Abstract
We report unrestrained, all-atom molecular dynamics simulations of HIV-1 protease that sample large conformational changes of the active site flaps. In particular, the unliganded protease undergoes multiple conversions between the “closed” and “semiopen” forms observed in crystal structures of inhibitor-bound and unliganded protease, respectively, including reversal of flap “handedness.” Simulations in the presence of a cyclic urea inhibitor yield stable closed flaps. Furthermore, we observe several events in which the flaps of the unliganded protease open to a much greater degree than observed in crystal structures and subsequently return to the semiopen state. Our data strongly support the hypothesis that the unliganded protease predominantly populates the semiopen conformation, with closed and fully open structures being a minor component of the overall ensemble. The results also provide a model for the flap opening and closing that is considered to be essential to enzyme function.
Keywords: AIDS, HIV protease, molecular dynamics simulations, protein dynamics
Due to its central role in processing viral polypeptide precursors, HIV-1 protease (HIV-PR) continues to be one of the primary targets of anti-AIDS drug discovery. A greater understanding of the mechanistic events associated with binding of HIV-PR substrates and inhibitors is critical for the design of more potent inhibitors of the enzyme. An extensive set of x-ray crystal structures of HIV-1 protease have been solved (1), revealing a C2 symmetric homodimer with a large substrate-binding pocket covered by two glycine-rich β-hairpins, or flaps (2–4). Consistent structural differences are present between the bound and free states of the protein (Fig. 1). In all of the liganded forms, the flaps are pulled in toward the bottom of the active site (“closed” form), whereas the structures for the unbound enzyme all adopt a “semiopen” conformation with the flaps shifted away from the dual Asp-25-Thr-26-Gly-27 catalytic triads but still substantially closed over the active site and in contact with each other. A more striking difference between the two forms is that the relative orientation (the “handedness”) of the β-hairpin flaps is reversed (Fig. 1). The nonflap residues show only slight variation. Although large-scale flap opening is presumably required for normal substrate access to the active site, no crystallographic structures representing such an open configuration have been reported. NMR experiments (5, 6) have established the flexibility of the flap region, suggesting that closed, semiopen, and fully open conformations of the protease are in dynamic equilibrium, with the semiopen form being prevalent for the free protease. The computational results presented here strongly support this hypothesis; the correct semiopen structure dominates our computational ensemble of unbound HIV-PR, but all three structurally distinct forms are observed to interconvert during the simulations.
Fig. 1.
The two experimentally determined conformations of HIV-PR. (A) Free HIV-PR with semiopen conformation of flaps (PDB ID code 1HHP). (B) Inhibitor-bound HIV-PR with closed flaps (PDB ID code 1HVR). Importantly, the handedness of the flaps changes in the two forms and is depicted above each structure. Color indicates distinct regions. Flaps: residues 43–58 and 43′-58′, red for free and blue for bound; flap tips: residues 49–52, yellow; flap elbow: residues 37–42, magenta; cantilever: residues 59–75, green; fulcrum: residues 10–23, orange; and interleaved β-strand motif forming the dimer interface: residues 1–4 and 96–99, blue/cyan.
Understanding the issues that govern HIV-PR flap mobility has profound implications for elucidating the detailed mechanism of this enzyme and in the design of new therapeutic agents, such as allosteric inhibitors intended to interfere with flap opening and thereby with enzymatic function. Thus, numerous prior computational studies have aimed at understanding flap opening dynamics. Collins et al. (7) reported results from activated molecular dynamics (MD) simulations in the gas phase that involved forcing the atomic coordinates for nonflap regions of a closed structure to the semiopen state. These simulations produced flap opening, but the use of restraints prevented investigation of coupling between flap and core domains (8). Scott and Schiffer (9) also observed irreversible flap opening after 3 ns of a 10-ns MD simulation in explicit water, but the extent of flap opening was not quantitatively described. Instead they focused on the flap tip regions, which “curled” back into the protein structure during the opening event, burying several hydrophobic residues. This flap curling was hypothesized to provide a key conformational trigger necessary for subsequent large-scale flap opening and therefore HIV-PR function. More recently, Hamelberg and McCammon (10) used activated dynamics to produce flap opening in HIV-PR. Perryman et al. (11) reported dynamics of unbound V82F/I84V mutant in which the closed form opened somewhat, but detailed analysis of flap conformation and handedness was not reported.
