Abstract
Understanding the genetic origins and demographic history of Indian populations is important both for questions concerning the early settlement of Eurasia and more recent events, including the appearance of Indo-Aryan languages and settled agriculture in the subcontinent. Although there is general agreement that Indian caste and tribal populations share a common late Pleistocene maternal ancestry in India, some studies of the Y-chromosome markers have suggested a recent, substantial incursion from Central or West Eurasia. To investigate the origin of paternal lineages of Indian populations, 936 Y chromosomes, representing 32 tribal and 45 caste groups from all four major linguistic groups of India, were analyzed for 38 single-nucleotide polymorphic markers. Phylogeography of the major Y-chromosomal haplogroups in India, genetic distance, and admixture analyses all indicate that the recent external contribution to Dravidian- and Hindi-speaking caste groups has been low. The sharing of some Y-chromosomal haplogroups between Indian and Central Asian populations is most parsimoniously explained by a deep, common ancestry between the two regions, with diffusion of some Indian-specific lineages northward. The Y-chromosomal data consistently suggest a largely South Asian origin for Indian caste communities and therefore argue against any major influx, from regions north and west of India, of people associated either with the development of agriculture or the spread of the Indo-Aryan language family. The dyadic Y-chromosome composition of Tibeto-Burman speakers of India, however, can be attributed to a recent demographic process, which appears to have absorbed and overlain populations who previously spoke Austro-Asiatic languages.
Keywords: agriculture, genetic origins, India, paternal lineages
Archaeological evidence advocates the settlement of India by modern humans, using Middle Palaeolithic tools, during the Late Pleistocene (1–5). The large number of deep-rooting, Indian-specific mtDNA lineages of macro haplogroups M and N, whose presence cannot be explained by a recent introduction from neighboring regions (6), is consistent with the archaeological data. These two lines of evidence suggest that the initial settlement, followed by local differentiation, has left a predominantly Late Pleistocene genetic signature in the maternal heritage of India (7–11). The initial settlement of South Asia, between 40,000 and 70,000 years ago, was most likely over the southern route from Africa because haplogroup M, which is the most frequent mtDNA component in India, is virtually absent in the Near East and Southwest Asia (6, 11–14).
Linguistically, the four main language families spoken in India have strong regional patterns, with the largest group, Indo-European (IE), prevalent in northern India. The second largest, the Dravidian (DR) family, covers the majority of the languages in the south. Most of the IE speakers belong to castes, whereas the majority of the tribal populations (>450) speak languages from the other three families (15). The existence of both IE-speaking tribal groups and DR castes indicates the complexity of historical interactions between Indian populations, and that there is no one-to-one correlation between language and mode of subsistence or social system. Even so, the trend of finding farming (castes) and IE languages grouped together has led some to suggest a major demic diffusion, associated with the spread of agriculture, from West Asia and/or Central Asia to India, (16). The situation in Northeast India is less intensely studied, but the proximity to related language families in East and Southeast Asia suggests possible origins for the Tibeto-Burman (TB) clade of the Sino-Tibetan family outside India. The Mundari group of Austro-Asiatic (AA) languages is currently found almost exclusively in East India. Khasi, a major tribe of Meghalaya, forms a notable exception, being surrounded by TB speakers in the northeast. Other members of the AA family are located in Southeast Asia, but the ultimate source of these languages is currently unresolved.
Several studies have argued that, in contrast to the relative uniformity of mtDNA, the Y chromosomes of Indian populations display relatively small genetic distances to those of West Eurasians (17), linking this finding to hypothetical migrations by Indo-Aryan speakers. Wells et al. (18) highlighted M17 (R1a) as a potential marker for one such event, as it demonstrates decreasing frequencies from Central Asia toward South India. Departing from the “one haplogroup equals one migration” scenario, Cordaux et al. (19) defined, heuristically, a package of haplogroups (J2, R1a, R2, and L) to be associated with the migration of IE people and the introduction of the caste system to India, again from Central Asia, because they had been observed at significantly lower proportions in South Indian tribal groups, with the high frequency of R1a among Chenchus of Andhra Pradesh (6) considered as an aberrant phenomenon (19). Conversely, haplogroups H, F*, and O2a, which were observed at significantly higher proportions among tribal groups of South India, led the same authors to single them out as having an indigenous Indian origin. Only O3e was envisaged as originating (recently) east of India (20), substantiating a linguistic correlation with the TB speakers of Southeast Asia.
