Abstract
The serotonergic system plays a key role in the regulation of brain states, and many of the known features of serotonergic neurons appear to match this function. Midbrain raphe nuclei provide a diffuse projection to all regions of the forebrain, and raphe neurons exhibit a slow metronome-like activity that sets the ambient levels of serotonin across the sleep–wake cycle. Serotonergic cells have also been implicated, however, in a variety of more specific functions that can hardly be related to their low-rate monotonous patterns of discharges. The amazing variety of serotonergic receptors and their type-specific distribution on cortical neurons also raise the possibility of a more intimate coordination between the activity of serotonergic neurons and their target cortical circuits. Here we report an unexpected diversity in the behavior of immunohistochemically identified serotonergic neurons. Two outstanding subpopulations were identified by using the in vivo juxtacellular recording and labeling technique. The first subpopulation of serotonergic cells exhibited the classic clock-like activity with no apparent short timescale interaction with the hippocampal electroencephalogram. The other subpopulation discharged action potentials that were phase-locked to the hippocampal theta rhythm, the oscillatory pattern associated with acquisition of information and memory formation. These results indicate that the ascending serotonergic system comprises cells involved in complex information processing beyond the regulation of state transitions. The heterogeneity of serotonergic neuron behavior can also help to explain the complexity of symptoms associated with serotonergic dysfunction.
Keywords: hippocampus, juxtacellular labeling, midbrain raphe, neuronal oscillations
The serotonergic system is a primary target of psychotherapy that stresses the importance of serotonin in normal and abnormal brain functioning. Serotonergic neurons of the midbrain raphe have been implicated in the control of affective and cognitive functions and in modulating the neural activities of networks across the sleep–wake cycle. Behaviorally related oscillations in the theta frequency range (4–8 Hz), which are most prominent in the hippocampus (1), are known to synchronize neuronal activity in a number of forebrain structures involved in these functions (2–4). Hippocampal theta oscillations occur selectively during exploratory behaviors and paradoxical sleep, whereas large-amplitude irregular activity is associated with quiet waking, consummatory behaviors, and slow-wave sleep (5, 6). As in sleep, hippocampal activity spontaneously alternates between theta and nontheta states under urethane anesthesia.
The majority of putative serotonergic neurons have been reported to be most active during waking, to decrease their firing rate during slow-wave sleep, and to cease firing during paradoxical sleep (7).∥ The silencing of serotonergic cells during paradoxical sleep is currently considered an important regulatory signal that contributes to the maintenance of the theta rhythm in this state. The role of serotonin in controlling theta during the waking state is less obvious, and it was proposed that serotonin might even promote (8) rather than suppress a certain type of theta rhythm characteristic for this state (5). In anesthetized animals, raphe stimulation disrupts spontaneous hippocampal theta rhythm, and pharmacological suppression of serotonergic neurons results in lasting theta in the hippocampus (9). Surprisingly, however, a negative correlation between hippocampal theta and raphe neuronal activity, which would be consistent with these findings, was found only in a minority (29%) of the slow-firing putative serotonergic cells (10). Moreover, the firing of a large population (up to 55%) of raphe neurons in both anesthetized (9–11) and freely behaving rats (12) appeared phase-locked to hippocampal field potential oscillations at theta frequency. However, raphe neurons with theta-related firing were identified only by indirect electrophysiological criteria, whose reliability was questioned in recent studies by using combined cellular electrophysiology and immunohistochemistry (13–15). Thus, the important question of how the seemingly homogeneous population of serotonergic neurons is able to participate in the wide range of brain activity remains unanswered.