One drawback to prior computational studies is that none have reported observing the return of these open states back to either of the crystallographic forms, and thus their relevance to the true dynamics of HIV-PR is unclear; the simulations may have been too short, or perhaps the observed opening was an artifact of force field inaccuracies or the impulse forces some studies used to aid sampling. A recent study by Meagher and Carlson (12) suggested that large conformational changes within the first few nanoseconds of HIV-PR simulation may simply be artifacts of improper equilibration. Likewise, the transition between closed and semiopen flap handedness that has been experimentally characterized has not been reported in unrestrained simulations.
These uncertainties reinforce the need for simulations of flap opening under reversible conditions that reproduce experimental structural data without imposing restraints or unphysical forces that may obscure dynamic correlations involved in true opening events. We report here a series of fully unrestrained standard molecular dynamics simulations that were initiated from both closed and semiopen forms of HIV-PR. NMR data suggest that these large-scale flap motions occur on the microsecond–millisecond timescale (13); previous simulations typically modeled only several nanoseconds of dynamics. To facilitate dynamic events on a computationally feasible timescale, we used a continuum solvent representation with a reduced viscosity. It has been shown previously that peptide folding rates are accelerated in low viscosity implicit solvent simulations (14).
Consistent with experiment, only the closed form is observed when the inhibitor is present. When the inhibitor is removed from this structure, the protease spontaneously converts to a more flexible and semiopen state with appropriate reversal of flap handedness. Similar to previous studies, we also observe spontaneous and large-scale opening of flaps in multiple simulations of unbound HIV-PR. Importantly, these open states are transient and subsequently convert back to the semiopen state. Our results are consistent with available x-ray and NMR experimental data and provide a model for the open state of the enzyme that is required for ligand entry to the active site and for the dynamic interconversion between this open state and the closed and semiopen forms that have been observed experimentally.
Results and Discussion
Difference Between Closed, Semiopen, and Open States of HIV-PR. Although the conformation of the flap region clearly differs in the closed and semiopen forms (Fig. 1), we desired a simple metric to monitor the structure during simulation. A comparative study of 73 HIV-PR complexes by Zoete and Karplus (15) showed that the structure is relatively insensitive to the identity of the ligand. We performed an analysis of that structure set along with crystal structures of unbound HIV-PR to investigate specific differences between closed and semiopen forms. Even though approximately one-third of the closed structures have a hydrogen bond between the flap tips, nearly the same number form a different flap tip hydrogen bond and the remainder, like the semiopen structures, have none. Likewise, the flap tip β-turn type is not a unique characteristic of the closed conformation. It is likely that the resolution of the crystallographic data, coupled with intrinsic flap tip flexibility, is insufficient to unambiguously define this region. We found that the most reliable measure of closed vs. semiopen form exploits the consistent difference in the handedness of the flaps (Fig. 1), which results in large rms deviation (RMSD) values when flap residue α-carbons are overlapped; this RMSD value between the closed and semiopen forms is 3.3 Å.
Unlike the closed and semiopen forms, a structure of the protease with fully open flaps has not yet been observed experimentally; thus, it is somewhat arbitrary to define which structures qualify as open. The open ensemble most likely consists of many structures with different relative positioning of both flaps. However, because the proposed mechanism of ligand binding requires extensive flap opening, the distance between flap tips (Ile-50Cα–Ile-50′Cα) is a reasonable metric to gauge the extent of opening. This distance is ≈4.3 Å in semiopen and ≈5.8 Å in closed forms (somewhat longer in the closed form, see Fig. 1).