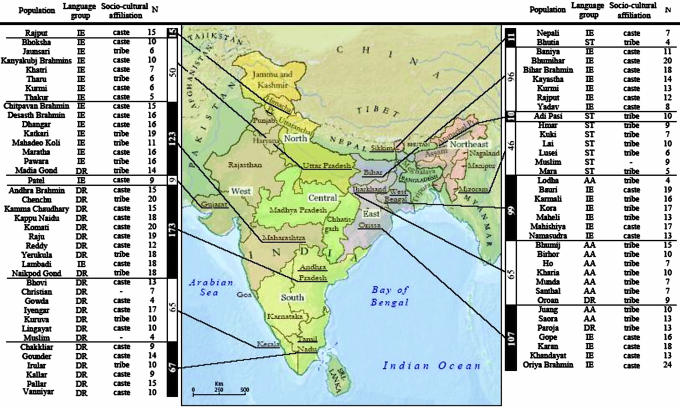
The present study significantly increases the available sample size for India by typing 936 individuals from 77 populations, representing all four major linguistic groups (Fig. 1). The increased range of informative SNPs typed permits more detailed resolution of geographic patterns and the identification of some region-specific subsets of lineages. These Y chromosomes are analyzed in the context of available data from West Asia, East Asia, Southeast Asia, Central Asia, Europe, the Near East, and Ethiopia. Measures of genetic distance, admixture, and factor analysis drawn from the Y-chromosome data are used to investigate three themes central to population genetics in India: demographic links to West and Central Asia, the genetic relationship between castes and tribes, and geographic versus linguistic grouping for the current populations of the Indian subcontinent.
Fig. 1.
Map of India showing sample locations. Regional groupings of populations as used in the text are highlighted in different colors.
Results
A total of 18 haplogroups were detected in 936 Indian Y chromosomes (Fig. 3A, which is published as supporting information on the PNAS web site). Together, haplogroups R1, R2, L, O, H, J2, and C characterize >90% of the Y-chromosomal variation in all socio-linguistic groups of India (Tables 2 and 3, which are published as supporting information on the PNAS web site). Both IE- and DR-speaking populations show a high combined frequency of haplogroups C*, L1, H1, and R2. The total frequency of these four haplogroups outside of India is marginally low. In turn, haplogroups E, I, G, J*, and R1* have a combined frequency of 53% in the Near East among the Turks and 24% in Central Asia, but they are rare or absent in India (0.86% in all populations and almost solely because of R1*). Similarly, haplogroups C3, D, N, and O specific to Central Asian (36%) and Southeast Asian populations (subclades of haplogroup O; 85%) are virtually absent in India (Fig. 3A). Only haplogroups J2 and R1a have interregional frequency patterns west of India with J2 being most common in Afro-Asiatic-speaking (and IE-speaking) populations of the Near East and Middle East, whereas R1a occurs at the highest frequencies in populations of India, East Europe, and Central Asia. The O2a and O3e subclades of haplogroup O in India also have interregional distributions, overlapping with those of Southeast Asia and East Asia.
Principal component analysis (Fig. 3B) investigates the phylogeography of the Y haplogroups with respect to each other, illustrating the associations of haplogroups, irrespective of regional or cultural categories. The first two components account for 75% of the variation observed, and within India delineate R*, R2, F*, and H, within the sphere of L, K, P*, and R1a. Of all of the R lineages, only R1* is separated from this grouping, forming a cluster together with G, I, and J, consistent with their common and widespread distribution throughout (Western) Europe. The O lineages fall out with C* and D (the latter tending to derive from Sino-Tibetan speakers). Once the third and fourth factors are considered, the ambiguity of A, B, and E (typically African in origin) is resolved, and the positions of C3 and N, also non-Indian in their distribution, are delineated to Central Asia.
By considering all haplogroup frequencies simultaneously, an indication of the relatedness between regions is obtained (Table 1). Here, for the sake of comparison only, the categories used by a previous study (19) are retained, but the tribal population is split into two because of the close association identified here between Hg O and tribal groups of the east and northeast of India (O2a represents 77% of AA speakers and 47% of TB speakers), which are combined to form the east and northeast tribes. In contrast to the earlier study (19), the caste populations of “north” and “south” India are not particularly more closely related to each other (average Fst value = 0.07) than they are to the tribal groups (average Fst value = 0.06). The multidimensional scaling plot of these values (Fig. 4, which is published as supporting information on the PNAS web site) demonstrates that the combined data set for the tribal peoples (derived from all regions of India, excluding those of the east and northeast) actually falls midway between those for northern and southern castes, whereas the tribal populations of the east and northeast are confirmed as a separate category. The position of the reduced tribal category, comprising groups from Southern, Northern, and Western India, is suggestive of geographical structuring north to south.