Results
In the present study, we used the juxtacellular labeling technique (16), which allows a detailed analysis of single-cell activity and a confirmation of their neurochemical and morphological identity by subsequent immunohistochemistry (13, 17). Extracellular single-neuron recordings were performed in rats anesthetized with urethane in two different states: one associated with large-amplitude irregular activity in the hippocampus and the other characterized by the presence of a sustained hippocampal theta rhythm (henceforth called nontheta and theta states, respectively). Twenty-one neurons were successfully labeled in 20 rats. These neurons were selected (see Materials and Methods) to be representatives of all major raphe cell groups reported in previous studies (10, 13, 14, 18).∥ Thus, the sample included both theta-on and theta-off units, i.e., cells activated or suppressed during theta and neurons with a wide range of firing rates (0.3–25 spikes per sec) and with a variety of spike waveforms. Fifteen of these cells were identified as serotonergic [5-hydroxytryptamine + (5-HT+)] or nonserotonergic (5-HT–) neurons; the remaining six cells, the neurochemical identity of which remained obscured, were excluded from this study.
Ten labeled cells were found immunoreactive for serotonin, whereas the remaining five cells were serotonin-immunonegative. None of these neurons were tyrosine hydroxylase (TH) immunoreactive. Serotonin-immunoreactive cells were exclusively located in the raphe nuclei. This and the fact that 5-HT– immunonegative cell bodies (e.g., neurobiotin labeled or TH+, Fig. 2 A) were also observed in the raphe further confirmed the specificity of the 5-HT immunolabeling. Cell bodies were located in the dorsal raphe, dorsal raphe interfascilular part, or median raphe nuclei (n = 9, 2, and 4, respectively). All reconstructed neurons had an axon that gave off local axon collaterals before exiting the raphe nuclei. The electrophysiological features widely used in previous studies to identify “putative” serotonergic and nonserotonergic neurons (i.e., firing rates, coefficient of variation, and spike waveform characteristics) were highly variable among both 5-HT+ and 5-HT– cells (13, 14). As a group, serotonergic neurons did not significantly differ from nonserotonergic raphe neurons in any of these parameters (Table 1).
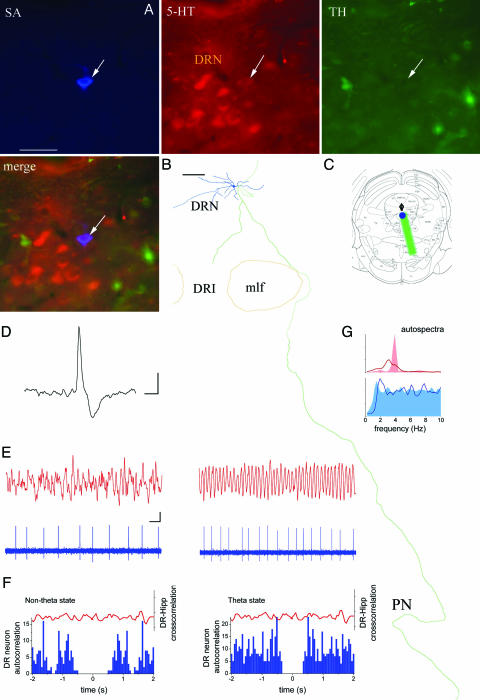
Fig. 2.
Firing pattern of a slow-firing serotonergic dorsal raphe neuron with clock-like activity. (A) The labeled neuron is located in the dorsal raphe nucleus (DRN). Blue staining shows the filled neurons visualized by streptavidin (SA), which expresses 5-HT (red) but not TH (green). Arrow indicates the position of the recorded and filled neuron. (B) Reconstruction of the dendritic (blue) and axonal (green) arborization. (C) General view of the soma location and axonal arborization (green) in the midbrain. (D) Wide (≈3.8-ms) asymmetric action potential of the cell. (E) Hippocampal EEG and DR neuron firing during nontheta and theta states. (F) Spike autocorrelation histogram (blue; bin = 0.05 s) with clock-like activity expressed more during nontheta states. Crosscorrelation (red) shows no relationship between DR unit and hippocampal field potentials. (G) Autospectra (0.24-Hz resolution) of hippocampal EEG (red) and DR unit activity (blue) during theta (filled area) and nontheta states (line). DRI, DRN interfascicular part; mlf, medial longitudinal fasciculus. [Scale bars: (A)50 μm; (B) 100 μm; (D) 2 ms and 2 mV; (E) 1 sec and 2 mV.]