MD Simulations of Inhibitor Bound HIV-PR. A 28-ns simulation of closed, inhibitor-bound HIV-PR at 300 K was initiated from the crystal structure of the complex and produced a stable trajectory. No large structural changes were observed; the flaps remained in their closed conformation throughout the entire run (see Fig. 3A). The hydrogen bond between the cyclic urea inhibitor's carbonyl oxygen and at least one of the Ile-50/50′ amides as well as the hydrogen bond between flap tips (Gly-51N·Ile-50′O) remained in place, likely contributing to the stability of the otherwise flexible glycine-rich flap tips. This behavior is typical of protein simulations on this timescale.
Fig. 3.
RMSD of flaps during simulation of bound and free HIV-PR, both starting from the closed state, overlapped on the crystal structures of semiopen (red line, PDB ID code 1HHP reference structure) and closed (blue line, PDB ID code 1HVR reference) HIV-PR. Low values for the blue line indicate that the flaps are in the closed state, and low values for the red line indicate semiopen flaps. (A) Inhibitor-bound HIV-PR flaps remain very close to the initial closed crystal structure (i.e., blue line remains at low RMSD values). (B) Unbound HIV-PR flaps that were initially closed (blue line has low initial RMSDs in the beginning) convert to semiopen, including reversal of handedness (low red values). The simulation later (≈35 ns) transiently samples the closed form before reverting back to semiopen.
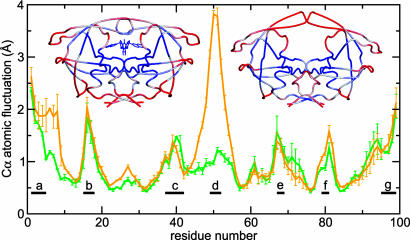
Whereas the global structure of the protease remained stable (Cα RMSD plateau < 2.0 Å) (Fig. 7, which is published as supporting information on the PNAS web site), certain regions exhibited higher flexibility (Fig. 2), including the termini, flap elbows (residues 37–42), cantilever β-turn region (residues 66–69), and two narrow regions around Gly-17 and Thr-80. Importantly, the flaps and flap tips did not show higher fluctuations than the core during simulation of the complex.
Fig. 2.
Cα atomic fluctuations from simulation of inhibitor-bound (green line) and free (orange line) HIV-PR averaged for both monomers, with error bars reflecting the difference between monomers. Regions: a, N-terminal β-strand; b, fulcrum loop; c, flap elbow; d, flap tips; e, cantilever β-turn; f, Thr-80 loop; g, C-terminal β-strand. Also shown are crystal structures of bound (left) and unbound (right) protease with color indicating low (blue) to high (red) fluctuations in simulations. The most significant difference between the two simulations is a large increase in flap tip flexibility for the unbound simulation.
MD Simulations of Ligand-Free HIV-PR. A 42-ns simulation of ligand-free HIV-PR at 300 K was also initiated from the crystallographic coordinates of the complex, that is, after removal of the inhibitor. This procedure permits use of the bound simulations as a direct control. The flaps spontaneously rearranged (Fig. 3B) from the closed to semiopen form near the beginning of the simulation (including reversal of flap handedness), and exhibited significantly increased fluctuations (Fig. 2, orange line). This finding is consistent with experimental data that indicate higher flexibility and predominance of the semiopen form in the absence of ligands. Of particular importance is the revisiting of closed flap conformations (Fig. 3B) before returning to the semiopen form and transient sampling of flap configurations that are neither semiopen nor closed. Thus a ligand-free HIV-PR simulation initiated from the closed crystal structure spontaneously converts to a more flexible ensemble that is dominated by the semiopen form yet also visits structures that are closed and others that are neither semiopen nor closed. This behavior is in sharp contrast with the stability of the same closed state under equivalent conditions in the presence of the inhibitor.