Table 1. Genetic distances between populations estimated from Y-haplogroup frequencies.
| North castes | South castes | Reduced tribes | Northeast-east tribes | Turkey | Central Asia | Mongols/Buryats | Southeast Asia | Iran | Iraq | East Europe | Russia | West Europe | Ethiopia | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| North castes | 0.00 | |||||||||||||
| South castes | 0.07 | 0.00 | ||||||||||||
| Reduced tribes | 0.06 | 0.05 | 0.00 | |||||||||||
| Northeast-east tribes | 0.21 | 0.20 | 0.19 | 0.00 | ||||||||||
| Turkey | 0.11 | 0.14 | 0.13 | 0.21 | 0.00 | |||||||||
| Central Asia | 0.07 | 0.12 | 0.10 | 0.19 | 0.05 | 0.00 | ||||||||
| Mongols/Buryats | 0.26 | 0.27 | 0.26 | 0.32 | 0.21 | 0.12 | 0.00 | |||||||
| Southeast Asia | 0.26 | 0.27 | 0.26 | 0.21 | 0.22 | 0.19 | 0.30 | 0.00 | ||||||
| Iran | 0.09 | 0.12 | 0.11 | 0.22 | 0.01 | 0.06 | 0.24 | 0.24 | 0.00 | |||||
| Iraq | 0.16 | 0.19 | 0.17 | 0.26 | 0.04 | 0.10 | 0.27 | 0.27 | 0.02 | 0.00 | ||||
| East Europe | 0.08 | 0.23 | 0.19 | 0.33 | 0.16 | 0.11 | 0.34 | 0.34 | 0.18 | 0.23 | 0.00 | |||
| Russia | 0.08 | 0.20 | 0.16 | 0.29 | 0.11 | 0.06 | 0.28 | 0.30 | 0.13 | 0.18 | 0.03 | 0.00 | ||
| West Europe | 0.26 | 0.29 | 0.25 | 0.35 | 0.14 | 0.17 | 0.36 | 0.36 | 0.22 | 0.23 | 0.28 | 0.19 | 0.00 | |
| Ethiopia | 0.31 | 0.33 | 0.32 | 0.39 | 0.21 | 0.27 | 0.40 | 0.39 | 0.21 | 0.24 | 0.40 | 0.37 | 0.42 | 0.00 |
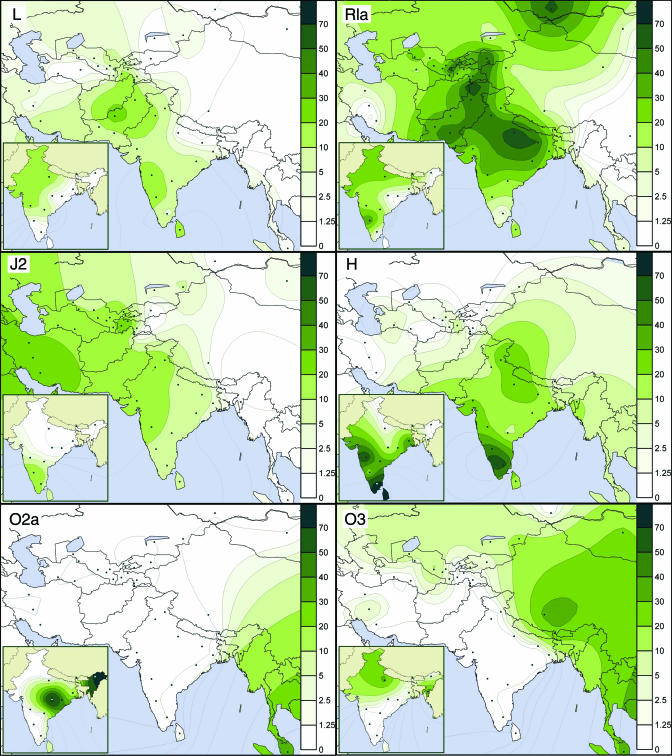
This geographical structure is displayed with greater precision by dividing the data set according to the regions of India presented in Fig. 1, except that the Punjab (caste only) is considered as a separate entity because of its isolation relative to the rest of the west (see Tables 2 and 3) and proximity to Central Asia. Considering individual haplogroup frequencies within each of these geographical regions, no consistent pattern (at the 95% level of certainty) was detected in the distribution of the Y haplogroups to distinguish either the castes from the tribes, or DRs from IEs (Fig. 5 B and C, which is published as supporting information on the PNAS web site). Therefore, it is appropriate to consider the distributions at the regional level, omitting Northeast India because of the dominance of haplogroup O there (Fig. 5A). The potential clines centered on North India (R1a), Northwest India (J2), South India (H), and East India (R2), identified in Fig. 5A, are illustrated by the distribution maps (Fig. 2 and Fig. 6, which is published as supporting information on the PNAS web site). These clines display distinct regional concentrations of J2, H, R1a, R2, O3, and O2a, confirming the primarily geographic nature of Y-chromosome frequency distribution in India.