Table 1. Spike-wave characteristics and firing rate of midbrain raphe neurons.
| Spike wave
|
Firing rate
|
||||
|---|---|---|---|---|---|
| Width of first and second peaks | Relative amplitude of first and second peaks | Theta | Nontheta | CoV | |
| 5-HT+ (n = 10) | 2.68 ± 0.30 | 2.20 ± 0.32 | 5.40 ± 1.95 | 4.69 ± 1.94 | 78 ± 11 |
| 5-HT- (n = 5) | 2.35 ± 0.17 | 1.50 ± 0.05 | 11.84 ± 5.59 | 4.90 ± 3.78 | 134 ± 40 |
| Broad (n = 4) | 3.63 ± 0.29 | 3.23 ± 0.15 | 0.90 ± 0.34 | 1.20 ± 0.21 | 54 ± 15 |
| Fast (n = 4) | 1.50 ± 0.04 | 1.01 ± 0.02 | 10.75 ± 4.60 | 8.50 ± 0.71 | 84 ± 19 |
| Others (n = 7) | 2.40 ± 0.37 | 1.51 ± 0.12 | 5.77 ± 8.05 | 1.37 ± 0.44 | 128 ± 93 |
| Bonferroni: | B—F, B—o | B—F, B—o | B—F | B—F, F—o | |
Bold letters indicate significant t test or significant effect of the group factor on the parameter (ANOVA P < 0.05); Bonferroni, pairs with significantly different means using post hoc Bonferroni test. CoV, coefficient of variance; B, broad; F, fast; o, others.
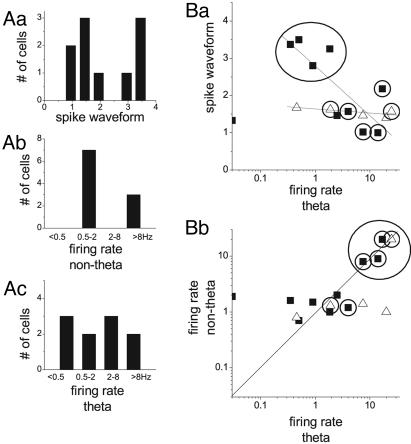
Serotonergic Neurons. The group of serotonergic neurons showed a substantial diversity in all examined parameters of firing rate, firing pattern, and spike shape. The firing rate varied in a wide range between 0.03 and 17 spikes per second during theta and showed a bimodal distribution during nontheta states (Fig. 1 Ab and Ac). The spike waveform characteristics, i.e., the spike duration and the relative amplitudes of the first and second peaks,∥ also showed clear bimodal distributions (Fig. 1 Aa). By using the multivariate distribution of these parameters (Fig. 1B and Table 1), two major serotonergic subgroups could be distinguished containing neurons with diagonally different discharge characteristics.
Fig. 1.
Diverse firing behavior of midbrain raphe neurons. (Aa–Ac) Distribution of the electrophysiological parameters of serotonergic neurons (n = 10) representing their spike waveform characteristics (relative amplitudes of the first and second peak of the action potential; Aa), and their average firing rates during nontheta (Ab) and theta states (Ac). Note bimodal distributions of the spike waveform parameter and of the nontheta firing rate and near-homogeneous wide distribution of firing rates during theta states. (B) Pairwise scatter plots showing the relationship between these parameters and also representing the 2D projections of the classes of raphe cells [10 serotonergic (filled squares) and 5 nonserotonergic neurons (open triangles)], identified by the multivariate distributions of the three electrophysiological parameters. Neurons with theta-related firing are marked with encircled symbols. (Ba) Separation of slow-firing serotonergic neurons (n = 4, all serotonergic) with clock-like activity (circled area). Note also significant negative correlation between spike waveform and firing rate during theta states for serotonergic cells (P = 0.01) and no correlation for nonserotonergic neurons (P = 0.22). (Bb) Separation of fast-firing neurons (three serotonergic, one nonserotonergic) with theta-rhythmic activity (circled area).
