R67A protein C mutant, proved to be excellent probes to understand how Ca2+ modulates the activation of protein C by thrombin in the absence and presence of TM.
Fig. 6.
Crystal structure of the catalytic domain of APC and the proposed model of protein C activation by thrombin in the absence and presence of Ca2+ and TM. (A) The three-dimensional positions of Arg-67 (blue) and Asp-82 (red) on two antiparallel β structures of APC are shown. The catalytic residue Ser-195 is shown in green. The coordinates (Protein Data Bank entry 1AUT) were used to prepare the figure (8). (B Left) In the proposed model, Arg-67 and Asp-82 of protein C (PC) are involved in electrostatic interactions in the absence of Ca2+. (B Center) The binding of Ca2+ to protein C disrupts this interaction, relocating Arg-67 to an inhibitory position for interaction with thrombin in the absence of TM. The metal ion also changes the conformation of the activation peptide (AP) of protein C. (B Right) TM binds to exosite-1 of thrombin via its EGF5 domain and to the basic exosite of the Ca2+-stabilized protein C (including Arg-67) via its EGF4 domain, thereby altering the conformation of the active-site/extended binding pocket of thrombin and facilitating the docking of the activation peptide of protein C in the catalytic groove of thrombin (see text for further discussion).
Analysis of the kinetic data presented above suggests that Arg-67 of protein C makes an inhibitory interaction with thrombin in the presence of Ca2+ and that overcoming this inhibitory effect partly accounts for the mechanism by which TM functions. In support of this hypothesis the elimination of the side chain of this residue in the Cys-67–82 protein C mutant or neutralization of its positive charge in the R67A protein C mutant resulted in a 40- to 50-fold improvement in the activation of the protein C mutants by thrombin in the presence of Ca2+. Our previous results have indicated that TM also overcomes the inhibitory function of Arg-35 of thrombin (13). Thus, the substitution of this residue with a Glu has led to a 20- to 25-fold improvement in the rate of wild-type protein C activation by R35E thrombin independent of Ca2+ (13). In the present study, the activation of both Cys-67–82 and R67A mutants of protein C by R35E thrombin was enhanced by approximately three orders of magnitude independent of TM and Ca2+. Thus, the accelerating effect of mutations in the zymogen and enzyme was additive. This rate enhancement is essentially identical to the accelerating effect of TM and Ca2+ in the wild-type system (1). These results clearly suggest that the primary function of TM in protein C activation by thrombin involves the alleviation of the inhibitory interactions of Arg-67 of protein C and Arg-35 of thrombin in the presence of Ca2+.
Based on these results, we propose a model (Fig. 6B) for protein C activation by thrombin and TM that supports and extends the model presented two decades ago by Esmon et al. (4, 12). They proposed that the Ca2+-altered conformation of protein C is not complementary for interaction with the activesite of thrombin alone but is complementary for interaction with the active site of the TM-altered conformation of the protease (1, 4, 12). Our model predicts that in the absence of Ca2+, the guanidyl group of Arg-67 of protein C is in a salt-bridge/hydrogen-bond contact with Asp-82, a residue immediately outside of the 70–80 loop of protein C. This proposal is consistent with the x-ray crystal structure of activated GD-PC, resolved in the absence of Ca2+ (8). The binding of Ca2+ to the 70–80 loop of protein C is associated with a conformational change, leading to disruption of the electrostatic interaction between Arg-67 and Asp-82 of the protein. A Ca2+-induced conformational change in protein C has been well documented (10, 11). The basic charge of Arg-67 in protein C is inhibitory for interaction with thrombin, nevertheless, this residue is also required for the ability of the zymogen to interact with TM4 of the cofactor in the activation complex. In support of this hypothesis, the elimination of the side chain of Arg-67 in the Cys-67–82 protein C mutant or its substitution with Ala in R67A protein C mutant results in a 40- to 50-fold improvement in activation rates of the mutants by thrombin in the presence of Ca2+. However, their activation rates are not stimulated by TM. The Ca2+-stabilized conformer of protein C (Arg-67 exposed) is not capable of docking into the active-site groove of thrombin, possibly due to the repulsive interaction with Arg-35 of thrombin with its guanidyl group pointing toward the active-site pocket of thrombin (6). The allosteric modulation of the 37 loop of thrombin by TM overcomes this inhibitory interaction. The observation that the activities of both R35E thrombin toward the Cys-67–82 protein C and R67A protein C mutants were independent of TM and Ca2+ is consistent with this model. It is likely that Arg-67 of protein C is surrounded by several other basic residues (Lys-62, Lys-63, and Lys-37–39), but it is not known whether the inhibitory effect is a direct repulsive one or an indirect one mediated through such adjacent residues. Finally, for TM to alleviate the inhibitory effect of Arg-35 of thrombin, protein C is required to interact with TM in the activation complex, because TM456 stimulated the activation of the D82K protein C mutant by thrombin but did not influence the activation of either Cys-67–82 or R67A protein C mutants.