Fig. 2.
Spatial frequency distribution maps of major Y-chromosome haplogroups in South Asia. For India, the data on tribal populations are shown in the inset maps and excluded from the main maps. The data for caste populations are averaged to the level of states in India. Because of different phylogenetic resolution different sets of published data are used for different haplogroup maps.
Admixture analysis (21) evaluates the potential parental contributions to northwestern castes (Punjab) and southern castes (Table 4, which is published as supporting information on the PNAS web site) and Central Asia (Table 5, which is published as supporting information on the PNAS web site). For South Indian populations this analysis revealed reciprocally high local admixture contributions for both caste and tribal populations (0.91 contribution of tribes to castes, SE 0.1; 0.98 contribution of castes to tribes, SE 0.11) over the contributions from outside of India. It should be stressed that these values do not necessarily reflect actual admixture proportions between the tribes and castes, as the algorithm that is used to estimate the admixture proportions divides the whole genetic composition of a hybrid between given parental populations. Rather, these findings confirm the results obtained above from the Fst analyses, that Southern castes and tribals are very similar to each other in their Y-chromosomal haplogroup compositions, and that their gene pool is significantly related to the castes of Northwest India (Fig. 5A), among whom a South Indian tribal contribution of 0.48 (SE 0.12) was observed. In contrast, the potential contribution from Central Asia to the Indian Y-chromosomal pool is minor. In the case of Northwest India, there is nothing to choose between two opposing scenarios: (i) the flow of Y chromosomes from Central Asia, and (ii) the flow of Y chromosomes in the opposite direction, to Central Asia from Northwest India. Meanwhile, the West Asian contribution to the Indian Y-chromosomal pool was significantly smaller in all three admixture tests.
Discussion
Leaving aside, for the moment, TB and AA speakers, the distributions of Y haplogroups between India and West and Central Asia display a clear patterning. J is the predominant Y-chromosome haplogroup in populations living west of India. The frequency and subgroup variation of J in West Asia, in the context of the complete absence of J1 and most J2 subgroups within the Indian sample, is consistent with an influx of a subset of J2 lineages to India from the Near East, followed by their subsequent diffusion from India's northwest toward the south and east. In contrast, within India, the complete absence of the derived C3 lineages, which represent >95% of haplogroup C variation in Central Asia (22), suggests that Indian C lineages cannot be ascribed to a recent admixture from the north.
Similarly, the proposition that a high frequency of R1a in India is caused by admixture with populations of Central Asian origin is difficult to substantiate, as the proposed source region does not meet the expectation of containing high frequencies of the other components of haplogroup R, with no examples of R* and generally low incidence of R2, which, unlike J2, does not show evidence of a recent diffusion throughout India from the northwest. Second, it is notable that the results from the admix2 program gave relatively high reciprocal admixture (0.3–0.35) proportions for Northwest Indian and Central Asian populations, despite the incompatibility of the respective haplogroup frequency pools; our Northwest Indian sample totally lacks haplogroups C3, DE, J*, I, G, N, and O, which cover almost half of the Central Asian Y chromosomes, whereas the Central Asian sample is poor in haplogroups C*, F*, H, L, and R2 (with a combined frequency of 10%). Hence, the admixture proportions are driven solely by the shared high frequency of R1a. In other words, if the source of R1a variation in India comes from Central Asia, as claimed by Wells et al. (18) and Cordaux et al. (19), then, under a recent gene flow scenario, one would expect to find the other Central Asian-derived NRY haplogroups (C3, DE, J*, I, G, N, O) in Northwest India at similarly elevated frequencies, but that is not the case.