Slow-firing serotonergic neurons with broad action potentials. This subgroup of four cells corresponded to the “putative” serotonergic neurons of previous studies (see example in Fig. 2). The distinction from the other serotonergic cells was confirmed by a clear separation of their spike waveform characteristics (Fig. 1 Aa) and the slow metronome-like firing mode, which was significantly different from the behavior of both the remaining serotonergic and the group of nonserotonergic neurons. Significant coherence with the hippocampal theta rhythm was not observed in the activity of any of these cells, but theta-related change in firing rate could be detected in each of them (three off, one on).
Fast-firing serotonergic neurons with theta rhythmic activity. The three cells of this subgroup showed the spike-waveform and firing characteristics previously attributed to nonserotonergic neurons either in the raphe or in other brain regions. These cells fired at high rates in both theta and nontheta states (Fig. 1Bb) and created a distinct peak in the bimodal nontheta distribution histogram (>8 Hz; Fig. 1 Ab). All three neurons in this subgroup fired phase-coupled to the hippocampal theta rhythm (coherence 0.59–0.88). Fig. 3 provides a representative example of the electrophysiological behavior and immunohistochemical characteristics of this class of serotonergic cells.
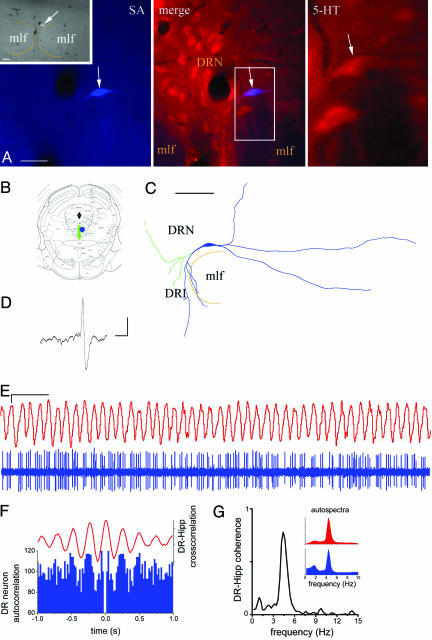
Fig. 3.
Firing pattern of a fast-firing serotonergic dorsal raphe neuron with theta-rhythmic activity. (A) The labeled neuron is located in the dorsal raphe nucleus (DRN). Inset shows a low-power image of the raphe nucleus. The recorded neuron (arrow) is 5-HT immunoreactive (red and merged images). (B) General view of the soma location and axonal arborization (green) in the midbrain. (C) Reconstruction of the filled cell showing the dendritic (blue) and axonal (green) arborization. (D) Narrow (≈1.5-ms) symmetric action potential of the cell. (E) Synchronized theta oscillations in the hippocampal EEG and DR unit spike train. (F) Spike autocorrelation histogram (blue; bin = 0.02 s) and hippocampus–raphe crosscorrelation (red) with regular peaks repeated at theta frequency (4.4 Hz). (G) Coherence spectrum with highly significant peak (0.78) at 4.4. Hz. (Inset) Autospectra (0.24-Hz resolution) of hippocampal EEG (red) and DR unit activity (blue) during theta state. DRI, DRN interfascicular part; mlf, medial longitudinal fasciculus; SA, streptavidin. [Scale bars: (A)50 μm; (Inset) 100 μm; (C) 100 μm; (D)2 ms and 2 mV; (E) 1 sec and 2 mV.]
The remaining three serotonergic cells fired at low rates (fewer than two spikes per sec; Fig. 1Bb) during nontheta states. These neurons were undistinguishable from the majority (four of five) of nonserotonergic neurons recorded in this study on the basis of electrophysiological parameters. Unlike the fast-firing serotonergic neurons, the discharge rates of these cells during the theta and nontheta states were not always related (Fig. 1Bb); i.e., neurons classified as slow firing during the nontheta states were either activated (theta-on) or suppressed (theta-off), associated with theta yielding a wide homogeneous distribution of firing rates (between 0.03 and four spikes per sec; Fig. 1Ba). One of these cells exhibited theta rhythmic firing (coherence, 0.54).
