Abstract
We have used NMR spectroscopy and x-ray crystallography to determine the three-dimensional structure of PF1378 (Pfu Pop5), one of four protein subunits of archaeal RNase P that shares a homolog in the eukaryotic enzyme. RNase P is an essential and ubiquitous ribonucleoprotein enzyme required for maturation of tRNA. In bacteria, the enzyme's RNA subunit is responsible for cleaving the single-stranded 5′ leader sequence of precursor tRNA molecules (pre-tRNA), whereas the protein subunit assists in substrate binding. Although in bacteria the RNase P holoenzyme consists of one large catalytic RNA and one small protein subunit, in archaea and eukarya the enzyme contains several (≥4) protein subunits, each of which lacks sequence similarity to the bacterial protein. The functional role of the proteins is poorly understood, as is the increased complexity in comparison to the bacterial enzyme. Pfu Pop5 has been directly implicated in catalysis by the observation that it pairs with PF1914 (Pfu Rpp30) to functionally reconstitute the catalytic domain of the RNA subunit. The protein adopts an α–β sandwich fold highly homologous to the single-stranded RNA binding RRM domain. Furthermore, the three-dimensional arrangement of Pfu Pop5's structural elements is remarkably similar to that of the bacterial protein subunit. NMR spectra have been used to map the interaction of Pop5 with Pfu Rpp30. The data presented permit tantalizing hypotheses regarding the role of this protein subunit shared by archaeal and eukaryotic RNase P.
Keywords: crystallography, NMR, ribonuclease P, ribonucleoprotein
RNase P is a ubiquitous and essential enzyme in all domains of life responsible for cleaving the single-stranded 5′ leader sequence of precursor tRNA (pre-tRNA), a vital reaction in the maturation of tRNA (1, 2). RNase P is a ribonucleoprotein complex that, in bacteria, consists of a large RNA (P RNA, ≈400 nt) responsible for mediating catalysis (3) and a small protein (≈120 residues) that is required in vivo (4) and appears to function by stabilizing the RNA subunit and/or increasing the enzyme's affinity for the substrate by binding to its typically single-stranded 5′ leader sequence (5–8). The bacterial RNA subunit is capable of cleaving its pre-tRNA substrate in vitro in the absence of its protein moiety at high concentrations of Mg2+; thus, RNase P is a ribozyme. The recent crystal structures of the bacterial RNA subunits from Thermotoga maritima (9) and Bacillus stearothermophilus (10) have provided a rich framework for understanding the function of this enzyme, while validating predictions based on biochemical and phylogenetic data (11, 12).
In eukaryotes RNase P is more complex, consisting of a smaller RNA and at least nine protein subunits (13). Although sequence and predicted secondary structural elements are conserved between the RNA subunits of eukaryal and bacterial RNase P (14), the eukaryal RNA has not been shown to be catalytically active in vitro. Despite similarity between the RNA components in Archaea and Eukarya, genome sequence analysis has failed to identify any eukaryotic homolog of the bacterial protein. Furthermore, the functional roles of the multiple protein subunits in eukaryotic RNase P remain to be elucidated.
Evidence suggests that the RNase P enzyme from archaea represents a mosaic of features of the eukaryal and bacterial enzymes. The archaeal enzyme contains an RNA that is more similar to the bacterial than to the eukaryal RNA (15), and RNA subunits from some archaea have been shown to be catalytically active in vitro under high salt conditions (16). On the other hand, archaeal RNase P is similar to the eukaryal enzyme in that it is comprised of multiple protein subunits (four; perhaps more), each of which has marked sequence similarity to protein subunits of eukaryal RNase P (17): Rpp21 (18), Rpp29 (19), Rpp30 (20), and Pop5 (21) [also known as Rpr2, Pop4 (22), Rpp1 (23), and hPop5, respectively]. The four archaeal proteins can be recombinantly expressed and combined in vitro with the corresponding in vitro-transcribed RNA subunit to yield an active enzyme, as demonstrated for the enzymes from Methanothermobacter thermoautotrophicus (24), Pyrococcus furiosus (H.-Y. Tsai and V. Gopalan, personal communication), and Pyrococcus horikoshii (25).
Although little is known about the arrangement of the proteins in the archaeal holoenzyme, yeast two-hybrid experiments using the protein subunits from Methanothermobacter thermoautotrophicus and Pyrococcus horikoshii have detected strong interactions between two pairs of proteins, Pop5+Rpp30 and Rpp21+Rpp29 (26, 27), mirroring similar findings with the eukaryotic proteins (28, 29). In vitro reconstitution experiments using Pyrococcus furiosus components (H.-Y. Tsai and V. Gopalan, personal communication) provide more evidence that the proteins function in pairs, because the Pop5+Rpp30 and Rpp21+Rpp29 subsets of proteins are each capable of forming an active enzyme in the presence of the intact RNA subunit. Furthermore, Pop5+Rpp30 can be functionally reconstituted with the catalytic domain of the RNA subunit. Detailed structural data on the RNase P components will greatly increase our understanding of how this ribonucleoprotein complex assembles and performs catalysis.
The three-dimensional structures of three archaeal RNase P proteins have been determined, each of which adopts a different fold: Rpp29 contains a beta-barrel fold (24, 30–32) reminiscent of archaeal Sm proteins (32); Rpp30 is a TIM barrel (33); Rpp21 contains a zinc ribbon (34). Here we report the crystal structure of Pyrococcus furiosus Pop5 (Pfu Pop5), a 120-residue protein conserved both within and between Archaea and Eukarya (Fig. 1), and present as a paralog in multicellular eukaryotes as Rpp14 (35). NMR chemical shift perturbations have allowed us to identify the site of interaction with Pfu Rpp30. Pfu Pop5 is unique among the archaeal RNase P proteins in that its α–β sandwich bears structural similarity to the bacterial RNase P protein. With the observation that the Pfu Pop5 structure is homologous to the ubiquitous single-stranded nucleic acid binding RNA recognition motif (RRM) domain (reviewed in ref. 36), these data provide clues regarding the evolution and function of RNase P.