Alternatively, although the simple admixture scenario does not hold, one could nevertheless argue that the other haplogroups were lost during a hypothetical bottleneck (lineage sorting among the early Indo-Aryans arriving to India). But in line with this scenario, one should expect to observe dramatically lower genetic variation among Indian R1a lineages. In fact, the opposite is true: the STR haplotype diversity on the background of R1a in Central Asia (and also in Eastern Europe) has already been shown to be lower than that in India (6). Rather, the high incidence of R1* and R1a throughout Central Asian and East European populations (without R2 and R* in most cases) is more parsimoniously explained by gene flow in the opposite direction, possibly with an early founder effect in South or West Asia. Note that the admixture method reports positive admixture proportions in cases where just one haplogroup is shared between populations (possibly because of shared deep common ancestry), even if other haplogroup frequencies strongly argue against a recent simple admixture scenario.
Even though more than one explanation could exist for genetic differentiation between castes and tribes in India, the Indo-Aryan migration scenario advocated in ref. 19 rested on the suggestion that all Indian caste groups are similar to each other while being significantly different from the tribes. Using a much more representative data set, numerically, geographically, and definitively, it was not possible to confirm any of the purported differentiations between the caste and tribal pools. Although differences could be found to occur within particular regions, between particular caste and tribal groups, consistent and statistically significant variations at the subcontinental scale were not detected. Although it is arguable that assimilation of tribal populations into the caste system could skew distributions in any particular region, it cannot explain the persistence and prevalence of those lineages put forward as being typical of incoming IEs (J2, R1a, R2, and L) among many of those populations who are still designated as tribals [see also the credibility gap in the groupings of Corduax et al. (19) illustrated in the factor analysis, Fig. 2]. Rather, taken together with the evidence from Fst values, the elements discussed so far (i.e., admixture, factor analysis, and frequency distributions) are more parsimoniously explained by a predominantly pre-IE, pre-Neolithic presence in India, for the majority of those Y lineages considered here (R1a, R2, L1), which occur together with strictly Indian-specific haplogroups and paragroups (C*, F*, H) among both caste and tribal groups.
The distribution of R2, with its concentration in Eastern and Southern India, is not consistent with a recent demographic movement from the northwest. Instead, its prevalence among castes in these regions might represent a recent population expansion, perhaps associated with the transition to agriculture, which may have occurred independently in South Asia (23). A pre-Neolithic chronology for the origins of Indian Y chromosomes is also supported by the lack of a clear delineation between DR and IE speakers. Again, although appeals to language change are plausible for explaining the appearance of supposedly tribe-specific Y lineages among incoming IE speakers, it is much harder to conceive of a systematic movement of external Y-chromosome types in the opposite direction, via the uptake of DR languages. The near absence of L lineages within the IE speakers from Bihar (0%), Orissa (0%), and West Bengal (1.5%) further suggests that the current distribution of Y haplogroups in India is associated primarily with geographic rather than linguistic or cultural determinants.
In contrast, the situation with the TB- and AA-speaking populations is rather intriguing and warrants further discussion. The AA groups have a very clear association with O2a Y-chromosome haplogroup, both in India and Southeast Asia (24, 25), whereas the close association between TB groups and the O3e lineage may indicate a second case where a Y haplogroup is linked to a cultural entity. The present-day distribution of haplogroup O argues for a Southeast Asian homeland for the AA speakers of India (Mundari group), in distinct contrast to the suggestions, based on mtDNA, that the Mundari speakers represent the earliest settlers of India (9, 26). Yet, the contemporary distribution within India of Y-chromosomal haplogroup O2a, on one hand, and AA speakers on the other, cautions against simplistic interpretations of either linguistic or genetic correlations. AA languages, besides being concentrated in East India, also appear as outliers in Madhya Pradesh (Central India) and Maharashtra (West India), whereas O2a is present, sporadically, within other linguistic groups in both South and East India.
Among TB speakers the share of mtDNAs typical of East Asia increases to nearly two-thirds (64%), inferred from ref. 27. This scenario would be consistent with a more recent migration event or the continued movement of women into India through the maintenance of social links. The near total absence of AA-speaking groups between East India and Southeast Asia has been interpreted as representing a recent (mid-Holocene) influx of TB populations, bearing O3e Y chromosomes, into this region (20). Cordaux et al. (20), when considering different scenarios for the prehistory of this area, favored the view that it was previously an unoccupied territory that had acted as a barrier to human migrations, possibly since the late Pleistocene. However, the presence of the AA-speaking Khasi in Meghalaya provides an alternative explanation, namely that there were previous inhabitants in this region who had been predominantly AA speakers. This explanation is favored by the presence of both O3e and O2a Y haplogroups within the TB populations reported here. The parsimonious explanation for this is that AA speakers were formerly distributed from Southeast Asia to India and intermixed with TB speakers as they migrated to the area. This scenario is supported by the widespread presence of East Asian mtDNA lineages among TB groups. So, paradoxically, it is in Northeastern India that there is evidence, from the Y chromosome, for both large-scale immigration (TB speakers) and language change (former AA speakers). One of the reasons this is still detectable is the relatively shallow time depth proposed for this “event,” a chronology that still covers the period proposed for the appearance of the caste system, the IE language family, and agriculture into India through the northwest (20).