Methods
Construction and Expression of Mutant Proteins. Wild-type protein C and the Arg-67/Asp-82 → 2Cys mutant in the Gla-domainless forms (GD-PC-Cys-67–82) were expressed in human embryonic kidney cells (HEK-293) by using the RSV-PL4 expression/purification vector system as described in ref. 19. The single protein C substitution mutants in which Arg-67 was substituted with Ala (R67A) and Asp-82 was substituted with a Lys (D82K) was prepared by PCR methods and expressed in the same expression/purification vector system. The expression and purification of the Arg-35 → Glu or Ala (R35E and R35A) mutants of thrombin (13), and the epidermal growth factor-like domains 456 of TM (TM456) has been described in ref. 19. Recombinant protein C inhibitor was prepared as described in ref. 21. Human plasma antithrombin was purchased from Haematologic Technologies (Essex Junction, VT), unfractionated heparin, DTNB, and DTT were purchased from Sigma, and Spectrozyme PCa was purchased from American Diagnostica (Greenwich, CT).
Protein C Activation. The initial rate of protein C activation by thrombin was measured in both the absence and presence of Ca2+, EDTA, and TM456 as described in ref. 13. In the absence of TM456, the time course of protein C (1 μM) activation by thrombin (1–50 nM) was studied in 0.1 M NaCl/0.02 M Tris·HCl, pH 7.5 (TBS) containing 1 mg/ml BSA, 0.1% PEG 8000 in the presence of either 2.5 mM Ca2+ (TBS/Ca2+) or 1 mM EDTA (TBS/EDTA) in 96-well assay plates. At different time intervals, thrombin activity was quenched by 500 nM AT in complex with 1 μM heparin, and the rate of protein C activation was measured from the cleavage rate of 200 μM Spectrozyme PCa (American Diagnostica) at 405 nm by a Vmax Kinetic Microplate Reader (Molecular Devices) as described in ref. 13. The experimental conditions in the presence of TM456 were the same with the exception that protein C activation by thrombin (0.5–1.0 nM) was carried out in the presence of a saturating concentration of TM456. The concentration of APC in the reaction mixture was determined by reference to a standard curve that was prepared by total activation of the zymogen with excess thrombin at the time of the experiments as described in ref. 13. The inhibitory effect of Ca2+ on protein C activation by thrombin alone was studied as a function of increasing concentrations of Ca2+ as described above. When a detailed kinetic analysis was done to determine apparent Km and kcat values, the initial rates of activation were measured as a function of the increasing concentrations of protein C by using 0.5 nM thrombin in complex with a saturating concentration of TM456 (200 nM) as described above. All reactions were carried out at room temperature, and it was ensured that <10% substrate was used in all activation reactions.
Analysis of Thiol Contents of the Cys-67–82 Protein C Mutant. The thiol contents of Cys-67–82 protein C was analyzed by spectrophotometric titrations with 0.1 mM DTNB in TBS (pH 8.0) containing 1 mM EDTA as described in ref. 25. The reactivity of the protein C mutant (1–10 μM) with DTNB was monitored at 412 nm for 10 min. Titrations were carried out by using both native and denatured proteins in 6 M urea. A standard curve with 1–10 μM DTT was generated, and BSA with a single thiol was used as a positive control.
Acknowledgments
We thank Audrey Rezaie for proofreading of the manuscript. The research discussed herein was supported by National Heart, Lung, and Blood Institute Grants HL 62565 and HL 68571 of the National Institutes of Health (to A.R.R.).
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: APC, activated protein C; DTNB, 5,5′-dithiobis (2-nitrobenzoic acid); GD-PC, Gla-domainless protein C; TM, thrombomodulin; TM456, TM fragment containing the epidermal growth factor-like domains 4, 5, and 6.