Unbound HIV-PR flap flexibility was mostly localized in the tips (Fig. 2); the flap β-hairpins themselves were not disrupted even during the rearrangement events (RMSD of residues 43–48 and 53–58 remained near 0.5 Å at all times). Intraflap hydrogen bonds were always present (Fig. 8, which is published as supporting information on the PNAS web site), with the exception of the tip hydrogen bonds between Gly-49 and Gly-52. This observation is in good agreement with cross-strand amide nuclear Overhauser effect data indicating that the flap β-hairpins are present in solution and are well ordered except at the tips (6). Backbone torsions underwent substantial changes in the tips and elbows (Figs. 9 and 10, which is published as supporting information on the PNAS web site). These torsion data imply that the flap β-hairpins each move as a rigid body with highly flexible tips, with the flap elbows acting as “hinges.” This flap motion generates a wide range of structures; yet, as can be seen from Fig. 3B, the overall conformation of the flap pair typically resembles either the semiopen or closed form, with other conformations forming a group of more open structures with flap tip distances of up to ≈17 Å. Although the flap tips are highly flexible, we did not observe any trans–cis isomerization of the Gly–Gly ω-bonds as was recently reported in HIV-PR simulations (10).
We performed another 3-ns simulation (Fig. 4) starting from the crystal structure of unliganded HIV-PR, which, like all free HIV-PR structures, adopts the semiopen flap conformation. As before, we observed high flap flexibility and transient conversion to a closed form with reversal of handedness, although it is apparent from Fig. 4 that adoption of the closed flap handedness need not be accompanied by downward movement of the flaps toward the catalytic residues as seen in the inhibitor-bound simulation and crystal structures. This simulation sampled a broad range of opening events, inducing flap tip separations as large as 30 Å. In each case, the protease returned to the semiopen form after the opening event. The large flap movements were primarily absorbed by the flap elbow, cantilever, and fulcrum regions.
Fig. 4.
Flaps RMSD and flap tips distance for free HIV-PR simulation started from a semiopen crystal structure. Snapshots (cartoon diagrams, side view) along the trajectory (cyan) are shown overlapped on the semiopen crystal structure (gray). Surface representations (top view) depict flap handedness and access to the active site, with the two flaps in green/orange and the catalytic Asp-25/25′ in yellow. The semiopen conformation is prevalent (low RMSDs for red line). Note that the transiently sampled closed structure (structure b) has the flap handedness characteristic of bound (closed) crystal structures, even though flaps do not become fully pulled into the active site in this simulations. Large flap openings are sampled (structures c–e), with flap tip distances reaching ≈30 Å and subsequently returning to the semiopen form (structures e–f).
Scott and Schiffer (9) proposed flap curling as a “trigger” for HIV-PR opening based on their observation of a hydrophobic cluster involving Ile-50 at the flap tip and the nearby Val-32, Pro-79, Thr-80, and Pro-81. We calculated the distance of Ile-50 Cβ to the center of mass of residues 32 and 79–81. Although some flap movements are accompanied by formation of this cluster, the more significant opening events are not; instead, they are associated with greater departure of flap tips away from the binding site (Fig. 11, which is published as supporting information on the PNAS web site). NMR relaxation experiments (6) indicate that flexibility of the flap tips (“flap curling”) is related to rapid (nanosecond timescale) interconversion between the members of the semiopen ensemble, whereas the actual opening events happen on a much slower (microseconds to milliseconds) timescale, and thus are likely not directly related to rapid flap tip fluctuations.
Comparison of Simulations with NMR Data on HIV-PR Dynamics. Torchia et al. (6, 13, 16) have proposed a model for flap dynamics in which the predominant structure in solution is semiopen with fast (≈1 ns) conformational changes in flap tips. The semiopen ensemble is in slower (≈100 μs) dynamic equilibrium with closed conformations as well as with less ordered open flap conformations (6, 13), which permit access to the active site as an outcome of a rare event (16). This picture is consistent with our simulations, in which flaps rapidly interconvert between a variety of flexible semiopen conformations, with infrequent opening events when flaps separate and become less ordered. The presence of a low population of closed flap conformations in the solution ensemble also is supported by our data; simulations of unbound protease transiently converted from the predominant semiopen form to the closed one, as indicated by the reversal of flap handedness. This model also is in accord with free energy calculations of Rick et al. (17), which indicated that the closed state is less favorable than the semiopen one in the absence of a ligand.