Conclusions
It is not necessary, based on the current evidence, to look beyond South Asia for the origins of the paternal heritage of the majority of Indians at the time of the onset of settled agriculture. The perennial concept of people, language, and agriculture arriving to India together through the northwest corridor does not hold up to close scrutiny. Recent claims for a linkage of haplogroups J2, L, R1a, and R2 with a contemporaneous origin for the majority of the Indian castes' paternal lineages from outside the subcontinent are rejected, although our findings do support a local origin of haplogroups F* and H. Of the others, only J2 indicates an unambiguous recent external contribution, from West Asia rather than Central Asia. The current distributions of haplogroup frequencies are, with the exception of the O lineages, predominantly driven by geographical, rather than cultural determinants. Ironically, it is in the northeast of India, among the TB groups that there is clear-cut evidence for large-scale demic diffusion traceable by genes, culture, and language, but apparently not by agriculture.
Materials and Methods
DNA Samples. DNA samples from 936 male individuals from 77 endogamous populations of India selected on the basis of their geographic distribution, language, and socio-cultural affinity were analyzed in this study. The studied populations belong to four major linguistic families of India: IE (n = 470), DR (n = 323), AA (n = 83), and TB (n = 60). DNA was extracted by using standard protocols (28) from blood samples collected from unrelated consenting individuals after approval of the ethical committee of the Central Forensic Science Laboratory. The genetic relationships of Indian Y chromosomes (present study and ref. 6) with world populations was carried out by using data on Turkey (29), Central Asia, Russia, and Mongolia (22), Southeast Asia (30), Iran and Iraq (31), Europe (32), and Ethiopia (33). The spatial frequency distribution maps were generated by the kriging procedure with the default settings of the surfer program of Golden Software (Golden, CO) and using data from refs. 6, 18, 22, 29–31, and 33–36.
Thirty-eight binary markers known to be specific to various paternal lineages within Europe, Central Asia, South Asia, and Southeast Asia were used to examine Y-chromosome haplogroups in India. The Y-SNPs included M168, M89, M9, RPS4Y711, M216, M217, YAP, M174, M201, M69, M52, Apt, M82, M170, p12f2, M172, M70, M147, M20, M11, M27, M5, M214, M231, M175, M95, M88, M122, M134, M242, 92R7, M45, M207, M173, SRY1532, M17, M269, and M124 and were examined hierarchically to provide an internal check on the reliability of typing. PCR-restriction fragment length polymorphism methodology was used to score YAP, 92R7, M9, RPS4Y711, M122, and SRY1532 markers as described (25, 37–39), whereas all other markers were typed by using primer pairs as described (40).
Statistical Analyses. Haplogroup frequency were estimated by a simple gene count method. Frequency charts were thrown in Microsoft excel, and 95% credible regions were calculated from the posterior distribution of the proportion of haplogroups in each population by using an adaptation of software kindly provided by Vincent Macaulay (University of Glasgow, Glasgow, Scotland). Principal component analysis was performed in statistica. Fst values were calculated on haplogroup frequencies by using arlequin (41) and multidimensional scaling produced in xl stat pro 7.5. Admixture proportions based on haplogroup frequencies were calculated by using admix2 software (21).
Supplementary Material
Acknowledgments
We thank Simon Ho (University of Oxford) for a critical reading of this manuscript. S.S., A.S., J.B., and S.G. thank the Directorate of Forensic Sciences, Ministry of Home Affairs for fellowship. G.H. and T.S. were supported by a fellowship from the Council of Scientific and Industrial Research, India. P.E. was funded by the National Environment Research Council of Great Britain. This research was supported by a financial grant to the Central Forensic Science Laboratory under the X Five-Year Plan of the Government of India.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: IE, Indo-European; DR, Dravidian; TB, Tibeto-Burman; AA, Austro-Asiatic.
References
- 1.Sankalia, H. D. (1988) Archaeology in Rajasthan (Sahitya Sansthan, Udaipur, India).
- 2.Paddaya, K. (1982) in The Transition from Lower to Middle Palaeolithic and the Origin of Modern Man, ed. Ronen, A. (British Archaeological Reports, Oxford, U.K.), pp. 257–264.