Nonserotonergic Neurons. These cells formed a heterogeneous group containing one fast-firing cell and four neurons with variable firing characteristics. The fast-firing neuron showed similar activity pattern and action potential waveform to the fast-firing serotonergic neurons. Significant theta coherence could also be detected in the case of this cell (coherence, 0.51). The remaining four neurons exhibited variable firing and spike shape undistinguishable from the third subgroup of serotonergic cells (Fig. 1B). During nontheta states, these neurons had low-frequency activity, which elevated or decreased further at the onset of hippocampal theta. One of these cells showed theta rhythmic activity (coherence, 0.35).
Discussion
As shown here, the diversity in the population of serotonergic neurons can be revealed only by combined analyses of their electrophysiological, immunochemical, and morphological properties. Electrophysiological criteria used to identify putative serotonergic neurons in previous studies may be appropriate for a gross classification of raphe cells, but they fail to identify subpopulations with atypical discharge properties. Recent intracellular (14, 15) and juxtacellular labeling (13) investigations reported that cell identification based on electrophysiological criteria alone yielded false-negative and -positive results. For example, Allers and Sharp (13) found that half of the slow-firing raphe neurons were nonserotonergic, and 2 of 10 fast-firing putative nonserotonergic neurons were in fact serotonergic. Although relatively small in numbers, these subsets represent real functional classes of cells, as indicated by their increase in number as the sample size increases [e.g., 18 of 180 fast-theta rhythmic neurons in the median raphe nucleus (10)]. In a recent study by Urbain et al.,∥ 117 of 531 (i.e., 22%) putative serotonergic neurons did not show the expected “classic” decrease in firing from waking to slow-wave sleep, and a significant number of such cells were active during paradoxical sleep.
The two activity patterns of serotonergic neurons found in this study may have different functions, which can be most effectively mediated by different types of receptors expressed in cortical networks. The slow clock-like activity seems optimal for maintaining state-dependent stable serotonin levels and requires the slow action of G protein-coupled serotonin 1A and 2A receptors. The extrasynaptically located inhibitory serotonin 1A receptor on pyramidal cell dendrites (19) can be activated by serotonin released not only through spillover from synaptic terminals but also from nonsynaptic varicosities, resulting in further reduction of the temporal and spatial precision of the serotonergic control. Activation of the excitatory G protein-coupled serotonin 2A receptors expressed by GABA-expressing (GABAergic) interneurons (20) was also shown to have a long-lasting effect on pyramidal neurons (21, 22).
The fast excitatory ion-channel-coupled serotonin 3 receptors are capable of following the theta rhythmic serotonergic input. These receptors are expressed by several classes of GABAergic interneurons (23–25), which have specific functions. Thus, cholecystokinin-containing basket cells, which are also endowed by nicotinic and cannabinoid receptors, are credited for conveying emotional-, mood-, and motivational-related inputs to the network of pyramidal cells (26, 27). Serotonin 3 receptors are also expressed on calbindin- and calretinin-containing GABAergic cells (28), which control not only pyramidal cells but other hippocampal inhibitory cells as well (29, 30). Serotonergic fibers do not contact the parvalbumin-containing GABAergic basket cells, which are responsible for generating the synchronized somatic membrane potential oscillations in pyramidal cells. Therefore, the rhythmic serotonergic input is unlikely to drive hippocampal theta field potentials. Rhythmically synchronized firing, however, may provide a precisely timed arrival of the information carried by this phasic input to the oscillating target networks not only in the hippocampus but also in other structures, including entorhinal, prefrontal cortex, and amygdala (2–4). Importantly, this serotonergic input is active during theta states when the bulk of the ascending serotonergic pathway is suppressed.∥
The large number and the variety of neurons exhibiting theta-related activity in the midbrain raphe (12) indicate that theta oscillation also plays an important role in organizing the neuronal activity within the raphe circuit. Rhythmically firing serotonergic neurons have local axon collaterals and can therefore participate in suppressing the activity of slow-firing neurons either through serotonin 1A autoreceptors or through a local serotonin 2 receptor-activated GABAergic circuit (31). The rhythmically firing nonserotonergic neurons may also comprise an essential part of this network (32, 33).