Fig. 1.
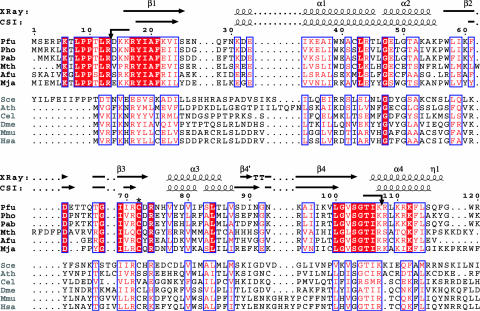
Sequence conservation and secondary structure of Pfu Pop5. Sequence alignment of Pfu Pop5 with sequences from Archaea (black) and Eukarya (gray). Sequences are from Pyrococcus furiosus (Pfu, NCBI code accession no. NP_579107), Pyrococcus horikoshii (Pho, NCBI code NP_143344), Pyrococcus abyssi (Pab, NCBI code NP_126368), Methanothermobacter thermoautotrophicus (Mth, NCBI code NP_275830), Archaeoglobus fulgidus (Afu, NCBI code NP_069325), Methanococcus jannaschii (Mja, NCBI code NP_247470), Saccharomyces cerevisiea (Sce, NCBI code NP_009369), Arabidopsis thaliana (Ath, NCBI code NP_683274), Caenorhabditis elegans (Cel, NCBI code NP_499477), Drosophila melanogaster (Dme, NCBI code NP_648955), Mus musculus (Mmu, NCBI code NP_080674), and Homo sapiens (Has, NCBI code NP_057002). Alignment was generated with clustalw (64) and colored according to similarity using a Risler scoring matrix (65); shaded residues indicate identity, and boxed residues indicate a global similarity score >0.7. Similarity scores within archaea were calculated separately. Secondary structure elements observed in the crystal structure as assigned by dssp (66) and elements predicted by chemical shift index (CSI) analysis (37) are indicated. Arrows indicate sites of trypsin cleavage as identified by digestion followed by ESI mass spectrometry (expected mass, 10,611 Da; observed, 10,610 Da). The figure was generated by using espript (67).
Results
Characterization. The Pfu Pop5 construct used consists of 120 wild-type residues (Fig. 1) with a single Cys to Ser point mutation at residue 72 that improved solution behavior as assayed by NMR and in vitro reconstitution experiments (H.-Y. Tsai, unpublished results). The N-terminal Met is removed during expression in E. coli as confirmed by ESI mass spectrometry (expected mass, 13,708 Da; observed, 13,707 Da). Dynamic light scattering data indicate that even the mutant protein tends to aggregate to approximately an octamer at 1 mM concentration, but behaves as a monomer at 300 μM concentration. Limited trypsin proteolysis experiments indicate that 12 and 13 residues at the N and C termini, respectively, are removed within 30 min, whereas the remaining core was not sensitive to digestion. Mass spectrometric analysis of the tryptic fragments revealed a mass of 10,610 Da, consistent with the 10,611 Da expected for residues 14–107, as indicated in Fig. 1.
NMR Spectroscopy and Resonance Assignments. Two-dimensional 1H-15N-correlated NMR spectra recorded at 22°C exhibited poor signal-to-noise and dispersion, indicating that the protein is poorly folded or aggregated at this temperature. Despite good dispersion and spectral quality in data collected at 55°C (see supporting information, which is published on the PNAS web site), relatively broad lines and dynamic light scattering experiments at elevated concentrations suggested that under those conditions Pfu Pop5 was aggregating, with an average mass of 117 kDa, not the expected 13.7 kDa monomer. Spectra of wild-type and C72S constructs were nearly identical, confirming that the overall fold of the mutant was unperturbed (data not shown). Backbone resonance assignments were obtained for 90% of the nonproline residues using standard triple resonance techniques. Only five residues could be assigned for the N-terminal 16 residues; heteronuclear 15N-{1H} NOE data suggested that the N terminus was dynamically disordered on time scales faster than overall tumbling, whereas the rest of the protein appeared to be well ordered, with an average NOE value of 0.81 (see supporting information); NOE values dipped slightly in the loops and the C-terminal helix. Analysis of C, N, HN, Cα, Cβ, and Hα chemical shifts (37, 38) predicted a βαββαβα secondary structure pattern for residues 18–115 (Fig. 1).
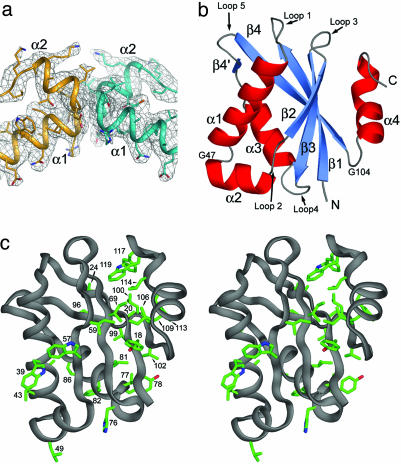
Crystal Structure. Crystals of Pfu Pop5 belong to the P41212 space group and have five monomers in the asymmetric unit. The crystal structure (Fig. 2) was determined by single isomorphous replacement from data collected on a crystal soaked with ethylmercuric phosphate, combined with solvent flattening and fivefold noncrystallographic symmetry averaging. The secondary structure pattern obtained from NMR analysis was used as a guide in tracing the backbone; there was excellent agreement between this predicted pattern and the observed fold, and a model was readily built into the electron density map by using o (39) (Fig. 2a; see supporting information). For each of the five molecules, interpretable density could be observed for residues 15–120. The absence of density at the N terminus is apparently due to local disorder, which is corroborated by the heteronuclear NOE and limited trypsin proteolysis data. The Pfu Pop5 structure was refined to 3.15 Å with Rworking and Rfree values of 22.90% and 25.66%, respectively.