- 3.Pappu, R. S. & Rao, J. V. P. (1983) Bull. Deccan College Res. Inst. 42, 119–130. [Google Scholar]
- 4.Possehl, G. L. (1994) Radiometric Dates for South Asian Archaeology (University of Pennsylvania, Philadelphia).
- 5.Misra, V. N. (2001) J. Biosci. 26, 491–531. [DOI] [PubMed] [Google Scholar]
- 6.Kivisild, T., Rootsi, S., Metspalu, M., Mastana, S., Kaldma, K., Reidla, M., Parik, J., Metspalu, M., Tolk, H.-V., Stepanov, V., et al. (2003) Am. J. Hum. Genet. 72, 313–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kivisild, T., Kaldma, K., Metspalu, M., Parik, J., Papiha, S. S. & Villems, R. (1999) in Genomic Diversity, eds. Papiha, S. S., Deka, R. & Chakraborty, R. (Kluwer, Boston), pp. 135–152.
- 8.Kivisild, T., Bamshad, M. J., Kaldma, K., Metspalu, M., Metspalu, E., Reidla, M., Laos, S., Parik, J., Watkins, W. S., Dixon, M. E., et al. (1999) Curr. Biol. 9, 1331–1334. [DOI] [PubMed] [Google Scholar]
- 9.Majumder, P. P. (2001) J. Biosci. 26, 533–545. [DOI] [PubMed] [Google Scholar]
- 10.Palanichamy, M., Sun, C., Agrawal, S., Bandelt, H.-J., Kong, Q.-P., Khan, F., Wang, C. Y., Chaudhuri, T. K., Palla, V. & Zhang, Y. P. (2004) Am. J. Hum. Genet. 75, 966–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Metspalu, M., Kivisild, T., Metspalu, E., Parik, J., Hudjashov, G., Kaldma, K., Serk, P., Karmin, M., Behar, D. M., Gilbert, M. T. P., et al. (2004) BMC Genet. 5, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quintana-Murci, L., Semino, O., Bandelt, H.-J., Passarino, G., McElreavey, K. & Santachiara-Benerecetti, A. S. (1999) Nat. Genet. 23, 437–441. [DOI] [PubMed] [Google Scholar]
- 13.Quintana-Murci, L., Chaix, R., Wells, S., Behar, D., Sayar, H., Scozzari, R., Rengo, C., Al-Zahery, N., Semino, O., Santachiara-Benerecetti, A. S., et al. (2004) Am. J. Hum. Genet. 74, 827–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richards, M., Macaulay, V., Hickey, E., Vega, E., Sykes, B., Guida, V., Rengo, C., Sellitto, D., Cruciani, F., Kivisild, T., et al. (2000) Am. J. Hum. Genet. 67, 1251–1276. [PMC free article] [PubMed] [Google Scholar]
- 15.Singh, K. S., ed. (1997) The Scheduled Tribes (Oxford Univ. Press, Oxford), Vol. III.
- 16.Cordaux, R., Deepa, E., Vishwanathan, H. & Stoneking, M. (2004) Science 304, 1125. [DOI] [PubMed] [Google Scholar]
- 17.Bamshad, M., Kivisild, T., Watkins, W. S., Dixon, M. E., Ricker, C. E., Rao, B. B., Mastan Naidu, J., Ravi Prasad, B. V., Govinda Reddy, P., Rasanayagam, A., et al. (2001) Genome Res. 11, 994–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wells, R. S., Yuldasheva, N., Ruzibakiev, R., Underhill, P. A., Evseeva, I., Blue-Smith, J., Jin, L., Su, B., Pitchappan, R., Shanmugalakshmi, S., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 10244–10249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cordaux, R., Aunger, R., Bentley, G., Nasidze, I., Sirajuddin, S. M. & Stoneking, M. (2004) Curr. Biol. 14, 231–235. [DOI] [PubMed] [Google Scholar]
- 20.Cordaux, R., Weiss, G., Saha, N. & Stoneking, M. (2004) Mol. Biol. Evol. 21, 1525–1533. [DOI] [PubMed] [Google Scholar]
- 21.Dupanloup, I. & Bertorelle, G. (2001) Mol. Biol. Evol. 18, 672–675. [DOI] [PubMed] [Google Scholar]
- 22.Karafet, T. M., Osipova, L. P., Gubina, M. A., Posukh, O. L., Zegura, S. L. & Hammer, M. F. (2002) Hum. Biol. 74, 761–789. [DOI] [PubMed] [Google Scholar]
- 23.Fuller, D. (2003) in Examining the Farming/Language Dispersal Hypothesis, eds. Bellwood, P. & Renfrew, C. (McDonald Institute for Archaeological Research, Cambridge, U.K.), pp. 191–213.