To compare the flexibility of HIV-PR inferred from MD simulations with experimental data (5, 6, 18), generalized order parameters S2 were computed for free and ligand-bound protease. The calculated results show very good qualitative accord with experiment; trends characterizing regions of high and low flexibility are reproduced quite well (Fig. 5). Flexible regions in the unliganded protease include residues 37–41 (flap elbows), 49–53 (flap tips), and Thr-80. The most pronounced difference between order parameters of free and bound protease is the loss of flexibility in the flap tip region upon formation of the complex (Fig. 5). Our simulations and experimental data match these trends very well. The fulcrum loop around Gly-17 also exhibits higher flexibility in our simulations. A protease complex with DMP323 and P9941 inhibitors does show low S2 values in this region (5), but more recent NMR data on the complex with DMP323 did not reproduce this trend (18). This discrepancy might by caused by the elevated temperature (34°C vs. 27°C) used in the earlier Nicholson et al. experiments. Increased flexibility in this region was also reported by others using simulations with quite different solvent models and protein force fields (9, 12, 19). The source of the discrepancy between simulations and experiment for Gly-17 (fulcrum loop) flexibility is thus unclear.
Fig. 5.
Backbone amide generalized-order parameters (S2) averaged for both monomers, with error bars reflecting the difference. (A) Experimental data (black) (6) and simulation (red) for unliganded protease. General trends of flexibility are reproduced, although simulated values are lower. (B) Order parameters for inhibitor-bound HIV-PR from simulation (blue) and from two experiments of HIV-PR bound to DMP323 [black triangles (5) and black circles (18)]. The loss of flexibility in the flap tip region (residues 49–52) as compared with that in A is readily observed in data from simulations and from experiments.
Absolute S2 values, however, are in general lower in the calculated values than in the experiment. It has been previously reported that quantitative comparison of calculated and experimental order parameters can be problematic, and higher flexibility from molecular dynamics simulations is often reported. Several issues have been discussed in detail (20); uncertainties in experimental values can arise from the assumptions applied in “model free” analysis and differences in procedures. Simulated values also depend on the force field used and convergence of the ensemble of structures sampled, leading to values that can be either too small or too large.
Simulations with an Explicit Water Model. Another possible explanation for lack of quantitative agreement of S2 values is that our use of low viscosity increased the amplitude of conformational fluctuations, in addition to the more rapid structural rearrangements that we desired. To evaluate this possibility, we performed additional HIV-PR simulations using an explicit solvent model. For the inhibitor-bound complex in explicit solvent (30 ns), the Cα atomic fluctuations were nearly identical to those obtained with the implicit water model (Fig. 12, which is published as supporting information on the PNAS web site), strongly suggesting that the range of motion sampled in our simulations is unlikely to be an artifact of the low viscosity, and the lack of quantitative agreement between simulated and experimental S2 arises from other sources. Consistent with timescales inferred from experiment, the closed unbound protease in a 45-ns explicit solvent simulation was unable to convert to semiopen or open states.
Conclusions
Using all-atom simulations with continuum solvation, we observed conversion between closed, semiopen, and fully open forms of HIV-PR, including the reversal of flap handedness between semiopen and closed forms as observed in all crystal structures. The preferred structure, as well as the extent of flexibility, depended strongly on the presence of an inhibitor. These results are in good agreement with existing NMR and crystallographic data, but the use of an artificially low solvent viscosity precludes the comparison of absolute timescales from our simulations to NMR relaxation experiments. However, this low viscosity permitted the observation of structural transitions on a computationally tractable timescale. Moreover, we do reproduce the trend of flap tip flexibility occurring on a much more rapid timescale than full opening. Future exploration of the events that we report but with potentially more accurate explicit solvent models would be highly desirable. Such studies would not only facilitate quantitative comparison of timescales but also provide useful insight into a possible structural role for water during HIV-PR dynamics. For example, a well ordered water molecule mediating interaction between the flap tips and peptide-based inhibitors is observed in multiple HIV-PR crystal structures (21, 22). Such interactions transcend bulk viscosity effects and may further reduce the likelihood of observing flap transitions in explicit solvent simulations.