Fig. 2.
Crystal structure of Pfu Pop5. (a)2Fo – Fc electron density map of the α-helical hairpin interface observed between neighboring molecules in the asymmetric unit, contoured at 1.0 σ. (b) Ribbon diagram of Pfu Pop5, colored according to secondary structure as assigned by DSSP (66). (c) Stereo view of Pfu Pop5 showing exposed aromatic and apolar side chains. Images were generated by using pymol (www.pymol.org).
Pfu Pop5 adopts an α-β sandwich fold comprised of a central, four-stranded antiparallel β-sheet surrounded by four α-helices (Fig. 2b). One face of the β-sheet packs with helices α1, α2, and α3 to form the hydrophobic core. The other face is more exposed to solvent, but features three exposed apolar side chains that loosely pack with six apolar side chains from helix α4 (Fig. 2c). At the end of helix α4, residues 114–116 adopt a 310 helix (η1) conformation followed by a short loop, allowing the side chains of Phe-117 and Trp-119 to be partially packed against the apolar side chains of the β-sheet (Fig. 2c). β-bulges perturb strands β2 and β4 from ideal β-strand geometry at residues 58–59 and 99–100, respectively. Poorly conserved loops (Fig. 1) connect secondary structure elements; the longest are Loop 1 (residues 24–32), which connects strand β1 to helix α1, and loop 5 (residues 87–96), connecting helix α3 to strand β4 and containing the two-residue strand β4′ (Fig. 2b).
Gly-47 is invariant throughout the Pop5 family (Fig. 1) and facilitates the sharp turn between helices α1 and α2 (Fig. 2a), with dihedral angles unfavored for non-glycine residues (φ = 79, ψ = 170). Loop 5 contains Gly-104, which is conserved between Archaea and Eukarya, sometimes present at the equivalent position 103 (Fig. 1). Strict conservation of this glycine does not seem to be due to backbone stereochemical restrictions, because its dihedral angles (φ = –57, ψ = –48) are within the most favored α-helical region of the Ramachandran plot. Many aromatic side chains are exposed to solvent on both faces of the central β sheet: Trp-39, Trp-57, His-76, Tyr-78, Phe-82, Phe-113, Phe-117, and Trp-119 (Fig. 2c).
The intermolecular interactions contributing to crystal matrix formation are noteworthy; every molecule in the asymmetric unit forms a small, symmetric dimer interface at the junction between helices α1 and α2 (residues 44–49) (Fig. 2a). Also, for all monomers in the asymmetric unit, another dimer interface is observed near helix α3 and strand β4, completely or mostly burying five hydrophobic residues (Tyr-78, Ile-81, Phe-82, Val-99, and Val-102) from helix α3 and strand β4 in each interacting monomer. The lowest B-factor values are found in helices α1, α2, and α3, and strand β4.
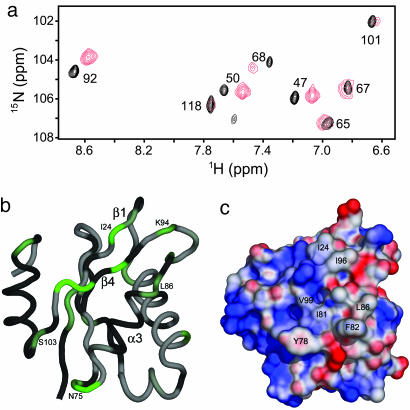
Interaction with Pfu Rpp30. Two-dimensional 1H-15N correlated NMR spectra of Pop5 in complex with Rpp30 were recorded to examine the interaction between these two proteins implied by yeast two-hybrid (26, 27) and partial reconstitution experiments (H.-Y. Tsai and V. Gopalan, personal communication). Large chemical shift perturbations for specific residues in Pfu Pop5 (see Fig. 4a) and complementary perturbations in the spectra of Pfu Rpp30 (unpublished data) are indicative of a high-affinity specific interaction between the two proteins. Although the details of the intermolecular interface remain to be elucidated, the most strongly perturbed amide resonances belonged to residues located principally in helix α3, strand β4 and the end of strand β1 (Fig. 4b).
Fig. 4.
Binding of Pfu Pop5 to protein partner Pfu Rpp30 as detected by NMR. (a) Overlay of an expanded region of two-dimensional 15N-1H correlated NMR spectra of Pfu Pop5 free (black) and in complex with Pfu Rpp30 (red). Perturbations of peak positions identify Pfu Pop5 residues involved in the interaction. (b) Binding-induced chemical shift perturbations mapped onto the Pop5 backbone (rotated 180° from the orientation in Fig. 2b), using a linear color ramp from white (less than the mean) to green (two standard deviations above the mean). Perturbation values were calculated by using scaled and summed differences in the 1H and 15N dimensions (68). Residues for which no information could be determined (because of spectral overlap or low peak intensity in the control spectrum) are shown in gray. (c) Electrostatic potential mapped onto the surface of the Pfu Pop5 structure, shown in the same orientation as b. The electrostatic potential was generated by using the apbs software (69) (100 mM monovalent cation; solvent dielectric constant, 80; protein dielectric constant, 20; probe radius, 1.4 Å; T = 310 K), with the surface colored by using a linear color ramp from –4.0 kT (red) to 4.0 kT (blue).