References
- 1.Esmon, C. T. (1993) Thromb. Haemostasis 70, 1–5. [PubMed] [Google Scholar]
- 2.Walker, F. J. & Fay, P. J. (1992) FASEB J. 6, 2561–2567. [DOI] [PubMed] [Google Scholar]
- 3.Davie, E. W., Fujikawa, K. & Kisiel, W. (1991) Biochemistry 30, 10363–10370. [DOI] [PubMed] [Google Scholar]
- 4.Johnson, A. E., Esmon, N. L., Laue, T. M. & Esmon, C. T. (1983) J. Biol. Chem. 258, 5554–5560. [PubMed] [Google Scholar]
- 5.Esmon, N. L., DeBault, L. E. & Esmon, C. T. (1983) J. Biol. Chem. 258, 5548–5553. [PubMed] [Google Scholar]
- 6.Bode, W., Mayr, I., Baumann, U., Huber, R., Stone, S. R. & Hofsteenge, J. (1989) EMBO J. 8, 3467–3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rezaie, A. R., Mather, T., Sussman, F. & Esmon, C. T. (1994) J. Biol. Chem. 269, 3151–3154. [PubMed] [Google Scholar]
- 8.Mather, T., Oganessyan, V., Hof, P., Huber, R., Foundling, S., Esmon, C. & Bode, W. (1996) EMBO J. 15, 6822–6831. [PMC free article] [PubMed] [Google Scholar]
- 9.Bode, W. & Schwager, P. (1975) J. Mol. Biol. 98, 693–717. [DOI] [PubMed] [Google Scholar]
- 10.Rezaie, A. R. & Esmon, C. T. (1995) Biochemistry 34, 12221–12226. [DOI] [PubMed] [Google Scholar]
- 11.Yang, L., Prasad, S., Di Cera, E. & Rezaie, A. R. (2004) J. Biol. Chem. 279, 38519–38524. [DOI] [PubMed] [Google Scholar]
- 12.Esmon, C. T. (1984) Prog. Hemost. Thromb. 7, 25–54. [PubMed] [Google Scholar]
- 13.Rezaie, A. R. & Yang, L. (2003) Proc. Natl. Acad. Sci. USA 100, 12051–12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuentes-Prior, P., Iwanaga, Y., Huber, R., Pagila, R., Rumennik, G., Seto, M., Morser, J., Light, D. R. & Bode, W. (2000) Nature 404, 518–525. [DOI] [PubMed] [Google Scholar]
- 15.Yang, L. & Rezaie, A. R. (2003) J. Biol. Chem. 278, 10484–10490. [DOI] [PubMed] [Google Scholar]
- 16.Knobe, K., Berntsdotter, A., Shen, L., Morser, J., Dahlbäck, B. & Villoutreix, B. O. (1999) Proteins 35, 218–234. [DOI] [PubMed] [Google Scholar]
- 17.Gale, A. J., Tsavaler, A. & Griffin, J. H. (2002) J. Biol. Chem. 277, 28836–28840. [DOI] [PubMed] [Google Scholar]
- 18.Manithody, C., Fay, P. J. & Rezaie, A. R. (2003) Blood 101, 4802–4807. [DOI] [PubMed] [Google Scholar]
- 19.Rezaie, A. R. & Esmon, C. T. (1992) J. Biol. Chem. 267, 26104–26109. [PubMed] [Google Scholar]
- 20.Miletich, J. P. & Broze, G. J. Jr., (1990) J. Biol. Chem. 265, 11397–11404. [PubMed] [Google Scholar]
- 21.Yang, L., Manithody, C. & Rezaie, A. R. (2002) Biochemistry 41, 6149–6157. [DOI] [PubMed] [Google Scholar]
- 22.Lu, G., Chhum, S. & Krishnaswamy, S. (2005) J. Biol. Chem. 280, 15471–15478. [DOI] [PubMed] [Google Scholar]
- 23.Rezaie, A. R. & Yang L. (2005) Biophys. Chem. 117, 255–261. [DOI] [PubMed] [Google Scholar]
- 24.Bode, W., Turk, D. & Karshikov, A. (1992) Protein Sci. 1, 426–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellman, G. L. (1959) Arch. Biochem. Biophys. 82, 70–77. [DOI] [PubMed] [Google Scholar]