Materials and Methods
Experiments were performed on male Sprague–Dawley rats anesthetized with urethane (1.2–1.5 g/kg), in accordance with National Institutes of Health guidelines.
Juxtacellular Recording and Labeling. Hippocampal electroencephalogram (EEG) was recorded with two pairs of twisted wires (125-μm stainless steel) separated by 1 mm at their tips and implanted in the CA1 region and at the hippocampal fissure, verified by the out-of-phase rhythmicity in the two recordings. The traces of hippocampal EEG along with their spectra and the power within the theta range (2–8 Hz) was continuously monitored during the experiment. Juxtacellular recordings were made with fine micropipettes (impedance, 30–60 MOhm) filled with 2% Neurobiotin in 0.5 M NaCl. Each neuron was recorded during control, associated with low-amplitude irregular activity in the hippocampus and during theta episodes that occurred either spontaneously or in response to sensory stimulation (tail pinch). The neuronal signal was amplified by a DC-coupled amplifier, filtered between 0.3 and 5 kHz, and, along with hippocampal EEG, digitized at 10 kHz. Once a unit had been isolated, the activity was recorded during theta and nontheta states followed by juxtacellular dye injection by using the application of positive current pulses (2–8 nA, 200 ms duration, 50% duty cycle) for 5–10 min (13, 16, 17).
Given the great variety of neurons in the midbrain raphe and the relatively low yield of juxtacellular labeling (one neuron per rat), the sampling was intentionally biased to collect at least a few representative units from all major cell groups reported in previous studies. Special effort was made to include units that, on visual inspection during the experiments, appeared to change their firing patterns in relation to the simultaneously recorded hippocampal EEG (10–12). Thus, the cells submitted for juxtacellular labeling were selected from a total of 162 neurons extracellularly recorded in 57 rats in the stereotaxic location of the midbrain raphe nuclei (7–9 mm caudal to bregma on the midline, between 5.0 and 8.3 mm from the surface of the brain). Juxtacellular labeling was attempted in 43 experiments. One neuron was filled with neurobiotin in each rat, except for two experiments, in which two neurons separated by ≈1.2 mm were labeled. In 14 rats, the labeled cell was outside the raphe or no cell was found. Another nine experiments were also excluded from the analysis due to ambiguous results, i.e., faint or multiple labeling (13, 16, 17).
Tissue Preparation and Immunohistochemistry. Rats were perfused with 0.9% saline followed by a fixative containing 4% paraformaldehyde in phosphate buffer (0.1 M, pH 7.4). Brains were removed, and sections were cut with a vibratome, washed with TBS (0.05 M, pH 7.4; Sigma-Aldrich), and blocked with a solution containing 2% BSA and 0.5% Triton X-100. Sections were incubated overnight in solution containing Marina blue-conjugated streptavidin (1:200; Molecular Probes) and analyzed under fluorescent microscope. Cell body- and primary dendrite-containing sections were incubated for 48 h at 4°C with primary antisera against serotonin (rat anti-5-HT; Chemicon, 1:100; ref. 34), TH (mouse anti-TH; Chemicon, 1:100), washed, and incubated in carbocyanine 3 (CY3)- or CY2-conjugated anti-rabbit, -mouse, or -rat secondary antibodies (1:500; The Jackson Laboratory). Sections were examined under fluorescence light microscope (Provis AX70; Olympus, Melville, NY) or confocal microscope (Zeiss LM 510). At the end of the experiment, sections were rinsed in TBS and incubated in avidin-biotinylated horseradish peroxidase complex (1:400; Vector Laboratories). The immunoperoxidase reaction was carried out by using Ni2+-intensified 3,3′-diaminobenzidine 4-HCl (Sigma-Aldrich) as a chromogen and 0.05% H2O2. Cells were reconstructed by using a drawing tube at ×400 magnification.