Discussion
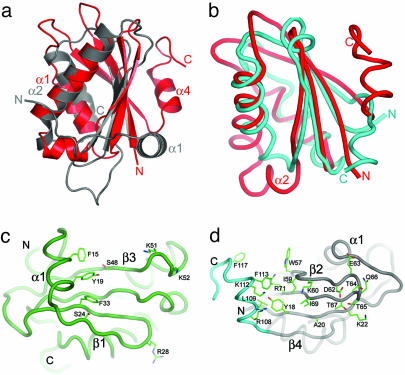
Pop5 Resembles the Bacterial RNase P Protein Subunit. Remarkably, the three dimensional arrangement of the structural elements that compose the α–β sandwich fold of Pfu Pop5 is similar to that of the bacterial RNase P proteins from Bacillus subtilis, Staphylococcus aureus, and Thermotoga maritima (40–42) (Fig. 3a). The similarity is particularly evident upon aligning helix α1 of Pop5 with helix α2 of the bacterial protein, which contains the conserved “RNR motif” and has been shown by tethered Fe-EDTA footprinting to be in close proximity to the bacterial catalytic RNA subunit (12). With this alignment, it can be seen that the principal difference in the three-dimensional organization of their structural elements is in the orientation of helix α4 of Pop5 and its corresponding helix α1 in the bacterial proteins; notably, this helix is poorly packed against the central sheet suggesting it could be readily rearranged. The high degree of structural similarity is particularly remarkable considering the proteins' differing primary sequence and secondary structural topology, βααββαβα in Pfu Pop5 versus αβββαβα in the bacterial RNase P proteins, suggesting that the two proteins have different evolutionary origins.
Fig. 3.
Similarity of Pfu Pop5 to the bacterial RNase P protein and the RRM motif. (a) Similarity of three-dimensional structures of Pfu Pop5 (red) and the bacterial RNase P protein (gray; Bacillus subtilis; Protein Data Bank code 1A6F) (40); the structures were superimposed by aligning residues 31–44 in helix α1 of Pop5 with residues 60–73 in helix α2 of 1A6F. Note that the secondary structural elements are arranged in a different order: βααββαβα in Pfu Pop5 versus αβββαβα in the bacterial RNase P proteins. (b) Ribbon diagrams of Pfu Pop5 (red) and Homo sapiens U1A RRM1 (cyan; 1NU4) (44). (c) The Staphylococcus aureus RNase P protein, with sticks shown for residues identified as being involved in RNA interactions by chemical shift perturbation or crosslinking (6, 41). (d) Speculative model of Pfu Pop5 with the C-terminal helix (cyan) reoriented as for the bacterial RNase P protein, revealing the putative RNA-binding surface of Pfu Pop5. Apolar and positively charged side chains protrude from the surface of the β-sheet. Such an orientation of helix α4 would allow access by single stranded RNA to the analogous aromatic and hydrophobic residues.
Pfu Pop5 Adopts the RRM Fold. Comparative fold analysis using dali (43) revealed that Pfu Pop5 is a structural homolog of one of the most abundant protein domains in eukarya, the RRM, also known as the ribonucleoprotein domain (RNP) or the RNA-binding domain. RRM domain-containing proteins are prevalent in all posttranscriptional events, including RNA processing, splicing, and editing (reviewed in ref. 36). DALI identified the N-terminal RRM domain of U1A (44) as a close structural homolog of Pfu Pop5 (Fig. 3b), with an rms deviation of 2.7 Å over 76 equivalent Cα atoms (Z score 5.9). In addition to a central β1α1β2β3α2β4 fold, the RRM domain is identified by conserved residues in the central strands β3 and β1, termed RNP1 and RNP2, respectively (45–47). The RRM α-β sandwich fold is often accompanied by N- and C-terminal extensions that are disordered in the free protein, yet capable of aiding nucleic acid binding (36). Residues 17–103 of Pfu Pop5 embody a classic RRM domain, with the exception of the insertion of the short helix α2 and the presence of longer β-strands than those of a typical RRM. Despite tertiary structure similarity, sequence analysis using blast (48) did not identify Pfu Pop5 as a putative RRM; indeed, when canonical residues in strands β3 and β1 are compared to the corresponding RNP1 and RNP2 consensus sequences, there is little similarity (see supporting information). Other RRM domains, such as the one in ALY (49) and the C-terminal RRM domain of La (50), show considerable divergence from the consensus sequences while maintaining typical RRM architecture.
A Putative Functional Role for Pop5. Based on its membership to the RRM family of single-stranded RNA-binding domain and structural similarity to bacterial RNase P protein subunit, we propose that Pop5 may function in precursor tRNA substrate binding. Single-stranded RNA binding by the RRM motif and the bacterial RNase P protein is accomplished by using aromatic and apolar side chains exposed from the central β-sheet (see supporting information) (6, 36, 41). Indeed, the exposed face of the β-sheet of Pfu Pop5 does form a tract populated with aromatic (Tyr-18, Trp-57) and hydrophobic residues (Ile-59, Ile-69) that compose a comparable RNA–RNA-binding surface (Fig. 3d). In the crystal, however, this putative RNA-binding surface is partially obscured by the C-terminal helix α4 (Fig. 2b), which would have to rearrange to allow RNA binding. Such conformational changes in an RRM domain are not without precedent, as the C-terminal residues of U1A RRM1 reorient in an induced fit manner upon binding to its RNA substrate (36, 51). Although the heteronuclear NOE values for residues in the C-terminal helix (see supporting information) indicate the helix is not highly flexible, propensity for conformational change by this helix is suggested by the observation that it, like the flexible N terminus, is readily severed at Lys-107 by limited trypsin proteolysis. A conserved glycine preceding this helix (Gly-104; Fig. 1) could serve as a hinge, allowing the helix to move, exposing the hydrophobic residues on the face of the β-sheet. Given this precedent, it is not unreasonable to imagine that, upon holoenzyme assembly, the C-terminal helix of Pfu Pop5 could adopt a position analogous to that of helix α1 of the bacterial RNase P protein (Fig. 3a), where the pre-tRNA substrate binds to functionally important hydrophobic residues in the central β sheet (5). Moreover, such a reorientation would position three aromatic side chains (Tyr-18, Phe-113, and Phe-117) similarly to those known to be involved in RNA binding in the Bsu RNase P protein (Tyr-34, Phe-20, and Phe-16, respectively) (Fig. 3d) (6, 7, 40).