Although the four available crystal structures of unbound HIV-PR all show the semiopen conformation, it has not been entirely clear whether this reflects the preferred flap conformation or crystal packing effects (21, 23). For the free protease, early MD simulations suggested that the closed form was predominant in solution (24), whereas several more recent computational studies showed irreversible opening of the flaps. Based on solution NMR data for the free protease, Torchia et al. have suggested (6, 13) that the ensemble of unbound structures is dominated by the semiopen family with subnanosecond timescale fluctuation in the flap tips and with closed structures being a minor component of the ensemble. The semiopen form is in slow equilibrium (≈100 μs) with a less structured, open form that exposes the binding site cavity. Our simulations of unliganded HIV-PR strongly support this latter model both in the types and relative populations of different structures.
Full flap opening in our simulations occurs through a concerted downward motion of the cantilever, fulcrum, and flap elbows that results in upwards motion of the flaps and, consequently, domain rotation around the region near the dimer interface and a shift of the catalytic Asp-25/25′ up from the bottom of the active site, which could be speculated to facilitate ligand binding (Fig. 6). This mechanism is further supported by detailed comparison of the crystal structures of inhibitor-bound and free protease (Fig. 6). Although the differences in the two structures are dominated by the flap rearrangement, concerted motions of flap elbows, fulcrum, and cantilever that are consistent with our model are indeed apparent. Interestingly, early dynamic analysis by Beveridge and coworkers (8, 25) also showed correlation between the flap and cantilever domains as well as between the two cantilever domains. The change in flap handedness upon ligand binding is clearly reproduced in our simulations, but its possible role in HIV-PR function remains unexplained.
Fig. 6.
Three forms of HIV-1 protease: closed (blue, PDB ID code 1HVR), semiopen (red, PDB ID code1HHP), and open (cyan, our model). We observe opening/closing wherein the flap elbows and exposed ends of fulcrum and cantilever move down and toward the terminal β-sheet dimer interface (indicated by yellow arrows). Examination of closed and semiopen crystal structures reveals differences in those regions that are smaller in magnitude but in agreement with the direction of changes seen during opening.
Methods
Simulations in amber (26) were initiated from crystallographic coordinates. The closed form used a structure of wild-type sequence in complex with the cyclic urea inhibitor XK263 (22) [Protein Data Bank (PDB) ID code 1HVR]. Semiopen coordinates were obtained from 1HHP (27), after changing two side chains (I3V and C95A) to match the sequence of 1HVR. Both catalytic Asp side chains were modeled in the protonated state. Hydrogen atoms were added by using the leap module in amber. FF99 (28, 29) was used for the protein, with backbone parameters (Table 1, which is published as supporting information on the PNAS web site) refit to ab initio energies for Ala-3 and Gly-3. The antechamber module and GAFF (30) with AM1-BCC charges (31) were used to obtain force field parameters for XK263 (Table 2, which is published as supporting information on the PNAS web site). Order parameters (S2) were calculated from a plateau region of the N–H internuclear vector autocorrelation functions (32), which were obtained with the ptraj module.
Aqueous solvation was modeled implicitly by using the Generalized Born approach (33) with modification for more accurate solvation of large proteins (34). Continuum solvent simulations of HIV-PR have been reported in the past (35). Temperature was maintained by using Langevin dynamics with a collision frequency of 1 ps–1, the timestep was 1 fs, and all bonds to hydrogen were constrained. All nonbonded interactions were calculated at each time step (i.e., no cutoff was used). The system was minimized and then gradually heated from 200K to 300K. Positional restraints were used first on all heavy atoms and then only for backbone atoms. Restraint force constants were decreased from 2 to 0.1 kcal/mol·Å2 (1 kcal = 4.18 kJ) in several stages. During the last step of equilibration, restraints were removed entirely, and all production simulations were at 300K and fully unrestrained. These simulations required ≈400 core processing unit hours on 1.5-GHz Itanium processors for each nanosecond of dynamics. Details for explicit solvent simulations are provided in the legend to Fig. 12.