Data Analysis. The digitized spike train from single-cell recordings with high signal-to-noise ratio was processed with a window discriminator and, together with the hippocampal EEG, was resampled at 250 Hz. The neuronal discharge was characterized by the average firing rate and the coefficients of variation calculated separately for theta and nontheta episodes and by two parameters of the spike waveshape, i.e., spike duration and the relative amplitudes of the first and second peaks of the spike. These latter two parameters strongly correlated (R = 0.81, ANOVA F[1,13] = 24.91, P < 0.001), and thus only one was used for presentation of the results. The neurons were classified by using the distribution of these parameters, and the groups were compared by ANOVA and post hoc Bonferroni test. Time and frequency domain analyses were used for statistical testing of significant relationships between EEG and unit signals at the theta frequency. Time domain analysis included autocorrelograms and unit–EEG crosscorrelogram. In the frequency domain, autospectra for both the unit and EEG signals along with their coherence function were calculated for 10–25 contiguous windows of 4-s duration by using the fast Fourier transform implemented in a customized program (12). The unit spikes, before the fast Fourier transform analysis, were subjected to digital low-pass filtering performed by convolving the sequence of standardized pulses representing the spike trains with a sinc function with parameters set so that the information in the spectra represented the interspike intervals rather than the shape of the standardized pulses.
Acknowledgments
We thank Dr. Martin Deschênes, György Buzsáki, and Nadia Urbain for critical reading of the manuscript and for helpful discussions; Philippe Lemieux and Dahlia Bursell for excellent technical assistance; and Guy Charette for cell reconstruction. This work was supported by the Canadian Institutes of Health Research (A.S.), the National Institute of Mental Health, and the Milton Fund of Harvard University (B.K.). All experimental procedures were approved by the Institutional Animal Care and Use Committee of Harvard Medical School.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TH, tyrosine hydroxylase; 5-HT, 5-hydroxytryptamine; EEG, electroencephalogram.
Footnotes
Urbain, N., Creamer, K. & Debonnel, G. (2004) Neurosci. Abstr. 34, 620.16.
References
- 1.Green, J. D. & Arduini, A. (1954) J. Neurophysiol. 17, 533–557. [DOI] [PubMed] [Google Scholar]
- 2.Siapas, A. G., Lubenov, E. V. & Wilson, M. A. (2005) Neuron 46, 141–151. [DOI] [PubMed] [Google Scholar]
- 3.Seidenbecher, T., Laxmi, T. R., Stork, O. & Pape, H. C. (2003) Science 301, 846–850. [DOI] [PubMed] [Google Scholar]
- 4.Lee, M. G., Hassani, O. K., Alonso, A. & Jones, B. E. (2005) J. Neurosci. 25, 4365–4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vanderwolf, C. H. (1969) Electroencephalogr. Clin. Neurophysiol. 26, 407–418. [DOI] [PubMed] [Google Scholar]
- 6.Buzsaki, G. (1989) Neuroscience 31, 551–570. [DOI] [PubMed] [Google Scholar]
- 7.McGinty, D. J. & Harper, R. M. (1976) Brain Res. 101, 569–575. [DOI] [PubMed] [Google Scholar]
- 8.Vanderwolf, C. H. & Baker, G. B. (1986) Brain Res. 374, 342–356. [DOI] [PubMed] [Google Scholar]