Of the known archaeal RNase P proteins, only Pop5 bears recognizable similarity to the highly conserved bacterial protein subunit, and yet the bacterial protein structure is highly conserved, even between organisms with highly divergent RNase P enzymes (40–42). Furthermore, the bacterial protein has been shown capable of hetero-reconstituting the activity of archaeal RNase P RNA (16). The highly conserved three-dimensional structures of the bacterial protein, despite variations in the RNA subunits, suggest that the conserved shape is dictated by its functional role of binding both the catalytic RNA and the substrate precursor-tRNA. Consequently, the global similarity between Pop5 and the bacterial protein allows us to postulate that, in archaeal and eukaryotic RNase P, Pop5 is fulfilling a role similar to that of its bacterial counterpart.
The structural similarity of Pfu Pop5 to the bacterial protein subunit suggests it functions in an analogous, if imperfect, manner. The divergent secondary structure topologies of the proteins, βααββαβα in Pfu Pop5 versus αβββαβα in the bacterial RNase P proteins, suggest that the archaeal and bacterial proteins have different evolutionary origins. Two plausible evolutionary scenarios are worth considering: in one, ancestral catalytic RNA subunits in bacteria and archaea could have independently recruited similarly shaped α–β sandwich proteins; alternatively, a Pop5-like protein with sufficiently similar shape may have been able to usurp the role of the single bacterial protein, promoting RNase P catalysis and proliferating throughout the archaeal and eukaryal domains. Plausible functional roles for the other protein subunits can be proposed by hypothesizing that the evolutionary increase in RNase P protein content from bacteria to archaea to eukarya is that the smaller RNA subunits found in archaea and eukarya depend on a concomitant increase in protein content to stabilize a catalytically active conformation.
Pop5, One of Several Protein Subunits of Archaeal and Eukaryotic RNase P. Yeast two-hybrid experiments with eukaryotic and archaeal homologs suggested that Pop5 interacts with Rpp30 (26–29). Furthermore, Pfu Pop5 and Pfu Rpp30 form a functional pair that together can partially reconstitute Pfu RNase P (in the absence of the other proteins, Rpp21 and Rpp29) (H.-Y. Tsai and V. Gopalan, personal communication). The finding that a truncated version of the RNase P RNA missing the specificity domain (P11, P12, and the loop regions) can also be reconstituted with this pair of proteins suggests that these proteins are associating with the catalytic portion of the RNase P RNA and/or are involved in substrate binding. A crosslinking study performed by using the homologous human components presents compelling evidence that Rpp30 binds the catalytic RNA subunit (52), although Pop5 did not crosslink to the central RNA when tested in the same study.
NMR chemical shift perturbations (Fig. 4a) indicate that the Rpp30-contacting surface can be located to a patch of hydrophobic residues constituted largely by helix α3, strand β4, and the end of strand β1, on the nearly opposite side of Pop5 as the putative single-stranded RNA-binding face (Fig. 4 b and c). This is not an unlikely binding interface; a cluster of solvent-exposed hydrophobic side chains (Tyr-78, Ile-81, Phe-82, Leu-86, Ile-96, and Val-99) on this surface of Pop5 might reasonably be expected to recognize a similar hydrophobic patch of Rpp30 in binding (Fig. 4c). This finding is consistent with the structural similarity of Pop5 to the bacterial proteins and their mode of interaction with the RNA subunit and substrate (6, 12, 40, 41). That is, assuming the single-stranded 5′ leader of the precursor tRNA is bound in the opened β sheet of Pop5 as in the bacterial protein, and that helix α1 contacts the catalytic RNA subunit (as is done by the analogous helix α2 in the bacterial protein), the only available surface for Rpp30 binding is that in which the largest chemical shift perturbations are observed in the spectra of Pop5 when Rpp30 binds to it.
Summary. With the elucidation of the Pfu Pop5 structure, a functional role for the protein can be hypothesized based on its structural similarity to the bacterial RNase P protein and the RRM. Additionally, NMR chemical shift perturbations have revealed the surface of Pfu Pop5 used in binding to its protein partner Pfu Rpp30. These data provide clues regarding the role of Pop5 in the archaeal and eukaryal RNase P enzymes, although further work will be necessary to determine how all of the protein subunits assemble onto the RNase P RNA and help it perform catalysis, ultimately providing insight into the structural and functional connections between the three domains of life of this conserved yet compositionally variable enzyme.
Materials and Methods
Sample Preparation. The Pfu Pop5 gene (PF1378 from Pyrococcus furiosus DSM 3638) was amplified by PCR with Pfu genomic DNA as the template and gene-specific primers and cloned into pET-33b. Clone authenticity was established by using automated DNA sequencing. Initial structural work with the wild-type protein was hindered by precipitation at ≈1 mM concentration. Mutation of cysteine residues 42 and 72 to serine singly and as a double mutant using a protocol modified from the QuikChange Site-Directed Mutagenesis kit (Stratagene) resulted in three mutants with improved solubility. The C72S mutant was used for all experiments and is referred to as “Pfu Pop5”; the wild-type protein is referred to explicitly when applicable. The protein was expressed in Escherichia coli Rosetta (DE3) cells (Novagen) and purified from both the soluble and insoluble fractions by selective denaturation and ion exchange chromatography (see supporting information). Trypsin (Sigma) was used to probe the protein for regions accessible to proteolysis; 10% (wt/wt) trypsin was added to 700 μM Pfu Pop5 at room temperature, and the reaction was terminated at 30 min by addition of 1 mg/ml Pefabloc (Roche) and frozen until analyzed by ESI mass spectrometry (Q-Tof II, Micromass, Manchester, U.K.). The diffusion coefficient of Pop5 was assayed at differing concentrations by using a DynaPro-801 dynamic light scattering/molecular sizing instrument (Protein Solutions).