Nomenclature for HIV-PR regions adopts the convention proposed by Harte and Beveridge (8).
Supplementary Material
Acknowledgments
We thank Ad Bax and Dennis Torchia for providing NMR data and Roberto Gomperts (Silicon Graphics, Mountain View, CA) for providing important Amber optimizations and Altix resources. This workwas supported by National Institutes of Health Grant GM6167803 and Department of Energy Grant DE-AC02–98CH10886 and by additional supercomputer time at the National Center for Supercomputing Applications under National Partnership for Advanced Computational Infrastructure MCA02N028. C.S. is a Cottrell Scholar of Research Corporation.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HIV-PR, HIV-1 protease; RMSD, rms deviation; MD, molecular dynamics; PDB, Protein Data Bank.
References
- 1.Vondrasek, J. & Wlodawer, A. (2002) Proteins Struct. Funct. Genet. 49, 429–431. [DOI] [PubMed] [Google Scholar]
- 2.Navia, M. A., Fitzgerald, P. M. D., McKeever, B. M., Leu, C. T., Heimbach, J. C., Herber, W. K., Sigal, I. S., Darke, P. L. & Springer, J. P. (1989) Nature 337, 615–620. [DOI] [PubMed] [Google Scholar]
- 3.Wlodawer, A., Miller, M., Jaskolski, M., Sathyanarayana, B. K., Baldwin, E., Weber, I. T., Selk, L. M., Clawson, L., Schneider, J. & Kent, S. B. H. (1989) Science 245, 616–621. [DOI] [PubMed] [Google Scholar]
- 4.Lapatto, R., Blundell, T., Hemmings, A., Overington, J., Wilderspin, A., Wood, S., Merson, J. R., Whittle, P. J., Danley, D. E., Geoghegan, K. F., et al. (1989) Nature 342, 299–302. [DOI] [PubMed] [Google Scholar]
- 5.Nicholson, L. K., Yamazaki, T., Torchia, D. A., Grzesiek, S., Bax, A., Stahl, S. J., Kaufman, J. D., Wingfield, P. T., Lam, P. Y. S., Jadhav, P. K., et al. (1995) Nat. Struct. Biol. 2, 274–280. [DOI] [PubMed] [Google Scholar]
- 6.Freedberg, D. I., Ishima, R., Jacob, J., Wang, Y. X., Kustanovich, I., Louis, J. M. & Torchia, D. A. (2002) Protein Sci. 11, 221–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins, J. R., Burt, S. K. & Erickson, J. W. (1995) Nat. Struct. Biol. 2, 334–338. [DOI] [PubMed] [Google Scholar]
- 8.Harte, W. E., Swaminathan, S., Mansuri, M. M., Martin, J. C., Rosenberg, I. E. & Beveridge, D. L. (1990) Proc. Natl. Acad. Sci. USA 87, 8864–8868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scott, W. R. P. & Schiffer, C. A. (2000) Structure (London) 8, 1259–1265. [DOI] [PubMed] [Google Scholar]
- 10.Hamelberg, D. & McCammon, J. A. (2005) J. Am. Chem. Soc., in press.