- 24.Su, B., Xiao, C., Deka, R., Seielstad, M. T., Kangwanpong, D., Xiao, J., Lu, D., Underhill, P. A., Cavalli-Sforza, L. L., Chakraborty, R., et al. (2000) Hum. Genet. 107, 582–590. [DOI] [PubMed] [Google Scholar]
- 25.Kayser, M., Brauer, S., Wiess, G., Schiefenhövel, W., Underhill, P., Shen, P., Oefner, P., Tommaseo-Ponzetta, M. & Stoneking, M. (2003) Am. J. Hum. Genet. 72, 281–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Basu, A., Mukherjee, N., Roy, S., Sengupta, S., Banerjee, S., Chakraborty, M., Dey, B., Roy, M., Roy, B., Bhattacharya, N. P., et al. (2003) Genome Res. 13, 2277–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cordaux, R., Saha, N., Bentley, G. R., Aunger, R., Sirajuddin, S. M. & Stoneking, M. (2003) Eur. J. Hum. Genet. 3, 253–264. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed.
- 29.Cinnioglu, C., King, R., Kivisild, T., Kalfoglu, E., Atasoy, S., Cavalleri, G. L., Lillie, A. S., Roseman, C. C., Lin, A. A., Prince, K., et al. (2004) Hum. Genet. 114, 127–148. [DOI] [PubMed] [Google Scholar]
- 30.Karafet, T., Xu, L., Du, R., Wang, W., Feng, S., Wells, R. S., Redd, A. J., Zegura, S. L. & Hammer, M. F. (2001) Am. J. Hum. Genet. 69, 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Al-Zahery, N., Semino, O., Benuzzi, G., Magri, C., Passarino, G., Torroni, A. & Santachiara-Benerecetti, A. S. (2003) Mol. Phylogenet. Evol. 28, 458–472. [DOI] [PubMed] [Google Scholar]
- 32.Semino, O., Passarino, G., Oefner, P. J., Lin, A. A., Arbuzova, S., Beckman, L. E., De Benedicitis, G., Francalacci, P., Kouvatsi, A., Limborska, S., et al. (2000) Science 290, 1155–1159. [DOI] [PubMed] [Google Scholar]
- 33.Semino, O., Santachiara-Benerecetti, A. S., Falaschi, F., Cavalli-Sforza, L. & Underhill, P. (2002) Am. J. Hum. Genet. 70, 265–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qamar, R., Ayub, Q., Mohyuddin, A., Helgason, A., Mazhar, K., Mansoor, A., Zerjal, T., Tyler-Smith, C. & Mehdi, S. Q. (2002) Am. J. Hum. Genet. 70, 1107–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zerjal, T., Wells, R., Yuldasheva, N., Ruzibakiev, R. & Tyler-Smith, C. (2002) Am. J. Hum. Genet. 71, 466–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deng, W., Shi, B., He, X., Zhang, Z., Xu, J., Li, B., Yang, J., Ling, L., Dai, C., Qiang, B., et al. (2004) J. Hum. Genet. 49, 339–348. [DOI] [PubMed] [Google Scholar]
- 37.Hammer, M. F. & Horai, S. (1995) Am. J. Hum. Genet. 56, 951–962. [PMC free article] [PubMed] [Google Scholar]
- 38.Hurles, M. E., Veitia, R., Arroyo, E., Armenteros, M., Bertranpetit, J., Perez-Lezaun, A., Bosch, E., Shlumukova, M., Cambon-Thomsen, A., McEl-reavey, K., et al. (1999) Am. J. Hum. Genet. 65, 1437–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kayser, M., Brauer, S., Wiess, G., Underhill, P., Roewer, L., Schiefenhövel, W. & Stoneking, M. (2000) Curr. Biol. 10, 1237–1246. [DOI] [PubMed] [Google Scholar]
- 40.Underhill, P. A., Passarino, G., Lin, A. A., Shen, P., Mirazon Lahr, M., Foley, R. A., Oefner, P. J. & Cavalli-Sforza, L. L. (2001) Ann. Hum. Genet. 65, 43–62. [DOI] [PubMed] [Google Scholar]
- 41.Schneider, S., Roessli, D. & Excoffier, L. (2000) arlequin: A Software for Population Genetics Data Analysis (University of Geneva, Geneva), Version 2.0.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.