Pfu Rpp30 (PF1914) was prepared by subcloning the proteincoding gene from pET33b into the pHMT vector for bacterial expression as a fusion to His6-tagged maltose binding protein (53), purified by immobilized metal-affinity chromatography and processed with tobacco etch virus protease (54) (see supporting information). The Pop5-Rpp30 complex was formed by titrating Pop5 into Rpp30 in 10 mM sodium acetate, pH 5.0/0.5 mM KCl to final protein concentrations of 260 μM for Pop5 and 280 μM (a slight excess) for Rpp30.
NMR Spectroscopy. NMR spectra were recorded at 55°C on 600- and 800-MHz Bruker (Billerica, MA) Avance DMX and DRX spectrometers equipped with triple-resonance probes and pulsed-field gradients. NMR data processing and analysis were performed with nmrpipe (55) and nmrview (56). NMR samples typically contained 300–600 μM Pfu Pop5 and 20 mM sodium acetate (pH 5.0), 0.5 mM KCl, 50 μM NaN3, and 95% H2O/5% D2O. Backbone resonance assignments were made by using standard triple resonance spectra [HNCACB, CBCA(CO)NH, and HNCO] (57, 58) recorded at 600 MHz. Chemical shift perturbations were analyzed by comparing peak positions in two-dimensional heteronuclear 1H-15N correlated and three-dimensional HNCO spectra of Pop5 in the absence and presence of Rpp30: the peaks in highly overlapped regions or peaks with weak intensity in the Rpp30-sample were not included in analysis; the peaks corresponding to residues 97, 99, and 100 unambiguously vanished upon formation of the complex and were assigned the largest perturbation value. Signals were referenced to the residual water signal and adjusted for statistical compliance (59).
Crystallization. Crystals of Pfu Pop5 C72S were grown at room temperature by hanging drop vapor diffusion. The reservoir solution consisted of 250 mM imidazole (pH 6.5) and the hanging drop was prepared by mixing 1 μl of reservoir solution with 1 μ l of 28 mg/ml protein in 10 mM sodium acetate (pH 5.0). A mercury derivative was prepared by adding 0.25 μl of 10 mM ethylmercuric phosphate to a 1-μl preequilibrated drop with a crystal (2 mM final concentration) and soaking for 24 h.
X-Ray Data Collection, Structure Determination, and Refinement. Crystals were mounted in 0.7-mm glass capillaries (Hampton Research). X-ray diffraction data for the native and mercury-soaked crystals were collected at room temperature using a Rigaku rotating anode generator and an R-AXIS-IV++ image plate detector. Reflection intensities were integrated and scaled using CrystalClear (Molecular Structure), and processed further by using the truncate module of the ccp4 program suite (60, 61) (see supporting information). Five heavy atom positions were located by using a direct methods heavy atom search procedure (RANTAN), and initial phases were calculated with mlphare. The phases were improved at 3.8-Å resolution by using the solvent flipping mode of solomon with a solvent fraction of 65%, and the resulting electron density map allowed a polyalanine trace to be built using the program o (39). After transforming the polyalanine model for the first monomer to the other four positions in the asymmetric unit, the phases were further improved and extended to 3.15-Å resolution by using solvent flattening and fivefold averaging protocols in DM. The resulting map was refined by using rigid-body, torsion-angle simulated annealing, and grouped B-factor protocols of cns (62). Tight fivefold noncrystallographic symmetry restraints were imposed; releasing the restraints led to an increase in the Rfree value. Little or no density was observed beyond Cβ of side chains of residues Arg-74, -108, -111, -120, Lys-15, -31, -60, -107, Gln-28, -116, and Glu-36, and their disorder is reflected in high B-factor values. Stereochemical quality of the model was assessed by using procheck (63).
Supplementary Material
Acknowledgments
We thank Venkat Gopalan (Ohio State University) and members of his group for stimulating discussions and invaluable resources, members of the Foster laboratory for stimulating discussions and assistance with NMR data collection and analysis, the Ohio State University (OSU) Campus Chemical Instrument Center staff for assistance with NMR and mass spectrometric instrumentation, and the OSU Plant–Microbe Genomics Facility for DNA sequencing. We particularly thank Michael W. W. Adams (University of Georgia, Athens), Jamie R. Williamson (The Scripps Research Institute, La Jolla, CA), and David S. Waugh (National Cancer Institute, Bethesda) for valuable reagents. This work was funded in part by National Institutes of Health Grant GM067807 (to M.P.F. and Venkat Gopalan). R.C.W. and C.J.B. acknowledge support from the National Science Foundation through Research Experiences for Undergraduates Grant MCB-0092962 (to M.P.F.) and from the Department of Biochemistry, and R.C.W. acknowledges support from National Institutes of Health Chemistry/Biology Interface Training Grant T32GM008512. C.E.B. acknowledges support from National Institutes of Health Grant GM067947.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: RRM, RNA recognition motif; Pfu, Pyrococcus furiosus.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID 2AV5). The NMR chemical shifts have been deposited in the BioMagResBank, www.bmrb.wisc.edu (accession no. 6917).