- 11.Perryman, A. L., Lin, J. H. & McCammon, J. A. (2004) Protein Sci. 13, 1108–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meagher, K. L. & Carlson, H. A. (2005) Proteins Struct. Funct. Bioinf. 58, 119–125. [DOI] [PubMed] [Google Scholar]
- 13.Ishima, R., Freedberg, D. I., Wang, Y. X., Louis, J. M. & Torchia, D. A. (1999) Structure (London) 7, 1047–1055. [DOI] [PubMed] [Google Scholar]
- 14.Zagrovic, B. & Pande, V. (2003) J. Comp. Chem. 24, 1432–1436. [DOI] [PubMed] [Google Scholar]
- 15.Zoete, V., Michielin, O. & Karplus, M. (2002) J. Mol. Biol. 315, 21–52. [DOI] [PubMed] [Google Scholar]
- 16.Katoh, E., Louis, J. M., Yamazaki, T., Gronenborn, A. M., Torchia, D. A. & Ishima, R. (2003) Protein Sci. 12, 1376–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rick, S. W., Erickson, J. W. & Burt, S. K. (1998) Proteins Struct. Funct. Bioinf. 32, 7–16. [DOI] [PubMed] [Google Scholar]
- 18.Tjandra, N., Wingfield, P., Stahl, S. & Bax, A. (1996) J. Biomol. NMR 8, 273–284. [DOI] [PubMed] [Google Scholar]
- 19.Piana, S., Carloni, P. & Rothlisberger, U. (2002) Protein Sci. 11, 2393–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Case, D. A. (2002) Acc. Chem. Res. 35, 325–331. [DOI] [PubMed] [Google Scholar]
- 21.Wlodawer, A. & Vondrasek, J. (1998) Annu. Rev. Biophys. Biomol. Struct. 27, 249–284. [DOI] [PubMed] [Google Scholar]
- 22.Lam, P. Y. S., Jadhav, P. K., Eyermann, C. J., Hodge, C. N., Ru, Y., Bacheler, L. T., Meek, J. L., Otto, M. J., Rayner, M. M., Wong, Y. N., et al. (1994) Science 263, 380–384. [DOI] [PubMed] [Google Scholar]
- 23.Miller, M., Schneider, J., Sathyanarayana, B. K., Toth, M. V., Marshall, G. R., Clawson, L., Selk, L., Kent, S. B. H. & Wlodawer, A. (1989) Science 246, 1149–1152. [DOI] [PubMed] [Google Scholar]
- 24.York, D. M., Darden, T. A. & Pedersen, L. G. (1993) J. Chem. Phys. 99, 8345–8348. [Google Scholar]
- 25.Swaminathan, S., Harte, W. E. & Beveridge, D. L. (1991) J. Am. Chem. Soc. 113, 2717–2721. [Google Scholar]
- 26.Case, D. A., Cheatham, T. E., III, Darden, T. A., Gohlke, H., Luo, R., Merz, K. M., Jr., Onufriev, A., Simmerling, C. L., Wang, B. & Woods, R. J. (2005) J. Comp. Chem. 26, 1668–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spinelli, S., Liu, Q. Z., Alzari, P. M., Hirel, P. H. & Poljak, R. J. (1991) Biochimie 73, 1391–1396. [DOI] [PubMed] [Google Scholar]
- 28.Wang, J. M., Cieplak, P. & Kollman, P. A. (2000) J. Comp. Chem. 21, 1049–1074. [Google Scholar]
- 29.Cornell, W. D., Cieplak, P., Bayly, C. I., Gould, I. R., Merz, K. M., Ferguson, D. M., Spellmeyer, D. C., Fox, T., Caldwell, J. W. & Kollman, P. A. (1995) J. Am. Chem. Soc. 117, 5179–5197. [Google Scholar]
- 30.Wang, J. M., Wolf, R. M., Caldwell, J. W., Kollman, P. A. & Case, D. A. (2004) J. Comp. Chem. 25, 1157–1174. [DOI] [PubMed] [Google Scholar]
- 31.Jakalian, A., Bush, B. L., Jack, D. B. & Bayly, C. I. (2000) J. Comp. Chem. 21, 132–146. [Google Scholar]
- 32.Lipari, G. & Szabo, A. (1982) J. Am. Chem. Soc. 104, 4546–4559. [Google Scholar]
- 33.Still, W. C., Tempczyk, A., Hawley, R. C. & Hendrickson, T. (1990) J. Am. Chem. Soc. 112, 6127–6129. [Google Scholar]
- 34.Onufriev, A., Bashford, D. & Case, D. A. (2000) J. Phys. Chem. B 104, 3712–3720. [Google Scholar]
- 35.David, L., Luo, R. & Gilson, M. K. (2000) J. Comp. Chem. 21, 295–309. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.