- 9.Vertes, R. P. & Kocsis, B. (1997) Neuroscience 81, 893–926. [DOI] [PubMed] [Google Scholar]
- 10.Viana Di Prisco, G., Albo, Z., Vertes, R. P. & Kocsis, B. (2002) Exp. Brain Res. 145, 383–394. [DOI] [PubMed] [Google Scholar]
- 11.Kocsis, B. & Vertes, R. P. (1996) NeuroReport 7, 2867–2872. [DOI] [PubMed] [Google Scholar]
- 12.Kocsis, B. & Vertes, R. P. (1992) J. Neurophysiol. 68, 1463–1467. [DOI] [PubMed] [Google Scholar]
- 13.Allers, K. A. & Sharp, T. (2003) Neuroscience 122, 193–204. [DOI] [PubMed] [Google Scholar]
- 14.Kirby, L. G., Pernar, L., Valentino, R. J. & Beck, S. G. (2003) Neuroscience 116, 669–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, Y. Q., Li, H., Kaneko, T. & Mizuno, N. (2001) Brain Res. 900, 110–118. [DOI] [PubMed] [Google Scholar]
- 16.Pinault, D. (1996) J. Neurosci. Methods 65, 113–136. [DOI] [PubMed] [Google Scholar]
- 17.Klausberger, T., Magill, P. J., Marton, L. F., Roberts, D. B., Cobden, P. M., Buzsaki, G. & Somogyi, P. (2003) Nature 421, 844–848. [DOI] [PubMed] [Google Scholar]
- 18.Aghajanian, G. K. & Vandermaelen, C. P. (1982) J. Neurosci. 2, 1786–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riad, M., Garcia, S., Watkins, K. C., Jodoin, N., Doucet, E., Langlois, X., el Mestikawy, S., Hamon, M. & Descarries, L. (2000) J. Comp. Neurol. 417, 181–194. [PubMed] [Google Scholar]
- 20.Morilak, D. A., Garlow, S. J. & Ciaranello, R. D. (1993) Neuroscience 54, 701–717. [DOI] [PubMed] [Google Scholar]
- 21.Piguet, P. & Galvan, M. (1994) J. Physiol. (London) 481, 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen, R. Y. & Andrade, R. (1998) J. Pharmacol. Exp. Ther. 285, 805–812. [PubMed] [Google Scholar]
- 23.Tecott, L. H., Maricq, A. V. & Julius, D. (1993) Proc. Natl. Acad. Sci. USA 90, 1430–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ropert, N. & Guy, N. (1991) J. Physiol. (London) 441, 121–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morales, M., Battenberg, E. & Bloom, F. (1998) J. Comp. Neurol. 402, 385–401. [PubMed] [Google Scholar]
- 26.Freund, T. F. (2003) Trends Neurosci. 26, 489–495. [DOI] [PubMed] [Google Scholar]
- 27.Klausberger, T., Marton, L. F., O'Neill, J., Huck, J. H., Dalezios, Y., Fuentealba, P., Suen, W. Y., Papp, E., Kaneko, T., Watanabe, M., et al. (2005) J. Neurosci. 25, 9782–9793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morales, M. & Bloom, F. E. (1997) J. Neurosci. 17, 3157–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freund, T. F. & Gulyas, A. I. (1997) Can. J. Physiol. Pharmacol. 75, 479–487. [PubMed] [Google Scholar]
- 30.Gulyas, A. I., Hajos, N. & Freund, T. F. (1996) J. Neurosci. 15, 3397–3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, R., Jolas, T. & Aghajanian, G. K. (2000) Brain Res. 873, 34–45. [DOI] [PubMed] [Google Scholar]
- 32.Li, S., Varga, V., Sik, A. & Kocsis, B. (2005) J. Neurophysiol. 94, 2561–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varga, V., Sik, A., Fritschy, J. M., Freund, T. M. & Kocsis, B. (2002) Neuroscience 109, 119–132. [DOI] [PubMed] [Google Scholar]
- 34.Rebsam, A., Seif, I. & Gaspar, P. (2002) J. Neurosci. 22, 8541–8552. [DOI] [PMC free article] [PubMed] [Google Scholar]