References
- 1.Altman, S. & Kirsebom, L. (1999) in The RNA World, eds. Gesteland, R. F., Cech, T. R. & Atkins, J. F. (Cold Spring Harbor Laboratory Press, Plainview, NY), pp. 351–380.
- 2.Harris, M. E., Frank, D. N. & Pace, N. R. (1998) in RNA Structure and Functions, eds. Simons, R. W. & Grunberg-Manago, M. (Cold Spring Harbor Laboratory Press, Plainview, NY), pp. 309–338.
- 3.Guerrier-Takada, C., Gardiner, K., Marsh, T., Pace, N. & Altman, S. (1983) Cell 35, 849–857. [DOI] [PubMed] [Google Scholar]
- 4.Schedl, P. & Primakoff, P. (1973) Proc. Natl. Acad. Sci. USA 70, 2091–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gopalan, V., Baxevanis, A. D., Landsman, D. & Altman, S. (1997) J. Mol. Biol. 267, 818–829. [DOI] [PubMed] [Google Scholar]
- 6.Niranjanakumari, S., Stams, T., Crary, S. M., Christianson, D. W. & Fierke, C. A. (1998) Proc. Natl. Acad. Sci. USA 95, 15212–15217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kurz, J. C., Niranjanakumari, S. & Fierke, C. A. (1998) Biochemistry 37, 2393–2400. [DOI] [PubMed] [Google Scholar]
- 8.Buck, A. H., Dalby, A. B., Poole, A. W., Kazantsev, A. V. & Pace, N. R. (2005) EMBO J. 24, 3360–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torres-Larios, A., Swinger, K. K., Krasilnikov, A. S., Pan, T. & Mondragon, A. (2005) Nature 437, 584–587. [DOI] [PubMed] [Google Scholar]
- 10.Kazantsev, A. V., Krivenko, A. A., Harrington, D. J., Holbrook, S. R., Adams, P. D. & Pace, N. R. (2005) Proc. Natl. Acad. Sci. USA 102, 13392–13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Massire, C., Jaeger, L. & Westhof, E. (1998) J. Mol. Biol. 279, 773–793. [DOI] [PubMed] [Google Scholar]
- 12.Tsai, H. Y., Masquida, B., Biswas, R., Westhof, E. & Gopalan, V. (2003) J. Mol. Biol. 325, 661–675. [DOI] [PubMed] [Google Scholar]
- 13.Xiao, S., Houser-Scott, F. & Engelke, D. R. (2001) J. Cell. Physiol. 187, 11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forster, A. C. & Altman, S. (1990) Cell 62, 407–409. [DOI] [PubMed] [Google Scholar]
- 15.Brown, J. W. (1999) Nucleic Acids Res. 27, 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pannucci, J. A., Haas, E. S., Hall, T. A., Harris, J. K. & Brown, J. W. (1999) Proc. Natl. Acad. Sci. USA 96, 7803–7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall, T. A. & Brown, J. W. (2002) RNA 8, 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jarrous, N., Reiner, R., Wesolowski, D., Mann, H., Guerrier-Takada, C. & Altman, S. (2001) RNA 7, 1153–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarrous, N., Eder, P. S., Wesolowski, D. & Altman, S. (1999) RNA 5, 153–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eder, P. S., Kekuda, R., Stolc, V. & Altman, S. (1997) Proc. Natl. Acad. Sci. USA 94, 1101–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Eenennaam, H., Lugtenberg, D., Vogelzangs, J. H., van Venrooij, W. J. & Pruijn, G. J. (2001) J. Biol. Chem. 276, 31635–31641. [DOI] [PubMed] [Google Scholar]
- 22.van Eenennaam, H., Pruijn, G. J. M. & van Venrooij, W. J. (1999) Nucleic Acids Res. 27, 2465–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stolc, V. & Altman, S. (1997) Genes Dev. 11, 2926–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boomershine, W. P., McElroy, C. A., Tsai, H. Y., Wilson, R. C., Gopalan, V. & Foster, M. P. (2003) Proc. Natl. Acad. Sci. USA 100, 15398–15403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kouzuma, Y., Mizoguchi, M., Takagi, H., Fukuhara, H., Tsukamoto, M., Numata, T. & Kimura, M. (2003) Biochem. Biophys. Res. Commun. 306, 666–673. [DOI] [PubMed] [Google Scholar]
- 26.Hall, T. A. & Brown, J. W. (2004) Archaea 1, 247–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kifusa, M., Fukuhara, H., Hayashi, T. & Kimura, M. (2005) Biosci. Biotechnol. Biochem. 69, 1209–1212. [DOI] [PubMed] [Google Scholar]
- 28.Jiang, T. & Altman, S. (2001) Proc. Natl. Acad. Sci. USA 98, 920–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Houser-Scott, F., Xiao, S., Millikin, C. E., Zengel, J. M., Lindahl, L. & Engelke, D. R. (2002) Proc. Natl. Acad. Sci. USA 99, 2684–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sidote, D. J. & Hoffman, D. W. (2003) Biochemistry 42, 13541–13550. [DOI] [PubMed] [Google Scholar]
- 31.Sidote, D. J., Heideker, J. & Hoffman, D. W. (2004) Biochemistry 43, 14128–14138. [DOI] [PubMed] [Google Scholar]
- 32.Numata, T., Ishimatsu, I., Kakuta, Y., Tanaka, I. & Kimura, M. (2004) RNA 10, 1423–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takagi, H., Watanabe, M., Kakuta, Y., Kamachi, R., Numata, T., Tanaka, I. & Kimura, M. (2004) Biochem. Biophys. Res. Commun. 319, 787–794. [DOI] [PubMed] [Google Scholar]
- 34.Kakuta, Y., Ishimatsu, I., Numata, T., Kimura, K., Yao, M., Tanaka, I. & Kimura, M. (2005) Biochemistry 44, 12086–12093. [DOI] [PubMed] [Google Scholar]
- 35.Koonin, E. V., Wolf, Y. I. & Aravind, L. (2001) Genome Res. 11, 240–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maris, C., Dominguez, C. & Allain, F. H. (2005) FEBS J. 272, 2118–2131. [DOI] [PubMed] [Google Scholar]
- 37.Wishart, D. S., Sykes, B. D. & Richards, F. M. (1992) Biochemistry 31, 1647–1651. [DOI] [PubMed] [Google Scholar]
- 38.Wishart, D. S. & Sykes, B. D. (1994) J. Biomol. NMR 4, 171–180. [DOI] [PubMed] [Google Scholar]
- 39.Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 40.Stams, T., Niranjanakumari, S., Fierke, C. A. & Christianson, D. W. (1998) Science 280, 752–755. [DOI] [PubMed] [Google Scholar]
- 41.Spitzfaden, C., Nicholson, N., Jones, J. J., Guth, S., Lehr, R., Prescott, C. D., Hegg, L. A. & Eggleston, D. S. (2000) J. Mol. Biol. 295, 105–115. [DOI] [PubMed] [Google Scholar]
- 42.Kazantsev, A. V., Krivenko, A. A., Harrington, D. J., Carter, R. J., Holbrook, S. R., Adams, P. D. & Pace, N. R. (2003) Proc. Natl. Acad. Sci. USA 100, 7497–7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holm, L. & Sander, C. (1995) Trends Biochem. Sci. 20, 478–480. [DOI] [PubMed] [Google Scholar]
- 44.Rupert, P. B., Xiao, H. & Ferre-D'Amare, A. R. (2003) Acta Crystallogr. D 59, 1521–1524. [DOI] [PubMed] [Google Scholar]
- 45.Adam, S., Nakagawa, T., Swanson, M., Woodruff, T. & G, D. (1986) Mol. Cell. Biol. 6, 2932–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swanson, M., Nakagawa, T., LeVan, K. & G, D. (1987) Mol. Cell. Biol. 7, 1731–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dreyfuss, G., Swanson, M. & Pinol-Roma, S. (1988) Trends Biochem. Sci. 13, 86–91. [DOI] [PubMed] [Google Scholar]
- 48.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 49.Perez-Alvarado, G. C., Martinez-Yamout, M., Allen, M. M., Grosschedl, R., Dyson, H. J. & Wright, P. E. (2003) Biochemistry 42, 7348–7357. [DOI] [PubMed] [Google Scholar]
- 50.Jacks, A., Babon, J., Kelly, G., Manolaridis, I., Cary, P. D., Curry, S. & Conte, M. R. (2003) Structure (Cambridge, U.K.) 11, 833–843. [DOI] [PubMed] [Google Scholar]
- 51.Avis, J. M., Allain, F. H., Howe, P. W., Varani, G., Nagai, K. & Neuhaus, D. (1996) J. Mol. Biol. 257, 398–411. [DOI] [PubMed] [Google Scholar]
- 52.Jiang, T., Guerrier-Takada, C. & Altman, S. (2001) RNA 7, 937–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kapust, R. B. & Waugh, D. S. (1999) Protein Sci. 8, 1668–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fox, J. D., Kapust, R. B. & Waugh, D. S. (2001) Protein Sci. 10, 622–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J. & Bax, A. (1995) J. Biomol. NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- 56.Johnson, B. A. & Blevins, R. A. (1994) J. Biomol. NMR 4, 603–614. [DOI] [PubMed] [Google Scholar]
- 57.Cavanagh, J., Fairbrother, W. J., Palmer, A. G., III, & Skelton, N. J. (1996) Protein NMR Spectroscopy: Principles and Practice (Academic, San Diego).
- 58.Sattler, M., Schleucher, J. & Griesinger, C. (1999) Progr. Nucl. Magn. Reson. Spectrosc. 34, 93–158. [Google Scholar]
- 59.Wang, L., Eghbalnia, H. R., Bahrami, A. & Markley, J. L. (2005) J. Biomol. NMR 32, 13–22. [DOI] [PubMed] [Google Scholar]
- 60.Collaborative Computational Project 4 (1994) Acta Crystallogr. D 50, 760–763.15299374 [Google Scholar]
- 61.Potterton, L., McNicholas, S., Krissinel, E., Gruber, J., Cowtan, K., Emsley, P., Murshudov, G. N., Cohen, S., Perrakis, A. & Noble, M. (2004) Acta Crystallogr. D 60, 2288–2294. [DOI] [PubMed] [Google Scholar]
- 62.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 63.Laskowski, R., MacArthur, M., Moss, D. & Thornton, J. (1993) J. Appl. Cryst. 26, 283–291. [Google Scholar]
- 64.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Risler, J. L., Delorme, M. O., Delacroix, H. & Henaut, A. (1988) J. Mol. Biol. 204, 1019–1029. [DOI] [PubMed] [Google Scholar]
- 66.Kabsch, W. & Sander, C. (1983) Biopolymers 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- 67.Gouet, P., Courcelle, E., Stuart, D. I. & Metoz, F. (1999) Bioinformatics 15, 305–308. [DOI] [PubMed] [Google Scholar]
- 68.Grzesiek, S., Bax, A., Clore, G. M., Gronenborn, A. M., Hu, J. S., Kaufman, J., Palmer, I., Stahl, S. J. & Wingfield, P. T. (1996) Nat. Struct. Biol. 3, 340–345. [DOI] [PubMed] [Google Scholar]
- 69.Baker, N. A., Sept, D., Joseph, S., Holst, M. J. & McCammon, J. A. (2001) Proc. Natl. Acad. Sci. USA 98, 10037–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.